In Vitro Susceptibility to Imipenem/Relebactam and Comparators in a Multicentre Collection of Mycobacterium abscessus Complex Isolates
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Bacterial Isolates
2.2. Antimicrobial Susceptibility Testing
2.3. Dual-Drug (Based on Imipenem/Relebactam) Susceptibility Testing by the Checkerboard Method and FIC Determination
2.4. Whole Genome Sequencing
3. Results
3.1. Distribution of the M. abscessus Complex in the Study
3.2. Antimicrobial Susceptibility
3.3. In Vitro Synergism of Combinations Based on Imipenem/Relebactam
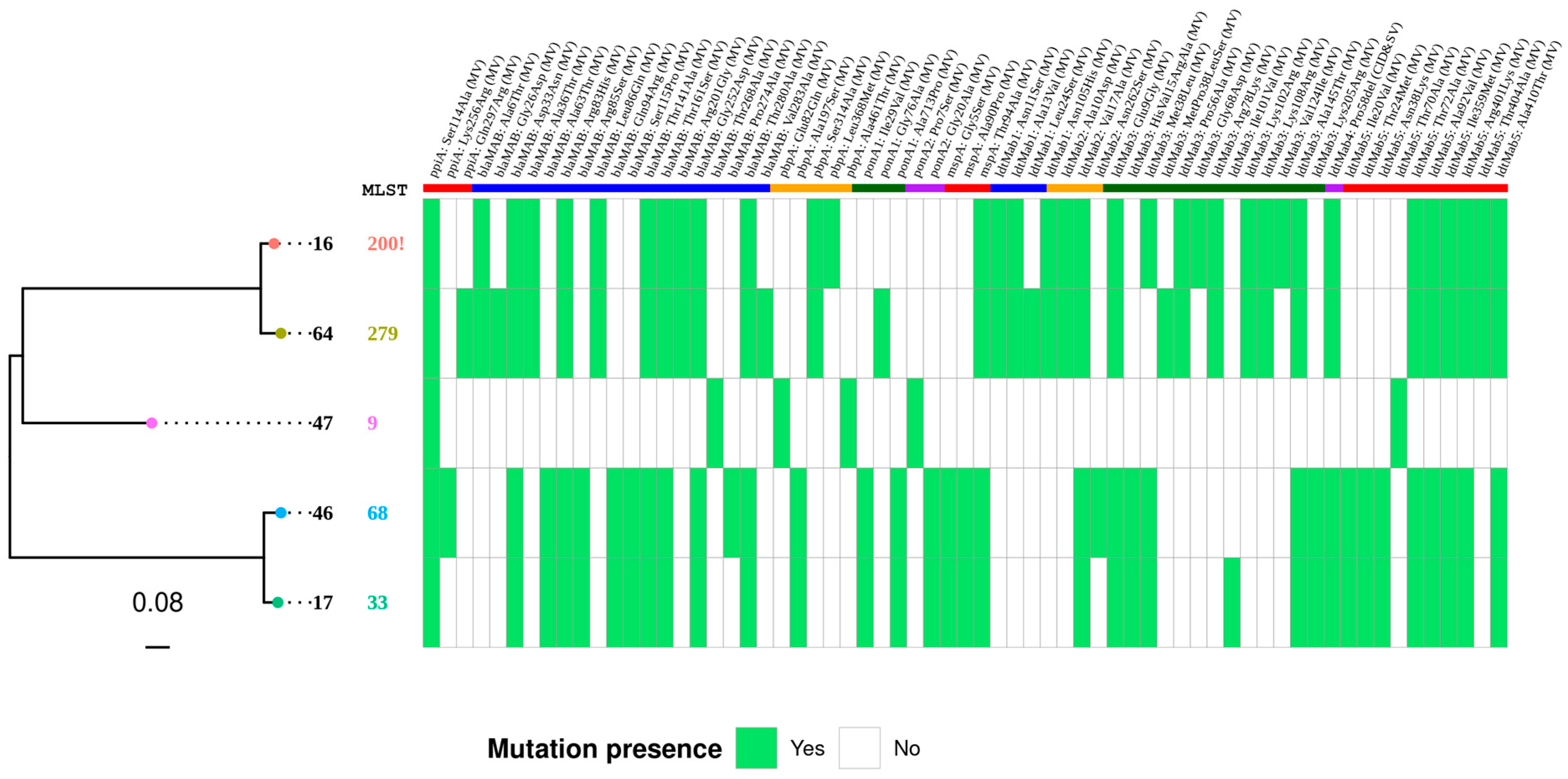
3.4. β-Lactam Resistance Determinants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, M.R.; Sheng, W.H.; Hung, C.C.; Yu, C.J.; Lee, L.N.; Hsueh, P.R. Mycobacterium abscessus Complex Infections in Humans. Emerg. Infect. Dis. 2015, 21, 1638–1646. [Google Scholar] [CrossRef]
- van Ingen, J.; Boeree, M.J.; van Soolingen, D.; Mouton, J.W. Resistance mechanisms and drug susceptibility testing of nontuberculous mycobacteria. Drug Resist. Updates 2012, 15, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Daley, C.L.; Iaccarino, J.M.; Lange, C.; Cambau, E.; Wallace, R.J.; Andrejak, C.; Böttger, E.C.; Brozek, J.; E Griffith, D.; Guglielmetti, L.; et al. Treatment of Nontuberculous Mycobacterial Pulmonary Disease: An Official ATS/ERS/ESCMID/IDSA Clinical Practice Guideline. Clin. Infect. Dis. 2020, 71, e1–e36, Erratum in Clin. Infect. Dis. 2020, 71, 3023. [Google Scholar] [CrossRef] [PubMed]
- Novosad, S.A.; Beekmann, S.E.; Polgreen, P.M.; Mackey, K.; Winthrop, K.L.; M. abscessus Study Team. Treatment of Mycobacterium abscessus Infection. Emerg. Infect. Dis. 2016, 22, 511–514. [Google Scholar] [CrossRef]
- Pasipanodya, J.G.; Ogbonna, D.; Ferro, B.E.; Magombedze, G.; Srivastava, S.; Deshpande, D.; Gumbo, T. Systematic Review and Meta-analyses of the Effect of Chemotherapy on Pulmonary Mycobacterium abscessus Outcomes and Disease Recurrence. Antimicrob. Agents Chemother. 2017, 61, 10-1128. [Google Scholar] [CrossRef]
- Burke, A.; Smith, D.; Coulter, C.; Bell, S.C.; Thomson, R.; Roberts, J.A. Clinical pharmacokinetic and pharmacodynamic considerations in the drug treatment of non-tuberculous mycobacteria in cystic fibrosis. Clin. Pharmacokinet. 2021, 60, 1081–1102. [Google Scholar] [CrossRef] [PubMed]
- Soroka, D.; Dubée, V.; Soulier-Escrihuela, O.; Cuinet, G.; Hugonnet, J.E.; Gutmann, L.; Mainardi, J.L.; Arthur, M. Characterization of broad-spectrum Mycobacterium abscessus class A β-lactamase. J. Antimicrob. Chemother. 2014, 69, 691–696. [Google Scholar] [CrossRef]
- Soroka, D.; Ourghanlian, C.; Compain, F.; Fichini, M.; Dubée, V.; Mainardi, J.L.; Hugonnet, J.E.; Arthur, M. Inhibition of β-lactamases of mycobacteria by avibactam and clavulanate. J. Antimicrob. Chemother. 2017, 72, 1081–1088. [Google Scholar] [CrossRef]
- Le Run, E.; Atze, H.; Arthur, M.; Mainardi, J.L. Impact of relebactam-mediated inhibition of Mycobacterium abscessus BlaMab β-lactamase on the in vitro and intracellular efficacy of imipenem. J. Antimicrob. Chemother. 2020, 75, 379–383. [Google Scholar] [CrossRef]
- Kaushik, A.; Ammerman, N.C.; Lee, J.; Martins, O.; Kreiswirth, B.N.; Lamichhane, G.; Parrish, N.M.; Nuermberger, E.L. In Vitro Activity of the New β-Lactamase Inhibitors Relebactam and Vaborbactam in Combination with β-Lactams against Mycobacterium abscessus Complex Clinical Isolates. Antimicrob. Agents Chemother. 2019, 63, 10-1128. [Google Scholar] [CrossRef]
- Bouzinbi, N.; Marcy, O.; Bertolotti, T.; Chiron, R.; Bemer, P.; Pestel-Caron, M.; Peuchant, O.; Guet-Revillet, H.; Fangous, M.S.; Héry-Arnaud, G.; et al. Evaluation of the GenoType NTM-DR assay performance for the identification and molecular detection of antibiotic resistance in Mycobacterium abscessus complex. PLoS ONE 2020, 15, e0239146. [Google Scholar] [CrossRef] [PubMed]
- CLSI. CLSI standard M24. In Susceptibility Testing of Mycobacteria, Nocardia spp., and Other Aerobic Actinomycetes, 3rd ed.; Clinical and Laboratory Standard Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Petrini, B. Mycobacterium abscessus: An emerging rapid growing potential pathogen. Apmis 2006, 114, 319–328. [Google Scholar] [CrossRef]
- Shen, G.H.; Wu, B.D.; Hu, S.T.; Lin, C.F.; Wu, K.M.; Chen, J.H. High efficacy of clofazimine and its synergistic effect with amikacin against rapidly growing mycobacteria. Int. J. Antimicrob. Agents 2010, 35, 400–404. [Google Scholar] [CrossRef]
- Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Lukjancenko, O.; Saputra, D.; Rasmussen, S.; Hasman, H.; Sicheritz-Pontén, T.; Aarestrup, F.M.; Ussery, D.W.; Lund, O. Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 2014, 52, 1529–1539. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7, 000685. [Google Scholar]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Robertson, J.; Nash, J.H.E. MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef]
- Seemann, T. Snippy: Fast Bacterial Variant Calling from NGS Reads. Available online: https://github.com/tseemann/snippy (accessed on 15 September 2024).
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.Y. Ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Rifat, D.; Chen, L.; Kreiswirth, B.N.; Nuermberger, E.L. Genome-Wide Essentiality Analysis of Mycobacterium abscessus by Saturated Transposon Mutagenesis and Deep Sequencing. mBio 2021, 12, e0104921. [Google Scholar] [CrossRef] [PubMed]
- Shoen, C.; Benaroch, D.; Sklaney, M.; Cynamon, M. In Vitro Activities of Omadacycline against Rapidly Growing Mycobacteria. Antimicrob. Agents Chemother. 2019, 63, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Sayed, A.R.M.; Shah, N.R.; Basso, K.B.; Kamat, M.; Jiao, Y.; Moya, B.; Sutaria, D.S.; Lang, Y.; Tao, X.; Liu, W.; et al. First Penicillin-Binding Protein Occupancy Patterns for 15 β-Lactams and β-Lactamase Inhibitors in Mycobacterium abscessus. Antimicrob. Agents Chemother. 2020, 65, 10-1128. [Google Scholar] [CrossRef]
- Lopeman, R.C.; Harrison, J.; Rathbone, D.L.; Desai, M.; Lambert, P.A.; Cox, J.A.G. Effect of Amoxicillin in combination with Imipenem-Relebactam against Mycobacterium abscessus. Sci. Rep. 2020, 10, 928. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, A.; Ammerman, N.C.; Martins, O.; Parrish, N.M.; Nuermberger, E.L. In Vitro Activity of New Tetracycline Analogs Omadacycline and Eravacycline against Drug-Resistant Clinical Isolates of Mycobacterium abscessus. Antimicrob. Agents Chemother. 2019, 63, 10-1128. [Google Scholar] [CrossRef]
- Story-Roller, E.; Maggioncalda, E.C.; Cohen, K.A.; Lamichhane, G. Mycobacterium abscessus and β-Lactams: Emerging Insights and Potential Opportunities. Front. Microbiol. 2018, 9, 2273. [Google Scholar] [CrossRef]
- Kumar, P.; Chauhan, V.; Silva, J.R.A.; Lameira, J.; d’Andrea, F.B.; Li, S.G.; Ginell, S.L.; Freundlich, J.S.; Alves, C.N.; Bailey, S.; et al. Mycobacterium abscessus l,d-transpeptidases are susceptible to inactivation by carbapenems and cephalosporins but not penicillins. Antimicrob. Agents Chemother. 2017, 61, 10-1128. [Google Scholar] [CrossRef]
- Le Run, E.; Tettelin, H.; Holland, S.M.; Zelazny, A.M. Evolution towards extremely high imipenem resistance in Mycobacterium abscessus outbreak strains. Antimicrob. Agents Chemother. 2024, 68, e00673-24. [Google Scholar] [CrossRef]
- Dousa, K.M.; Kurz, S.G.; Taracila, M.A.; Bonfield, T.; Bethel, C.R.; Barnes, M.D.; Selvaraju, S.; Abdelhamed, A.M.; Kreiswirth, B.N.; Boom, W.H.; et al. Insights into the L,D-Transpeptidases and D,D-Carboxypeptidase of Mycobacterium abscessus: Ceftaroline, Imipenem, and Novel Diazabicyclooctane Inhibitors. Antimicrob. Agents Chemother. 2020, 64, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Burke, A.; Carter, R.; Tolson, C.; Congdon, J.; Duplancic, C.; Bursle, E.; Bell, S.C.; Roberts, J.A.; Thomson, R. In vitro susceptibility testing of imipenem-relebactam and tedizolid against 102 Mycobacterium abscessus isolates. Int. J. Antimicrob. Agents 2023, 62, 106938. [Google Scholar] [CrossRef] [PubMed]
- Story-Roller, E.; Maggioncalda, E.C.; Lamichhanea, G. Select β-Lactam Combinations Exhibit Synergy against Mycobacterium abscessus In Vitro. Antimicrob. Agents Chemother. 2019, 63, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Bitar, M.; Le Moigne, V.; Herrmann, J.L.; Arthur, M.; Mainardi, J.L. In vitro, intracellular and in vivo synergy between amoxicillin, imipenem and relebactam against Mycobacterium abscessus. J. Antimicrob. Chemother. 2025, 80, 1560–1567. [Google Scholar] [CrossRef]
| MIC Distribution for MABc Isolates (n = 175) | MIC50 | MIC90 | %R | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | >128 | ||||
| Cefepime | 3 | 18 | 146 | >128 | >128 | c | |||||||||||
| Ceftaroline | 1 | 2 | 14 | 33 | 117 | >128 | >128 | c | |||||||||
| Meropenem | 5 | 44 | 77 | 28 | 13 | 64 | 128 | c | |||||||||
| Imipenem | 1 | 19 | 58 | 72 | 17 | 16 | 32 | 10.2 | |||||||||
| Imipenem/relebactam | 1 | 10 | 48 | 79 | 25 | 4 | 8 | 16 | 2.4 | ||||||||
| Linezolid | 14 | 27 | 18 | 38 | 63 | 7 | 8 | 16 | 4.2 | ||||||||
| Moxifloxacine | 3 | 10 | 51 | 62 | 41 | 8 | 16 | 92.2 | |||||||||
| Clarithromycin 72 h a | 73 | 35 | 28 | 18 | 5 | 2 | 2 | 4 | 0.25 | 1 | 3.6 | ||||||
| Clarithromycin 14d b | 25 | 24 | 16 | 12 | 4 | 72 | 14 | 32 | 32 | 51.5 | |||||||
| Omadacycline | 3 | 14 | 49 | 66 | 35 | 1 | 2 | c | |||||||||
| Eravacycline | 22 | 42 | 49 | 37 | 17 | 0.25 | 1 | c | |||||||||
| Amikacine | 1 | 4 | 16 | 42 | 67 | 28 | 7 | 2 | 8 | 16 | 1.2 | ||||||
| Tigecyclina | 1 | 2 | 21 | 24 | 35 | 51 | 31 | 2 | 0.5 | 2 | 0 | ||||||
| Clofazimine | 3 | 24 | 63 | 45 | 28 | 3 | 1 | 0.25 | 1 | 0.6 | |||||||
| IMI/REL—Meropenem | IMI/REL—Ceftaroline | IMI/REL—Amikacin | IMI/REL—Clarithromycin | IMI/REL—Tigecycline | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate ID | A | a | B | b | FICI | A | a | B | b | FICI | A | a | B | b | FICI | A | a | B | b | FICI | A | a | B | b | FICI |
| 8 | 32 | 16 | 128 | 32 | 0.75 | 32 | 16 | 128 | 64 | 1 | 32 | 16 | 8 | 2 | 0.75 | 32 | 16 | 0.3 | 0.1 | 1 | 32 | 8 | 0.1 | 0.1 | 0.75 |
| 16 | 32 | 16 | 128 | 64 | 1 | 32 | 16 | 128 | 64 | 1 | 32 | 16 | 8 | 4 | 1 | 32 | 16 | 32 | 16 | 1 | 32 | 16 | 0.3 | 0.1 | 0.8 |
| 17 | 8 | 2 | 64 | 8 | 0.38 | 8 | 2 | 64 | 8 | 0.38 | 8 | 2 | 16 | 4 | 0.5 | 8 | 4 | 32 | 4 | 0.6 | 8 | 4 | 0.5 | 0.3 | 1 |
| 29 | 16 | 4 | 32 | 8 | 0.5 | 16 | 4 | 128 | 16 | 0.38 | 16 | 4 | 16 | 2 | 0.38 | 16 | 4 | 64 | 16 | 0.5 | 16 | 4 | 0.1 | 0 | 0.37 |
| 47 | 32 | 16 | 128 | 64 | 1 | 32 | 16 | 128 | 16 | 0.63 | 32 | 8 | 16 | 4 | 0.5 | 32 | 16 | 32 | 8 | 0.8 | 32 | 16 | 1 | 0.5 | 1 |
| 56 | 2 | 0.5 | 32 | 2 | 0.31 | 2 | 0.5 | 128 | 16 | 0.38 | 2 | 0.5 | 4 | 0.5 | 0.38 | 2 | 0.5 | 32 | 8 | 0.5 | 2 | 0.5 | 2 | 0.5 | 0.5 |
| 62 | 8 | 2 | 64 | 16 | 0.5 | 8 | 2 | 64 | 16 | 0.5 | 8 | 1 | 64 | 4 | 0.19 | 8 | 2 | 32 | 8 | 0.5 | 8 | 2 | 2 | 0.5 | 0.5 |
| 64 | 32 | 16 | 128 | 32 | 0.75 | 32 | 16 | 128 | 32 | 0.75 | 32 | 8 | 4 | 0.5 | 0.38 | 32 | 16 | 32 | 16 | 1 | 32 | 8 | 1 | 0.5 | 0.75 |
| 75 | 16 | 4 | 64 | 8 | 0.38 | 16 | 4 | 64 | 16 | 0.5 | 16 | 4 | 32 | 1 | 0.28 | 16 | 4 | 64 | 16 | 0.5 | 16 | 4 | 2 | 0.5 | 0.5 |
| 90 | 8 | 2 | 128 | 32 | 0.5 | 8 | 2 | 64 | 8 | 0.38 | 8 | 2 | 8 | 1 | 0.38 | 8 | 2 | 64 | 4 | 0.3 | 8 | 2 | 2 | 0.5 | 0.5 |
| 110 | 8 | 2 | 64 | 8 | 0.38 | 8 | 2 | 64 | 8 | 0.38 | 8 | 2 | 4 | 1 | 0.5 | 8 | 4 | 2 | 0.5 | 0.8 | 8 | 4 | 2 | 1 | 1 |
| 121 | 4 | 1 | 64 | 8 | 0.38 | 4 | 1 | 32 | 8 | 0.5 | 4 | 1 | 8 | 1 | 0.38 | 4 | 1 | 32 | 4 | 0.4 | 4 | 1 | 1 | 0.1 | 0.37 |
| 137 | 16 | 4 | 128 | 32 | 0.5 | 16 | 8 | 64 | 16 | 0.75 | 16 | 4 | 2 | 0.5 | 0.5 | 16 | 8 | 32 | 16 | 1 | 16 | 8 | 1 | 0.5 | 1 |
| 145 | 4 | 1 | 32 | 8 | 0.5 | 4 | 1 | 128 | 32 | 0.5 | 4 | 1 | 32 | 4 | 0.38 | 4 | 1 | 32 | 8 | 0.5 | 4 | 2 | 2 | 1 | 1 |
| 158 | 8 | 1 | 128 | 16 | 0.25 | 8 | 4 | 64 | 16 | 0.75 | 8 | 1 | 8 | 1 | 0.25 | 8 | 2 | 2 | 0.25 | 0.4 | 8 | 2 | 0.5 | 0.1 | 0.37 |
| 162 | 16 | 4 | 64 | 8 | 0.38 | 16 | 4 | 64 | 4 | 0.31 | 16 | 4 | 4 | 0.5 | 0.38 | 16 | 8 | 1 | 0.5 | 1 | 16 | 4 | 1 | 0.1 | 0.31 |
| IMI—Meropenem | IMI—Ceftaroline | IMI—Amikacin | IMI—Clarithromycin | IMI—Tigecycline | |||||||||||||||||||||
| Isolate ID | A | a | B | b | FICI | A | a | B | b | FICI | A | a | B | b | FICI | A | a | B | b | FICI | A | a | B | b | FICI |
| 8 | 32 | 16 | 128 | 16 | 0.63 | 32 | 16 | 128 | 64 | 1 | 32 | 16 | 8 | 2 | 0.8 | 32 | 8 | 0.3 | 0.1 | 0.8 | 32 | 16 | 0.1 | 0 | 0.75 |
| 16 | 32 | 16 | 128 | 16 | 0.63 | 32 | 16 | 128 | 64 | 1 | 32 | 32 | 8 | 4 | 1.5 | 32 | 4 | 32 | 32 | 1.13 | 32 | 16 | 0.3 | 0.1 | 0.74 |
| 17 | 16 | 8 | 64 | 16 | 0.8 | 16 | 16 | 64 | 2 | 1.03 | 16 | 8 | 16 | 4 | 0.8 | 16 | 8 | 32 | 8 | 0.8 | 16 | 8 | 0.5 | 0.1 | 0.74 |
| 29 | 16 | 16 | 32 | 4 | 1.13 | 16 | 16 | 128 | 4 | 1.03 | 16 | 4 | 16 | 8 | 0.8 | 16 | 4 | 64 | 32 | 0.8 | 16 | 16 | 0.1 | 0 | 1.13 |
| 47 | 32 | 16 | 128 | 64 | 1 | 32 | 16 | 128 | 64 | 1 | 32 | 32 | 16 | 2 | 1.13 | 32 | 8 | 32 | 16 | 0.8 | 32 | 32 | 1 | 0.3 | 1.25 |
| 56 | 8 | 4 | 32 | 16 | 1 | 8 | 2 | 128 | 64 | 0.8 | 8 | 8 | 4 | 0.3 | 1.06 | 8 | 1 | 32 | 32 | 1.13 | 8 | 8 | 2 | 0.3 | 1.13 |
| 62 | 8 | 4 | 64 | 32 | 1 | 8 | 8 | 64 | 2 | 1.03 | 8 | 8 | 64 | 4 | 1.06 | 8 | 1 | 32 | 32 | 1.13 | 8 | 8 | 2 | 0.1 | 1.06 |
| 64 | 32 | 16 | 128 | 16 | 0.63 | 32 | 32 | 128 | 4 | 1.03 | 32 | 32 | 4 | 0.5 | 1.13 | 32 | 16 | 32 | 16 | 1 | 32 | 32 | 1 | 0.1 | 1.06 |
| 75 | 32 | 16 | 64 | 8 | 0.63 | 32 | 32 | 64 | 2 | 1.03 | 32 | 16 | 32 | 4 | 0.63 | 32 | 16 | 64 | 32 | 1 | 32 | 16 | 2 | 0.5 | 0.75 |
| 90 | 16 | 8 | 128 | 32 | 0.8 | 16 | 16 | 64 | 2 | 1.03 | 16 | 16 | 8 | 0.5 | 1.06 | 16 | 8 | 64 | 8 | 0.63 | 16 | 16 | 2 | 0.3 | 1.13 |
| 110 | 16 | 8 | 64 | 16 | 0.8 | 16 | 16 | 64 | 2 | 1.03 | 16 | 16 | 4 | 0.5 | 1.13 | 16 | 2 | 2 | 2 | 1.13 | 16 | 16 | 2 | 0.3 | 1.13 |
| 121 | 8 | 2 | 64 | 32 | 0.8 | 8 | 8 | 32 | 1 | 1.03 | 8 | 8 | 8 | 1 | 1.13 | 8 | 4 | 32 | 4 | 0.63 | 8 | 8 | 1 | 0.1 | 1.12 |
| 137 | 32 | 32 | 128 | 8 | 1.06 | 32 | 32 | 64 | 2 | 1.03 | 32 | 32 | 2 | 0.3 | 1.13 | 32 | 4 | 32 | 32 | 1.13 | 32 | 32 | 1 | 0.1 | 1.12 |
| 145 | 8 | 4 | 32 | 8 | 0.8 | 8 | 8 | 128 | 4 | 1.03 | 8 | 8 | 32 | 4 | 1.13 | 8 | 2 | 32 | 16 | 0.8 | 8 | 8 | 2 | 0.3 | 1.13 |
| 158 | 16 | 8 | 128 | 32 | 0.8 | 16 | 16 | 64 | 2 | 1.03 | 16 | 16 | 8 | 0.5 | 1.06 | 16 | 8 | 2 | 0.5 | 0.8 | 16 | 16 | 0.5 | 0 | 1.06 |
| 162 | 16 | 8 | 64 | 32 | 1 | 16 | 16 | 64 | 2 | 1.03 | 16 | 16 | 4 | 0.1 | 1.03 | 16 | 2 | 1 | 1 | 1.13 | 16 | 8 | 1 | 0.5 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seoane-Estévez, A.; Aja-Macaya, P.; Garcia-Pose, A.; López-Roa, P.; Ruedas-López, A.; Gonzalez-Galán, V.; Esteban, J.; Arca-Suárez, J.; Pampín, M.; Beceiro, A.; et al. In Vitro Susceptibility to Imipenem/Relebactam and Comparators in a Multicentre Collection of Mycobacterium abscessus Complex Isolates. Antibiotics 2025, 14, 682. https://doi.org/10.3390/antibiotics14070682
Seoane-Estévez A, Aja-Macaya P, Garcia-Pose A, López-Roa P, Ruedas-López A, Gonzalez-Galán V, Esteban J, Arca-Suárez J, Pampín M, Beceiro A, et al. In Vitro Susceptibility to Imipenem/Relebactam and Comparators in a Multicentre Collection of Mycobacterium abscessus Complex Isolates. Antibiotics. 2025; 14(7):682. https://doi.org/10.3390/antibiotics14070682
Chicago/Turabian StyleSeoane-Estévez, Alejandro, Pablo Aja-Macaya, Andrea Garcia-Pose, Paula López-Roa, Alba Ruedas-López, Verónica Gonzalez-Galán, Jaime Esteban, Jorge Arca-Suárez, Martín Pampín, Alejandro Beceiro, and et al. 2025. "In Vitro Susceptibility to Imipenem/Relebactam and Comparators in a Multicentre Collection of Mycobacterium abscessus Complex Isolates" Antibiotics 14, no. 7: 682. https://doi.org/10.3390/antibiotics14070682
APA StyleSeoane-Estévez, A., Aja-Macaya, P., Garcia-Pose, A., López-Roa, P., Ruedas-López, A., Gonzalez-Galán, V., Esteban, J., Arca-Suárez, J., Pampín, M., Beceiro, A., Oviaño, M., Bou, G., & on behalf of the GEIM-SEIMC Study Group. (2025). In Vitro Susceptibility to Imipenem/Relebactam and Comparators in a Multicentre Collection of Mycobacterium abscessus Complex Isolates. Antibiotics, 14(7), 682. https://doi.org/10.3390/antibiotics14070682







