Growth-Phase-Dependent Modulation of Quorum Sensing and Virulence Factors in Pseudomonas aeruginosa ATCC 27853 by Sub-MICs of Antibiotics
Abstract
1. Introduction
2. Results
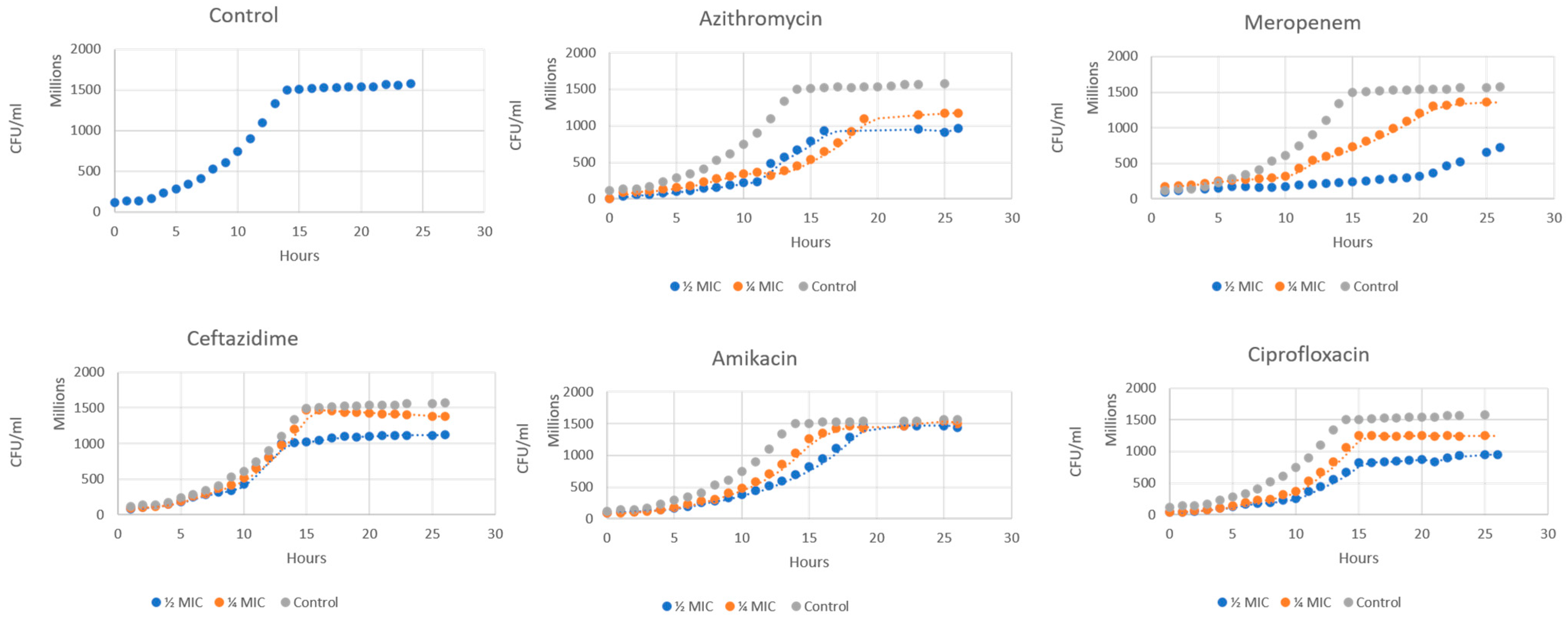
2.1. Growth Curve Analysis
2.2. Pyocyanin Production
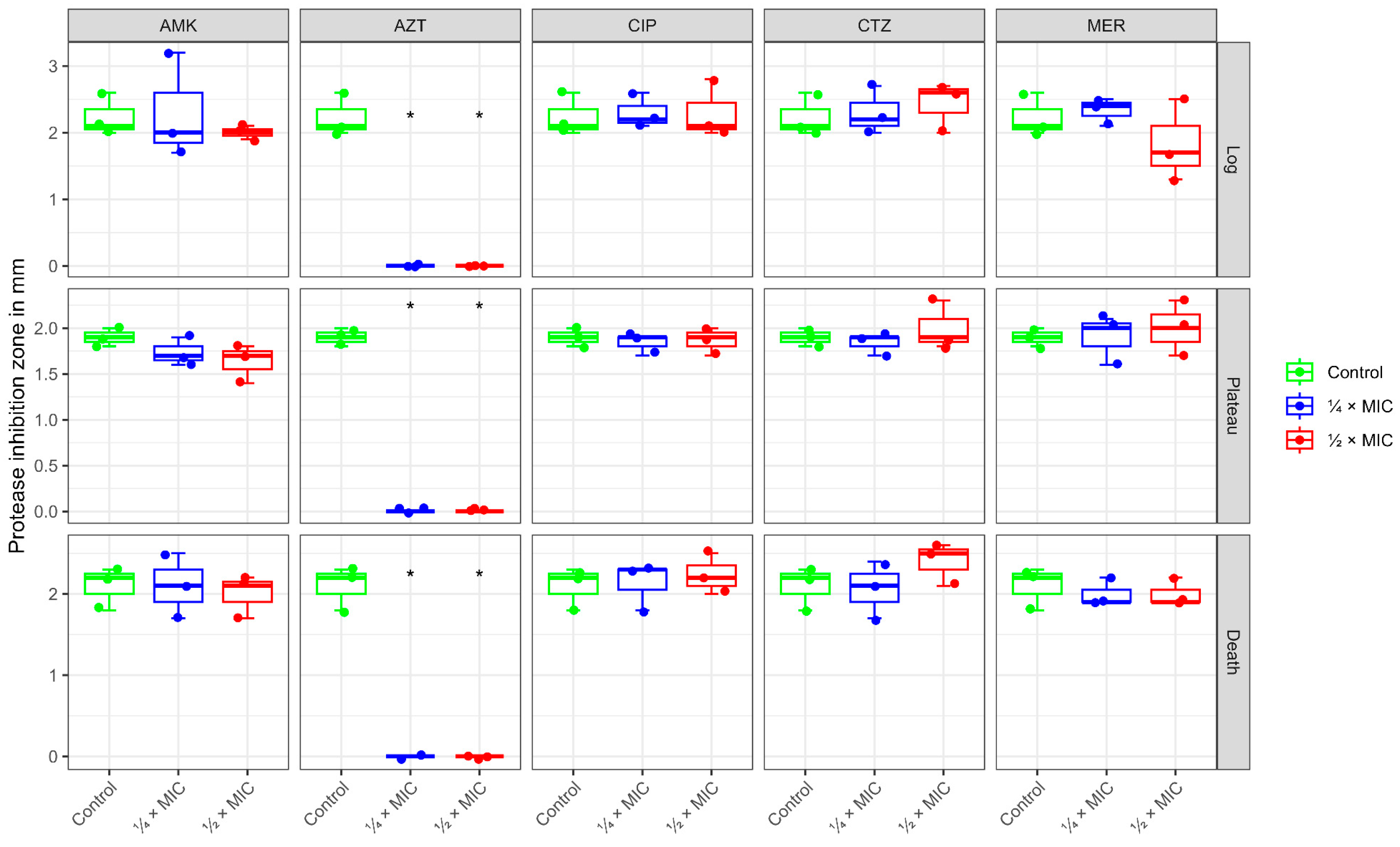
2.3. Protease Activity
2.4. Gene Expression Profile
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain
4.2. Determination of Minimum Inhibitory Concentration of Studied Antibiotics
4.3. Growth Curve Determination
4.4. Phenotypic Detection of Pyocyanin Production
4.5. Phenotypic Detection of Protease Activity
4.6. Quantitative Reverse Transcription PCR Investigating the Expression of QS Genes and a Pyocyanin Gene
4.7. Statistical Analyses
5. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AHL | Acyl-homoserine lactone |
| AMK | Amikacin |
| ANOVA | Analysis of variance |
| AZT | Azithromycin |
| cDNA | Complementary DNA |
| CFU | Colony-forming unit |
| CIP | Ciprofloxacin |
| CLSI | Clinical and Laboratory Standards Institute |
| CTZ | Ceftazidime |
| DNA | Deoxyribonucleic acid |
| HHQ | 2-Heptylquinol |
| HSD | Honest significant difference |
| MER | Meropenem |
| MHB | Mueller-Hinton broth |
| MIC | Minimum inhibitory concentration |
| mRNA | Messenger RNA |
| NCBI | National Center for Biotechnology Information |
| OD | Optical density |
| PBS | Phosphate-buffered saline |
| PBP | Penicillin-binding protein |
| PCR | Polymerase chain reaction |
| qRT-PCR | Quantitative reverse transcription PCR |
| QS | Quorum sensing |
| QSI | QS inhibitor |
| RNA | Ribonucleic acid |
| ROS | Reactive oxygen species |
| SD | Standard deviation |
| sub-MIC | Sub-inhibitory concentration |
Appendix A
| Antibiotics | Growth Phase | Pyocyanin OD | Control | ¼ MIC | ½ MIC | p |
|---|---|---|---|---|---|---|
| CTZ | Log | Mean ± SD | 0.561 ± 0.038 a,b | 0.753 ± 0.055 a | 0.767 ± 0.076 b | 0.009 * |
| Plateau | Mean ± SD | 1.612 ± 0.050 | 1.823 ± 0.142 | 1.603 ± 0.089 | 0.062 | |
| Death | Mean ± SD | 0.592 ± 0.071 | 0.594 ± 0.038 | 0.662 ± 0.037 | 0.241 | |
| AMK | Log | Mean ± SD | 0.561 ± 0.038 | 0.551 ± 0.060 | 0.611 ± 0.031 | 0.284 |
| Plateau | Mean ± SD | 1.612 ± 0.050 | 1.692 ± 0.055 | 1.751 ± 0.178 | 0.371 | |
| Death | Mean ± SD | 0.592 ± 0.071 | 0.596 ± 0.127 | 0.504 ± 0.023 | 0.382 | |
| AZT | Log | Mean ± SD | 0.561 ± 0.038 b | 0.739 ± 0.086 a,b | 0.508 ± 0.017 a | 0.005 * |
| Plateau | Mean ± SD | 1.612 ± 0.050 a | 1.865 ± 0.146 a | 1.706 ± 0.067 | 0.049 * | |
| Death | Mean ± SD | 0.592 ± 0.071 | 0.574 ± 0.038 | 0.504 ± 0.037 | 0.166 | |
| MER | Log | Mean ± SD | 0.561 ± 0.038 b | 0.637 ± 0.061 a | 1.833 ± 0.136 a,b | <0.001 * |
| Plateau | Mean ± SD | 1.612 ± 0.050 | 1.568 ± 0.121 | 2.113 ± 0.703 | 0.275 | |
| Death | Mean ± SD | 0.592 ± 0.071 | 0.637 ± 0.080 | 0.846 ± 0.400 | 0.432 | |
| CIP | Log | Mean ± SD | 0.561 ± 0.038 | 0.605 ± 0.022 | 0.546 ± 0.031 | 0.124 |
| Plateau | Mean ± SD | 1.612 ± 0.050 | 1.550 ± 0.126 | 1.519 ± 0.068 | 0.46 | |
| Death | Mean ± SD | 0.592 ± 0.071 | 0.520 ± 0.107 | 0.565 ± 0.062 | 0.582 |
| Antibiotics | Growth Phase | Protease Inhibition Zone in mm | ¼ MIC | ½ MIC | Control | p |
|---|---|---|---|---|---|---|
| CTZ | Log | Mean ± SD | 2.300 ± 0.361 | 2.433 ± 0.379 | 2.233 ± 0.321 | 0.788 |
| Plateau | Mean ± SD | 1.833 ± 0.115 | 2.000 ± 0.265 | 1.900 ± 0.100 | 0.542 | |
| Death | Mean ± SD | 2.067 ± 0.351 | 2.400 ± 0.265 | 2.100 ± 0.265 | 0.377 | |
| AMK | Log | Mean ± SD | 2.300 ± 0.794 | 2.000 ± 0.100 | 2.233 ± 0.321 | 0.751 |
| Plateau | Mean ± SD | 1.733 ± 0.153 | 1.633 ± 0.208 | 1.900 ± 0.100 | 0.200 | |
| Death | Mean ± SD | 2.100 ± 0.400 | 2.000 ± 0.265 | 2.100 ± 0.265 | 0.906 | |
| AZT | Log | Mean ± SD | 0.000 ± 0.000 a | 0.000 ± 0.000 b | 2.233 ± 0.32 a,b | <0.001 * |
| Plateau | Mean ± SD | 0.000 ± 0.000 a | 0.000 ± 0.000 b | 1.900 ± 0.100 a,b | <0.001 * | |
| Death | Mean ± SD | 0.000 ± 0.000 a | 0.000 ± 0.000 b | 2.100 ± 0.265 a,b | <0.001 * | |
| MER | Log | Mean ± SD | 2.333 ± 0.208 | 1.833 ± 0.611 | 2.233 ± 0.321 | 0.361 |
| Plateau | Mean ± SD | 1.900 ± 0.265 | 2.000 ± 0.300 | 1.900 ± 0.100 | 0.842 | |
| Death | Mean ± SD | 2.000 ± 0.173 | 2.000 ± 0.173 | 2.100 ± 0.265 | 0.801 | |
| CIP | Log | Mean ± SD | 2.300 ± 0.265 | 2.300 ± 0.436 | 2.233 ± 0.321 | 0.964 |
| Plateau | Mean ± SD | 1.833 ± 0.115 | 1.867 ± 0.153 | 1.900 ± 0.100 | 0.813 | |
| Death | Mean ± SD | 2.133 ± 0.289 | 2.233 ± 0.252 | 2.100 ± 0.265 | 0.824 |
| Genes | Antibiotics | Fold Change | Log | Plateau | Death | p |
|---|---|---|---|---|---|---|
| lasI | CTZ | Mean ± SD | 0.521 ± 0.073 | 1.352 ± 0.020 | 0.587 ± 0.376 | 0.056 |
| AZT | Mean ± SD | 1.133 ± 0.426 | 0.865 ± 0.017 | 1.984 ± 1.726 | 0.584 | |
| MER | Mean ± SD | 1.279 ± 0.038 | 2.578 ± 0.672 | 2.523 ± 0.473 | 0.117 | |
| CIP | Mean ± SD | 0.548 ± 0.152 b | 0.622 ± 0.257 a | 2.586 ± 0.127 a,b | 0.003 * | |
| AMK | Mean ± SD | 0.739 ± 0.425 | 0.720 ± 0.401 | 2.396 ± 1.579 | 0.286 | |
| lasR | CTZ | Mean ± SD | 0.623 ± 0.142 a | 1.171 ± 0.059 a | 1.012 ± 0.141 | 0.042 * |
| AZT | Mean ± SD | 1.179 ± 0.363 b | 1.131 ± 0.149 a | 2.474 ± 0.068 a,b | 0.016 * | |
| MER | Mean ± SD | 1.475 ± 0.054 a | 1.879 ± 0.207 | 2.136 ± 0.110 a | 0.039 * | |
| CIP | Mean ± SD | 0.564 ± 0.073 b | 0.653 ± 0.071 a | 2.478 ± 0.035 a,b | <0.001 * | |
| AMK | Mean ± SD | 0.864 ± 0.228 | 0.784 ± 0.045 | 2.030 ± 0.700 | 0.102 | |
| phzA | CTZ | Mean ± SD | 2.596 ± 2.370 | 2.576 ± 0.427 | 1.745 ± 0.171 | 0.799 |
| AZT | Mean ± SD | 1.765 ± 0.432 | 1.215 ± 0.254 | 2.131 ± 0.281 | 0.148 | |
| MER | Mean ± SD | 1.442 ± 0.364 | 1.632 ± 0.238 | 1.232 ± 1.552 | 0.914 | |
| CIP | Mean ± SD | 1.048 ± 0.247 | 0.612 ± 0.396 | 2.825 ± 1.356 | 0.142 | |
| AMK | Mean ± SD | 1.163 ± 0.145 | 0.812 ± 0.393 | 1.328 ± 0.680 | 0.582 | |
| pqsA | CTZ | Mean ± SD | 0.804 ± 0.415 | 0.982 ± 0.224 | 0.741 ± 0.938 | 0.921 |
| AZT | Mean ± SD | 1.937 ± 0.100 | 0.857 ± 0.163 | 1.599 ± 0.396 | 0.051 | |
| MER | Mean ± SD | 2.216 ± 0.663 | 1.888 ± 0.586 | 1.527 ± 0.153 | 0.499 | |
| CIP | Mean ± SD | 1.008 ± 0.346 | 0.409 ± 0.497 | 1.822 ± 0.045 | 0.061 | |
| AMK | Mean ± SD | 1.156 ± 0.365 | 0.409 ± 0.006 | 1.508 ± 0.434 | 0.092 | |
| pqsR | CTZ | Mean ± SD | 0.354 ± 0.053 | 0.505 ± 0.098 | 1.096 ± 0.732 | 0.323 |
| AZT | Mean ± SD | 1.917 ± 0.248 | 1.709 ± 0.614 | 2.409 ± 1.188 | 0.691 | |
| MER | Mean ± SD | 2.386 ± 0.556 b | 0.166 ± 0.225 a,b | 1.887 ± 0.189 a | 0.018 * | |
| CIP | Mean ± SD | 0.896 ± 0.232 | 0.348 ± 0.052 | 2.036 ± 1.978 | 0.433 | |
| AMK | Mean ± SD | 1.458 ± 0.826 | 0.944 ± 0.229 | 0.812 ± 1.141 | 0.734 | |
| rhlI | CTZ | Mean ± SD | 0.641 ± 0.337 | 1.151 ± 0.113 | 0.985 ± 0.220 | 0.246 |
| AZT | Mean ± SD | 1.159 ± 0.240 | 1.276 ± 0.168 | 1.996 ± 0.708 | 0.269 | |
| MER | Mean ± SD | 1.603 ± 0.204 | 1.516 ± 0.235 | 1.010 ± 0.180 | 0.117 | |
| CIP | Mean ± SD | 0.744 ± 0.035 | 0.578 ± 0.506 | 0.710 ± 0.501 | 0.916 | |
| AMK | Mean ± SD | 0.985 ± 0.041 | 0.349 ± 0.063 | 1.160 ± 0.519 | 0.143 | |
| rhlR | CTZ | Mean ± SD | 0.720 ± 0.244 | 0.922 ± 0.090 | 1.916 ± 1.235 | 0.346 |
| AZT | Mean ± SD | 1.489 ± 0.113 | 0.889 ± 0.035 | 1.816 ± 0.484 | 0.102 | |
| MER | Mean ± SD | 1.340 ± 0.363 a | 0.169 ± 0.236 a,b | 1.738 ± 0.047 b | 0.017 * | |
| CIP | Mean ± SD | 0.945 ± 0.316 a | 0.642 ± 0.160 b | 2.474 ± 0.182 a,b | 0.008 * | |
| AMK | Mean ± SD | 1.034 ± 0.013 | 0.442 ± 0.273 | 1.863 ± 1.644 | 0.438 |
| Primer Name | Primer Sequence | Annealing |
|---|---|---|
| rpod F | 5′CGAACTGCTTGCCGACTT3′ | 53 °C |
| rpod R | 5′GCGAGAGCCTCAAGGATAC3′ | |
| lasI F | 5′CGCACATCTGGGAACTCA3′ | 52 °C |
| lasI R | 5′CGGCACGGATCATCATCT3′ | |
| lasR F | 5′CTGTGGATGCTCAAGGACTAC3′ | 50 °C |
| lasR R | 5′AACTGGTCTTGCCGATGG 3′ | |
| rhlI F | 5′CTGTGGATGCTCAAGGACTAC3′ | 53 °C |
| rhlI R | 5′AACTGGTCTTGCCGATGG3′ | |
| rhlR F | 5′GTAGCGGGTTTGCGGATG3′ | 54 °C |
| rhlR R | 5′CGGCATCAGGTCTTCATCG3′ | |
| pqsA F | 5′GACCGGCTGTATTCGATTC3′ | 51 °C |
| pqsA R | 5′GCTGAACCAGGGAAAGAAC3′ | |
| pqsR F | 5′CTGATCTGCCGGTAATTGG3′ | 51 °C |
| pqsR R | 5′ATCGACGAGGAACTGAAGA3′ | |
| phzA F | 5′TGAACGGTCAGCGGTACAGGG3′ | 57 °C |
| phzA R | 5′TCCACCGTGGCGCGATTCAG3′ |
References
- O’Loughlin, C.T.; Miller, L.C.; Siryaporn, A.; Drescher, K.; Semmelhack, M.F.; Bassler, B.L. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. USA 2013, 110, 17981–17986. [Google Scholar] [CrossRef] [PubMed]
- Dubern, J.-F.; Halliday, N.; Cámara, M.; Winzer, K.; Barrett, D.A.; Hardie, K.R.; Williams, P. Growth rate and nutrient limitation as key drivers of extracellular quorum sensing signal molecule accumulation in Pseudomonas aeruginosa. Microbiology 2023, 169, 001316. [Google Scholar] [CrossRef] [PubMed]
- Vadakkan, K.; Ngangbam, A.K.; Sathishkumar, K.; Rumjit, N.P.; Cheruvathur, M.K. A review of chemical signaling pathways in the quorum sensing circuit of Pseudomonas aeruginosa. Int. J. Biol. Macromol. 2024, 254, 127861. [Google Scholar] [CrossRef] [PubMed]
- El-Mowafy, S.A.; Abd El Galil, K.H.; Habib, E.-S.E.; Shaaban, M.I. Quorum sensing inhibitory activity of sub-inhibitory concentrations of β-lactams. Afr. Health Sci. 2017, 17, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Nolan, C.; Behrends, V. Sub-inhibitory antibiotic exposure and virulence in Pseudomonas aeruginosa. Antibiotics 2021, 10, 1393. [Google Scholar] [CrossRef] [PubMed]
- Aleanizy, F.S.; Alqahtani, F.Y.; Eltayb, E.K.; Alrumikan, N.; Almebki, R.; Alhossan, A.; Almangour, T.A.; AlQahtani, H. Evaluating the effect of antibiotics sub-inhibitory dose on Pseudomonas aeruginosa quorum sensing dependent virulence and its phenotypes. Saudi J. Biol. Sci. 2021, 28, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhang, X.; Ding, X.; Wang, Y.; Zhu, G. Regulatory mechanisms of sub-inhibitory levels antibiotics agent in bacterial virulence. Appl. Microbiol. Biotechnol. 2021, 105, 3495–3505. [Google Scholar] [CrossRef] [PubMed]
- Pesci, E.C.; Pearson, J.P.; Seed, P.C.; Iglewski, B.H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1997, 179, 3127–3132. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Abu-Sini, M.; Al-Kafaween, M.A.; Al-Groom, R.M.; Hilmi, A.B.M. Comparative in vitro activity of various antibiotic against planktonic and biofilm and the gene expression profile in Pseudomonas aeruginosa. AIMS Microbiol. 2023, 9, 313. [Google Scholar] [CrossRef] [PubMed]
- Mojsoska, B.; Ghoul, M.; Perron, G.G.; Jenssen, H.; Alatraktchi, F.A. Changes in toxin production of environmental Pseudomonas aeruginosa isolates exposed to sub-inhibitory concentrations of three common antibiotics. PLoS ONE 2021, 16, e0248014. [Google Scholar] [CrossRef] [PubMed]
- Glen, K.A.; Lamont, I.L. Penicillin-binding protein 3 sequence variations reduce susceptibility of Pseudomonas aeruginosa to β-lactams but inhibit cell division. J. Antimicrob. Chemother. 2024, 79, 2170–2178. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sanz-García, F.; Hernando-Amado, S.; Martínez, J.L. Evolution under low antibiotic concentrations: A risk for the selection of Pseudomonas aeruginosa multidrug-resistant mutants in nature. Environ. Microbiol. 2022, 24, 1279–1293. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M.; Dutta, D.; Willcox, M.D.P. Activity of Antimicrobial Peptides and Ciprofloxacin against Pseudomonas aeruginosa Biofilms. Molecules 2020, 25, 3843. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Goh, E.-B.; Yim, G.; Tsui, W.; McClure, J.; Surette, M.G.; Davies, J. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. USA 2002, 99, 17025–17030. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Lee, J.-W.; Javaid, A.; Park, S.-K.; Kim, Y.-M. Inhibition of biofilm and virulence properties of Pseudomonas aeruginosa by sub-inhibitory concentrations of aminoglycosides. Microb Pathog. 2020, 146, 104249. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xia, L.; Chen, J.; Lian, Y.; Dandekar, A.A.; Xu, F.; Wang, M. Resistance elicited by sub-lethal concentrations of ampicillin is partially mediated by quorum sensing in Pseudomonas aeruginosa. Environ. Int. 2021, 156, 106619. [Google Scholar] [CrossRef] [PubMed]
- Deshamukhya, C.; Das, B.J.; Paul, D.; Dhar, D.; Bhattacharjee, A. Imipenem and meropenem influence the Las/Rhl quorum-sensing systems in clinical isolates of Pseudomonas aeruginosa. Lett. Appl. Microbiol. 2023, 76, ovad084. [Google Scholar] [CrossRef] [PubMed]
- Cirz, R.T.; O’Neill, B.M.; Hammond, J.A.; Head, S.R.; Romesberg, F.E. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J. Bacteriol. 2006, 188, 7101–7110. [Google Scholar] [CrossRef] [PubMed]
- Deryabin, D.; Inchagova, K. Inhibitory effect of aminoglycosides and tetracyclines on quorum sensing in Chromobacterium violaceum. Microbiology 2018, 87, 1–8. [Google Scholar] [CrossRef]
- Lang, M.; Carvalho, A.; Baharoglu, Z.; Mazel, D. Aminoglycoside uptake, stress, and potentiation in Gram-negative bacteria: New therapies with old molecules. Microbiol. Mol. Biol. Rev. 2023, 87, e00036-22. [Google Scholar] [CrossRef] [PubMed]
- Kai, T.; Tateda, K.; Kimura, S.; Ishii, Y.; Ito, H.; Yoshida, H.; Kimura, T.; Yamaguchi, K. A low concentration of azithromycin inhibits the mRNA expression of N-acyl homoserine lactone synthesis enzymes, upstream of lasI or rhlI, in Pseudomonas aeruginosa. Pulm. Pharmacol. Ther. 2009, 22, 483–486. [Google Scholar] [CrossRef] [PubMed]
- Skindersoe, M.E.; Alhede, M.; Phipps, R.; Yang, L.; Jensen, P.O.; Rasmussen, T.B.; Bjarnsholt, T.; Tolker-Nielsen, T.; Høiby, N.; Givskov, M. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2008, 52, 3648–3663. [Google Scholar] [CrossRef] [PubMed]
- Kukurugya, M.A.; Mendonca, C.M.; Solhtalab, M.; Wilkes, R.A.; Thannhauser, T.W.; Aristilde, L. Multi-omics analysis unravels a segregated metabolic flux network that tunes co-utilization of sugar and aromatic carbons in Pseudomonas putida. J. Biol. Chem. 2019, 294, 8464–8479. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gupta, P.; Chhibber, S.; Harjai, K. Subinhibitory concentration of ciprofloxacin targets quorum sensing system of Pseudomonas aeruginosa causing inhibition of biofilm formation & reduction of virulence. Indian J. Med. Res. 2016, 143, 643–651. [Google Scholar] [PubMed]
- Kumar, L.; Brenner, N.; Brice, J.; Klein-Seetharaman, J.; Sarkar, S.K. Cephalosporins interfere with quorum sensing and improve the ability of Caenorhabditis elegans to survive Pseudomonas aeruginosa infection. Front. Microbiol. 2021, 12, 598498. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.; Brenner, N.; Brice, J.; Klein-Seetharaman, J.; Sarkar, S.K. Cephalosporins target quorum sensing and suppress virulence of Pseudomonas aeruginosa in Caenorhabditis elegans infection model. bioRxiv 2020. [Google Scholar] [CrossRef]
- Keegan, N.R.; Colón Torres, N.J.; Stringer, A.M.; Prager, L.I.; Brockley, M.W.; McManaman, C.L.; Wade, J.T.; Paczkowski, J.E. Promoter selectivity of the RhlR quorum-sensing transcription factor receptor in Pseudomonas aeruginosa is coordinated by distinct and overlapping dependencies on C4-homoserine lactone and PqsE. PLoS Genet. 2023, 19, e1010900. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Wu, B.; Li, J.; Hong, M.; Liu, Y.; Deng, Y.; Ye, J.; Wang, Y.; Zhu, H. Synthesis and active evaluation of LecB-targeted cyclic dipeptide quorum sensing inhibitors. J. Mol. Struct. 2025, 1340, 142533. [Google Scholar] [CrossRef]
- Husain, F.M.; Ahmad, I.; Baig, M.H.; Khan, M.S.; Khan, M.S.; Hassan, I.; Al-Shabib, N.A. Broad-spectrum inhibition of AHL-regulated virulence factors and biofilms by sub-inhibitory concentrations of ceftazidime. RSC Adv. 2016, 6, 27952–27962. [Google Scholar] [CrossRef]
- Shin, B.; Park, C.; Park, W. Stress responses linked to antimicrobial resistance in Acinetobacter species. Appl. Microbiol. Biotechnol. 2020, 104, 1423–1435. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Naga, N.G.; El-Badan, D.E.; Rateb, H.S.; Ghanem, K.M.; Shaaban, M.I. Quorum Sensing Inhibiting Activity of Cefoperazone and Its Metallic Derivatives on Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2021, 11, 716789. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhou, J.-W.; Chen, T.-T.; Tan, X.-J.; Sheng, J.-Y.; Jia, A.-Q. Can the quorum sensing inhibitor resveratrol function as an aminoglycoside antibiotic accelerant against Pseudomonas aeruginosa? Int. J. Antimicrob. Agents. 2018, 52, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Boban, T.; Nadar, S.; Tauro, S. Breaking down bacterial communication: A review of quorum quenching agents. Future J. Pharm. Sci. 2023, 9, 77. [Google Scholar] [CrossRef]
- Dalhoff, A. Selective toxicity of antibacterial agents—Still a valid concept or do we miss chances and ignore risks? Infection 2021, 49, 29–56. [Google Scholar] [CrossRef] [PubMed]
- Blasi, F.; Mantero, M.; Aliberti, S. Antibiotics as immunomodulant agents in COPD. Curr. Opin. Pharmacol. 2012, 12, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Naga, N.G.; El-Badan, D.E.; Mabrouk, M.E.; Rateb, H.S.; Ghanem, K.M.; Shaaban, M.I. Innovative application of ceftriaxone as a quorum sensing inhibitor in Pseudomonas aeruginosa. Sci. Rep. 2025, 15, 5022. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ji, T.; Tang, W.; Wu, Q.; Saunders, J.R. Bacterial foraging algorithm with varying population. Biosystems 2010, 100, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Nalca, Y.; Jänsch, L.; Bredenbruch, F.; Geffers, R.; Buer, J.; Häussler, S. Quorum-sensing antagonistic activities of azithromycin in Pseudomonas aeruginosa PAO1: A global approach. Antimicrob. Agents Chemother. 2006, 50, 1680–1688. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Park, H.-Y.; Park, S.J.; Park, S.-J.; Kim, S.-K.; Ha, C.; Im, S.-J.; Lee, J.-H. Growth phase-differential quorum sensing regulation of anthranilate metabolism in Pseudomonas aeruginosa. Mol. Cells 2011, 32, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Tezel, U.; Pavlostathis, S.G. Quaternary ammonium disinfectants: Microbial adaptation, degradation and ecology. Curr. Opin. Biotechnol. 2015, 33, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Ke, Y.; Bai, F. Active efflux in dormant bacterial cells–new insights into antibiotic persistence. Drug Resist. Updates 2017, 30, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, A.; Laureti, L.; Crussard, S.; Abida, H.; Rodríguez-Rojas, A.; Blázquez, J.; Baharoglu, Z.; Mazel, D.; Darfeuille, F.; Vogel, J.; et al. β-Lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat. Commun. 2013, 4, 1610. [Google Scholar] [CrossRef] [PubMed]
- Martins, D.; McKay, G.; Sampathkumar, G.; Khakimova, M.; English, A.M.; Nguyen, D. Superoxide dismutase activity confers (p) ppGpp-mediated antibiotic tolerance to stationary-phase Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2018, 115, 9797–9802. [Google Scholar] [CrossRef] [PubMed]
- Wayne, P. CLSI Supplements M100; CLSI Performance Standards for Antimicrobial Susceptibility Testing; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- Elsheredy, A.; El-Soudany, I.; Elsherbini, E.; Metwally, D.; Ghazal, A. Effect of azithromycin and phenylalanine-arginine beta-naphthylamide on quorum sensing and virulence factors in clinical isolates of Pseudomonas aeruginosa. Iran. J. Microbiol. 2021, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- İnat, G.; Sırıken, B.; Başkan, C.; Erol, İ.; Yıldırım, T.; Çiftci, A. Quorum sensing systems and related virulence factors in Pseudomonas aeruginosa isolated from chicken meat and ground beef. Sci. Rep. 2021, 11, 15639. [Google Scholar] [CrossRef] [PubMed]
- El-Shaer, S.; Shaaban, M.; Barwa, R.; Hassan, R. Control of quorum sensing and virulence factors of Pseudomonas aeruginosa using phenylalanine arginyl β-naphthylamide. J. Med. Microbiol. 2016, 65, 1194–1204. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amer, A.N.; Attia, N.; Baecker, D.; Mansour, R.E.; El-Soudany, I. Growth-Phase-Dependent Modulation of Quorum Sensing and Virulence Factors in Pseudomonas aeruginosa ATCC 27853 by Sub-MICs of Antibiotics. Antibiotics 2025, 14, 731. https://doi.org/10.3390/antibiotics14070731
Amer AN, Attia N, Baecker D, Mansour RE, El-Soudany I. Growth-Phase-Dependent Modulation of Quorum Sensing and Virulence Factors in Pseudomonas aeruginosa ATCC 27853 by Sub-MICs of Antibiotics. Antibiotics. 2025; 14(7):731. https://doi.org/10.3390/antibiotics14070731
Chicago/Turabian StyleAmer, Ahmed Noby, Nancy Attia, Daniel Baecker, Rasha Emad Mansour, and Ingy El-Soudany. 2025. "Growth-Phase-Dependent Modulation of Quorum Sensing and Virulence Factors in Pseudomonas aeruginosa ATCC 27853 by Sub-MICs of Antibiotics" Antibiotics 14, no. 7: 731. https://doi.org/10.3390/antibiotics14070731
APA StyleAmer, A. N., Attia, N., Baecker, D., Mansour, R. E., & El-Soudany, I. (2025). Growth-Phase-Dependent Modulation of Quorum Sensing and Virulence Factors in Pseudomonas aeruginosa ATCC 27853 by Sub-MICs of Antibiotics. Antibiotics, 14(7), 731. https://doi.org/10.3390/antibiotics14070731







