Genomic and Functional Characterization of Multidrug-Resistant E. coli: Insights into Resistome, Virulome, and Signaling Systems
Abstract
1. Introduction
2. Results
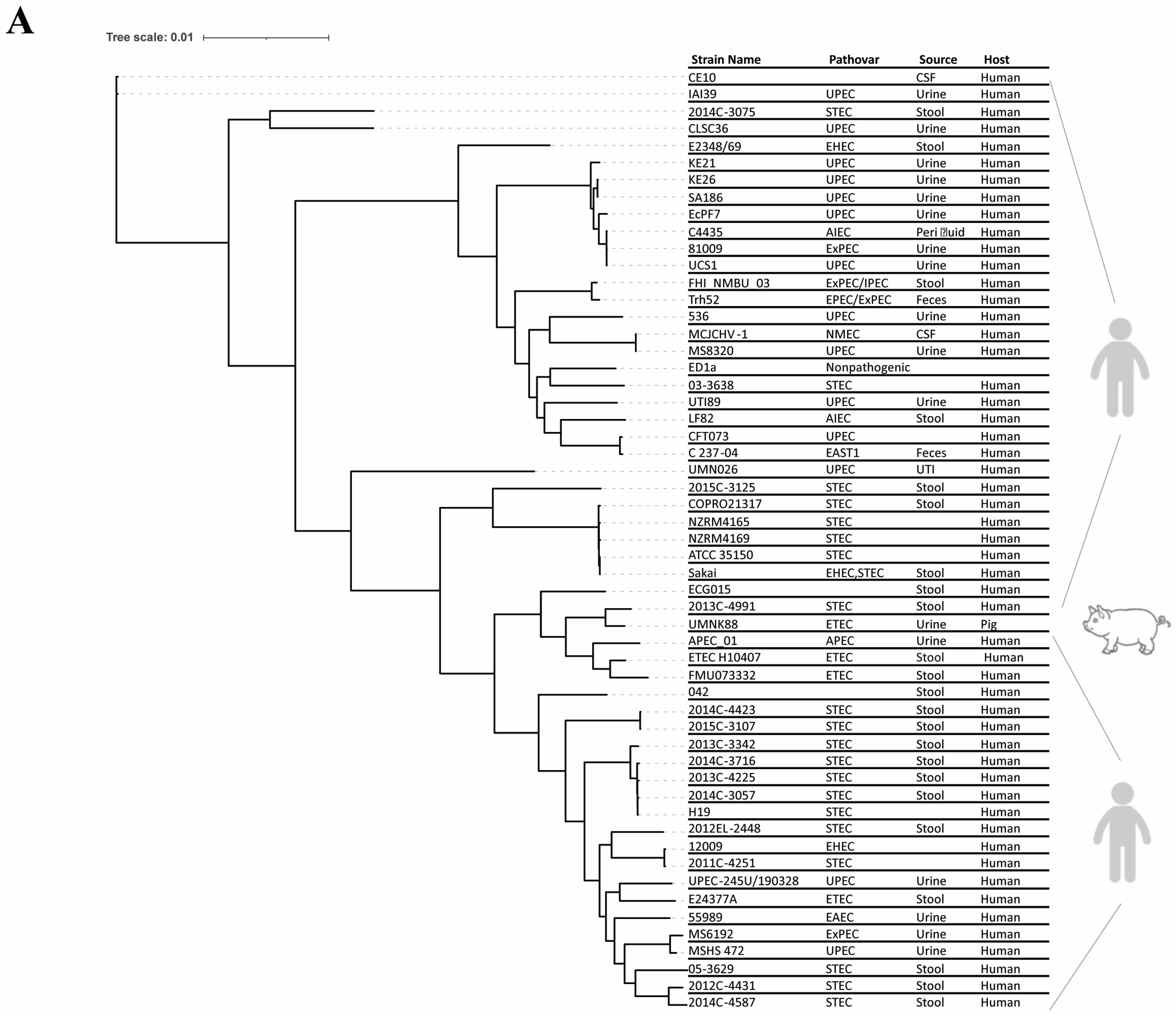
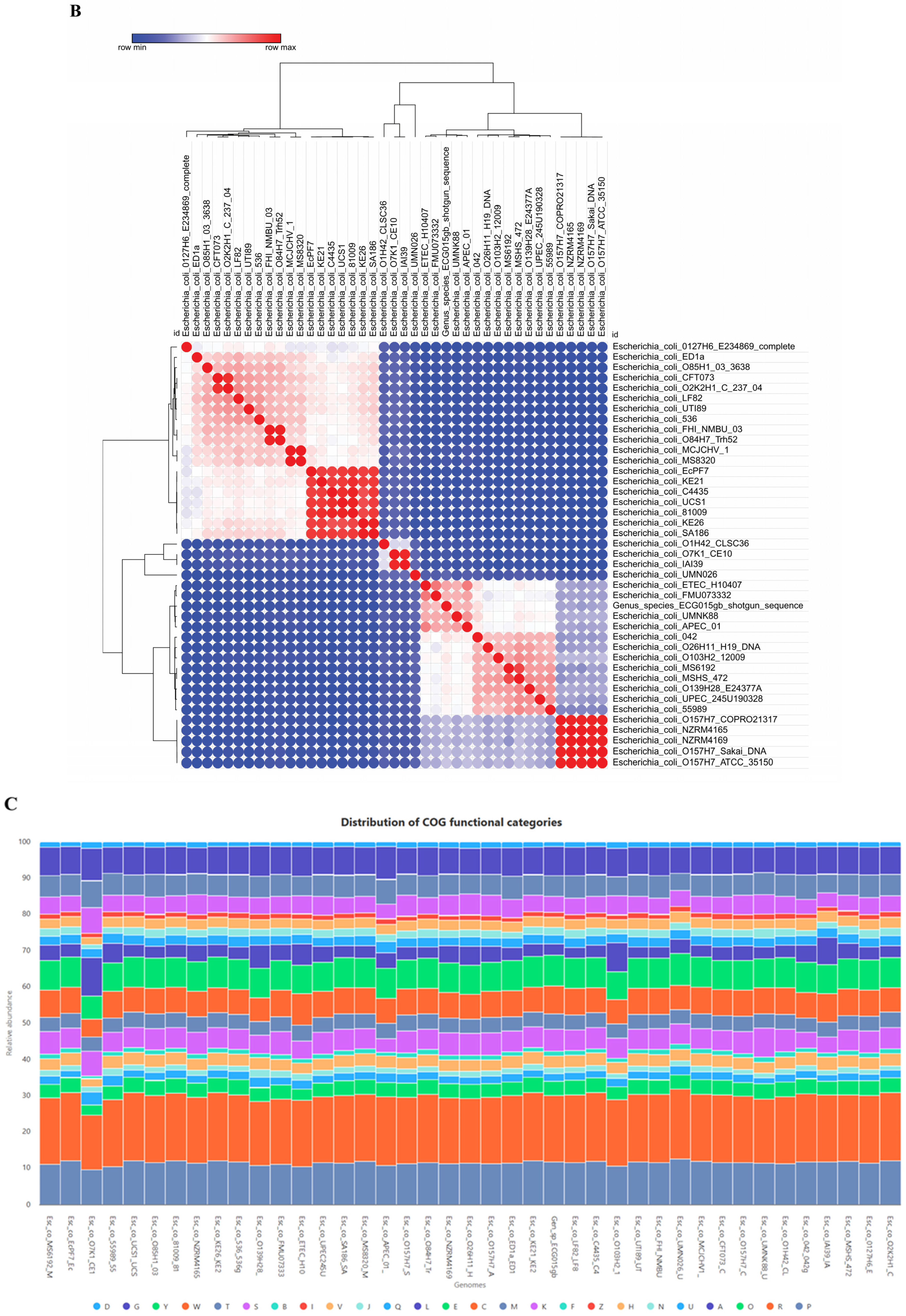
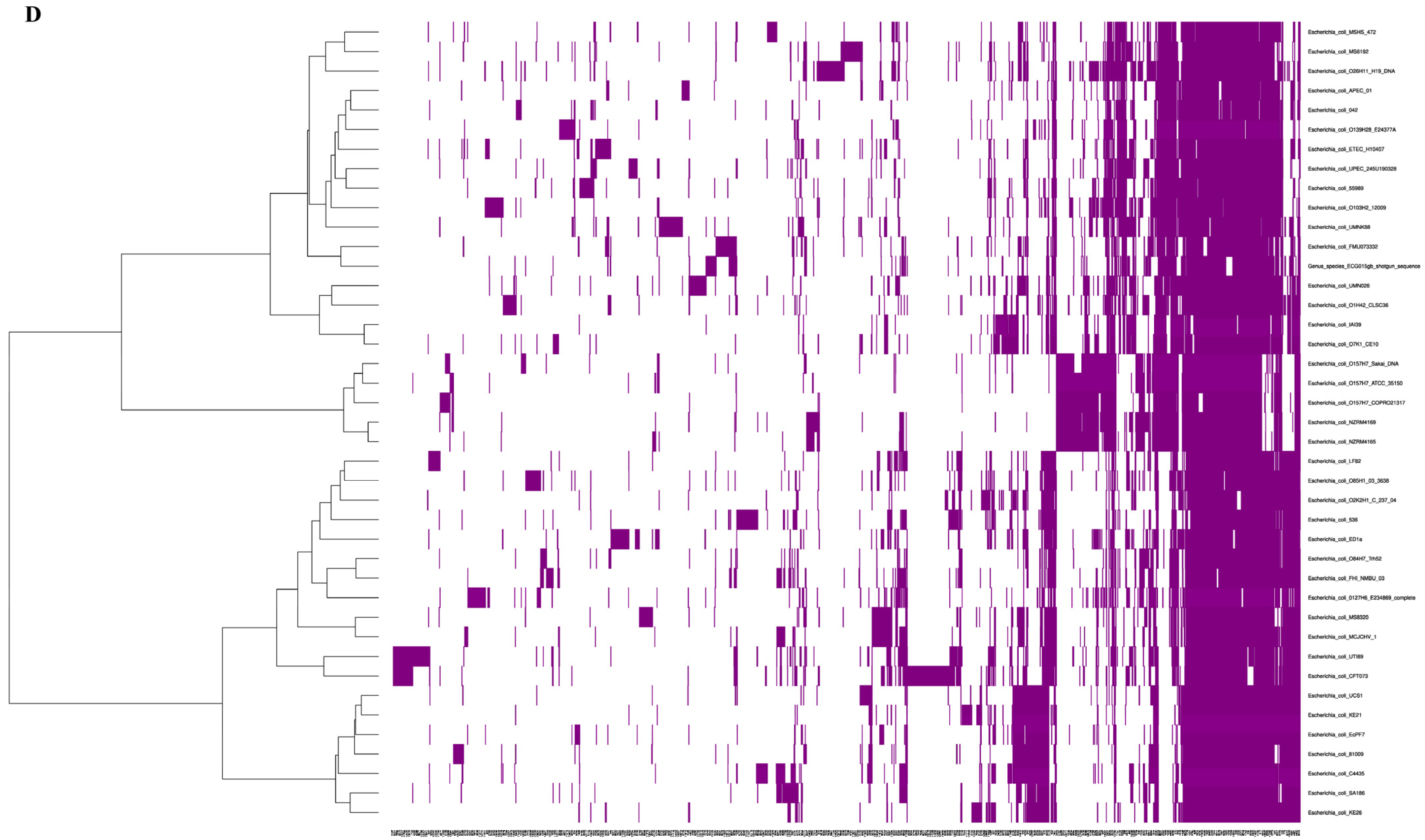
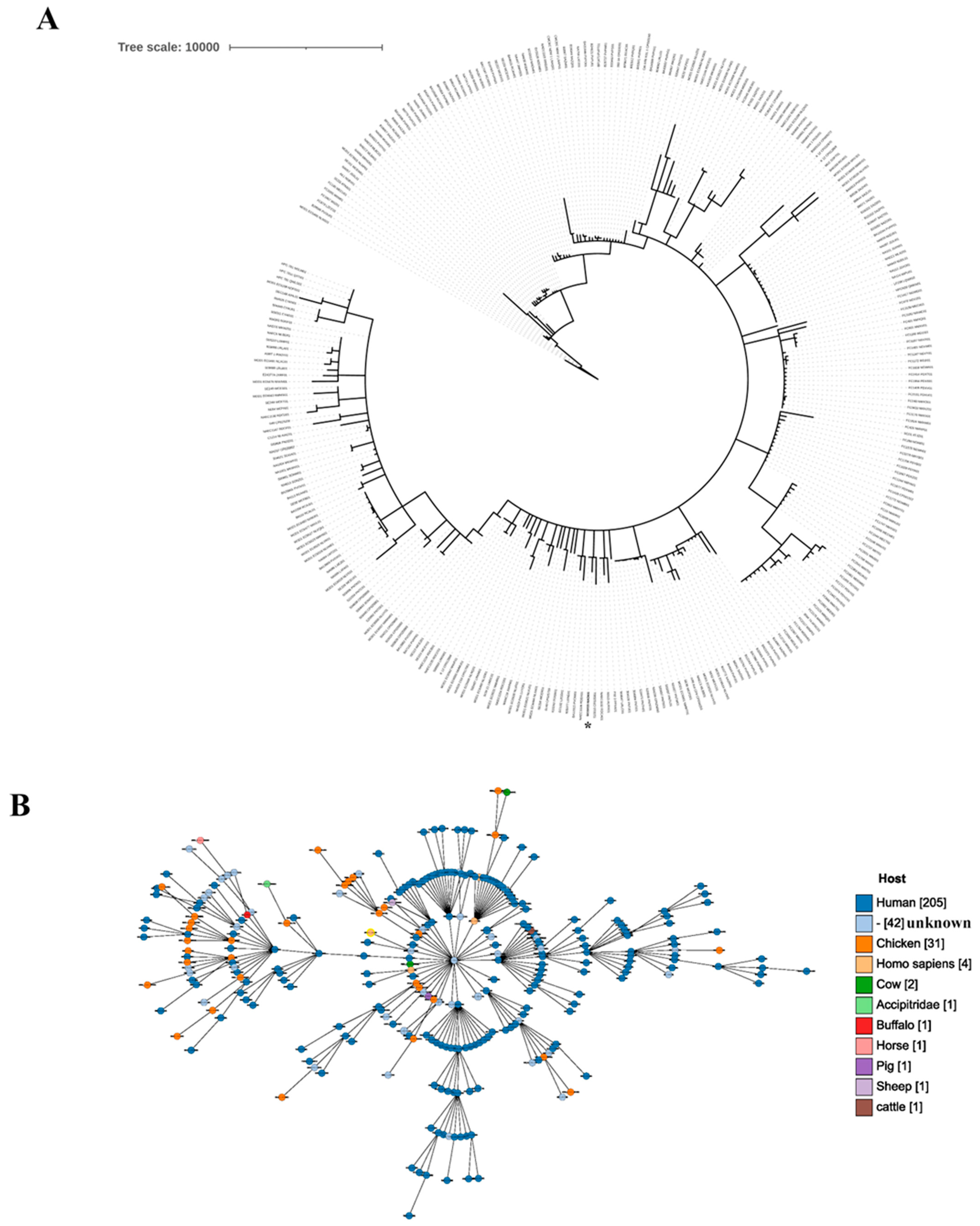
2.1. Genomic Characteristics of the Gut Isolate ECG015
2.2. Sequence Comparison of Etk and Wzc Tyrosine Kinases
2.3. Prevalence of Virulence Genes in the Gut Isolate ECG015
2.4. Genes Associated with Antibiotic Resistance Found in the E. coli Isolate ECG015
2.5. Multidrug Efflux Pumps Identified in the Isolate ECG015
2.6. Signaling Systems Detected in the E. coli Strain ECG015
2.7. Homology Modeling and Docking
2.8. CpxR, the Master Regulator of AMR and Programmed Cell Death
3. Discussion
4. Materials and Methods
4.1. Genome Draft Sequence, Annotation, and Analysis
4.2. Multiple-Sequence Alignment and Phylogenetic Tree
4.3. Homology Modelling, Molecular Docking, and DNA–Protein Complex
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Mendelson, M.; Matsoso, M.P. The World Health Organization Global Action Plan for antimicrobial resistance. S. Afr. Med. J. 2015, 105, 325. [Google Scholar] [CrossRef] [PubMed]
- Pulingam, T.; Parumasivam, T.; Gazzali, A.M.; Sulaiman, A.M.; Chee, J.Y.; Lakshmanan, M.; Chin, C.F.; Sudesh, K. Antimicrobial resistance: Prevalence, economic burden, mechanisms of resistance and strategies to overcome. Eur. J. Pharm. Sci. 2022, 170, 106103. [Google Scholar] [CrossRef]
- Monari, C.; Onorato, L.; Coppola, N.; Raviglione, M.C.B.; Gon, G. Burden of Antimicrobial Resistance Among Women with Post-Partum Infections in Low-Middle Income Countries: A Systematic Review. J. Epidemiol. Glob. Health 2024, 14, 274–290. [Google Scholar] [CrossRef]
- Kumar, S.B.; Arnipalli, S.R.; Ziouzenkova, O. Antibiotics in Food Chain: The Consequences for Antibiotic Resistance. Antibiotics 2020, 9, 688. [Google Scholar] [CrossRef]
- Ashiru-Oredope, D.; Hopkins, S. Antimicrobial resistance: Moving from professional engagement to public action. J. Antimicrob. Chemother. 2015, 70, 2927–2930. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.E.; Joshi, L.T. Knocking Out Antimicrobial Resistance editorial: Reflections on the United Nations General Assembly high-level meeting on antimicrobial resistance. J. Med. Microbiol. 2024, 73, 001937. [Google Scholar] [CrossRef]
- Jesudason, T. WHO publishes updated list of bacterial priority pathogens. Lancet Microbe 2024, 5, 100940. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- WHO Country Office for India; Department of Biotechnology, Government of India. Indian Priority Pathogen List. 2021. Available online: https://cdn.who.int/media/docs/default-source/searo/india/antimicrobial-resistance/ippl_final_web.pdf?sfvrsn=9105c3d1_6 (accessed on 12 June 2025).
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. WHO Pathogens Priority List Working Group. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Tomczyk, S.; Taylor, A.; Brown, A.; de Kraker, M.E.A.; El-Saed, A.; Alshamrani, M.; Hendriksen, R.S.; Jacob, M.; Löfmark, S.; Perovic, O.; et al. WHO AMR Surveillance and Quality Assessment Collaborating Centres Network. Impact of the COVID-19 pandemic on the surveillance, prevention, and control of antimicrobial resistance: A global survey. J. Antimicrob. Chemother. 2021, 76, 3045–3058. [Google Scholar] [CrossRef] [PubMed]
- Dutescu, I.A.; Hillier, S.A. Encouraging the Development of New Antibiotics: Are Financial Incentives the Right Way Forward? A Systematic Review and Case Study. Infect. Drug Resist. 2021, 14, 415–434. [Google Scholar] [CrossRef] [PubMed]
- Sartelli, M.; CHardcastle, T.; Catena, F.; Chichom-Mefire, A.; Coccolini, F.; Dhingra, S.; Haque, M.; Hodonou, A.; Iskandar, K.; Labricciosa, F.M.; et al. Antibiotic Use in Low and Middle-Income Countries and the Challenges of Antimicrobial Resistance in Surgery. Antibiotics 2020, 9, 497. [Google Scholar] [CrossRef]
- Veeraraghavan, B.; Walia, K. Antimicrobial susceptibility profile & resistance mechanisms of Global Antimicrobial Resistance Surveillance System (GLASS) priority pathogens from India. Indian J. Med. Res. 2019, 149, 87–96. [Google Scholar] [CrossRef]
- Veeraraghavan, B.; Shankar, C.; Karunasree, S.; Kumari, S.; Ravi, R.; Ralph, R. Carbapenem resistant Klebsiella pneumoniae isolated from bloodstream infection: Indian experience. Pathog. Glob. Health 2017, 111, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Walia, K.; Madhumathi, J.; Veeraraghavan, B.; Chakrabarti, A.; Kapil, A.; Ray, P.; Singh, H.; Sistla, S.; Ohri, V.C. Establishing Antimicrobial Resistance Surveillance & Research Network in India: Journey so far. Indian J. Med. Res. 2019, 149, 164–179. [Google Scholar] [CrossRef]
- Bertagnolio, S.; Dobreva, Z.; Centner, C.M.; Olaru, I.D.; Donà, D.; Burzo, S.; Huttner, B.D.; Chaillon, A.; Gebreselassie, N.; Wi, T.; et al. WHO Research Agenda for AMR in Human Health Collaborators. WHO global research priorities for antimicrobial resistance in human health. Lancet Microbe 2024, 5, 100902. [Google Scholar] [CrossRef]
- Vijay, S.; Sharma, M.; Misri, J.; Shome, B.R.; Veeraraghavan, B.; Ray, P.; Ohri, V.C.; Walia, K. An integrated surveillance network for antimicrobial resistance, India. Bull. World Health Organ. 2021, 99, 562–571. [Google Scholar] [CrossRef]
- Mendelson, M.; Røttingen, J.A.; Gopinathan, U.; Hamer, D.H.; Wertheim, H.; Basnyat, B.; Butler, C.; Tomson, G.; Balasegaram, M. Maximising access to achieve appropriate human antimicrobial use in low-income and middle-income countries. Lancet 2016, 387, 188–198. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Matsoso, P.; Pant, S.; Brower, C.; Røttingen, J.A.; Klugman, K.; Davies, S. Access to effective antimicrobials: A worldwide challenge. Lancet 2016, 387, 168–175. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling drug-resistant infections globally: Final report and recommendations. Rev. Antimicrob. Resist. 2016. Available online: https://amr-review.org/Publications.html (accessed on 12 June 2025).
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.; Wertheim, H.F.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef] [PubMed]
- Theuretzbacher, U.; Outterson, K.; Engel, A.; Karlén, A. The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 2020, 18, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, M.A.; Walsh, C.T. Antibiotics for emerging pathogens. Science 2009, 325, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Silver, L.L. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 2011, 24, 71–109. [Google Scholar] [CrossRef]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007, 6, 29–40. [Google Scholar] [CrossRef]
- Brown, E.D.; Wright, G.D. Antibacterial drug discovery in the resistance era. Nature 2016, 529, 336–343. [Google Scholar] [CrossRef]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Lewis, K. Persister cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef]
- Wright, G.D. Antibiotic resistance in the environment: A link to the clinic? Curr. Opin. Microbiol. 2010, 5, 589–594. [Google Scholar] [CrossRef]
- WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance. Geneva: World Health Organization. 2024. Available online: https://iris.who.int/bitstream/handle/10665/376776/9789240093461-eng.pdf?sequence=1 (accessed on 27 May 2025).
- Jesser, K.J.; Levy, K. Updates on defining and detecting diarrheagenic Escherichia coli pathotypes. Curr. Opin. Infect. Dis. 2020, 33, 372. [Google Scholar] [CrossRef] [PubMed]
- Singhal, A.; Sharma, R.; Jain, M.; Vyas, L. Hospital and Community Isolates of Uropathogens and their Antibiotic Sensitivity Pattern from a Tertiary Care Hospital in North West India. Ann. Med. Health Sci. Res. 2014, 4, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Modgil, V.; Kaur, H.; Mohan, B.; Taneja, N. Molecular, phylogenetic and antibiotic resistance analysis of enteroaggregative Escherichia coli/uropathogenic Escherichia coli hybrid genotypes causing urinary tract infections. Indian J. Med. Microbiol. 2020, 38, 421–429. [Google Scholar] [CrossRef]
- Ba, X.; Guo, Y.; Moran, R.A.; Doughty, E.L.; Liu, B.; Yao, L.; Li, J.; He, N.; Shen, S.; Li, Y.; et al. Global emergence of a hypervirulent carbapenem-resistant Escherichia coli ST410 clone. Nat. Commun. 2024, 15, 494. [Google Scholar] [CrossRef]
- Huang, J.; Lv, C.; Li, M.; Rahman, T.; Chang, Y.F.; Guo, X.; Song, Z.; Zhao, Y.; Li, Q.; Ni, P.; et al. Carbapenem-resistant Escherichia coli exhibit diverse spatiotemporal epidemiological characteristics across the globe. Commun. Biol. 2024, 7, 51. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wu, Z.; Wang, Y.; Zhang, R.; Zhou, H.W.; Wang, S.; Lei, L.; Li, M.; Cai, J.; Tyrrell, J.; et al. Heterogeneous and Flexible Transmission of mcr-1 in Hospital-Associated Escherichia coli. mBio 2018, 9, e00943-18. [Google Scholar] [CrossRef]
- Ludden, C.; Raven, K.E.; Jamrozy, D.; Gouliouris, T.; Blane, B.; Coll, F.; de Goffau, M.; Naydenova, P.; Horner, C.; Hernandez-Garcia, J.; et al. One Health Genomic Surveillance of Escherichia coli Demonstrates Distinct Lineages and Mobile Genetic Elements in Isolates from Humans versus Livestock. mBio 2019, 10, e02693-18. [Google Scholar] [CrossRef]
- Thanh Duy, P.; Thi Nguyen, T.N.; Vu Thuy, D.; Chung The, H.; Alcock, F.; Boinett, C.; Dan Thanh, H.N.; Thanh Tuyen, H.; Thwaites, G.E.; Rabaa, M.A.; et al. Commensal Escherichia coli are a reservoir for the transfer of XDR plasmids into epidemic fluoroquinolone-resistant Shigella sonnei. Nat. Microbiol. 2020, 5, 256–264. [Google Scholar] [CrossRef]
- Da Silva, G.J.; Mendonça, N. Association between antimicrobial resistance and virulence in Escherichia coli. Virulence 2012, 3, 18–28. [Google Scholar] [CrossRef]
- Bertelli, C.; Greub, G. Rapid bacterial genome sequencing: Methods and applications in clinical microbiology. Clin. Microbiol. Infect. 2013, 19, 803–813. [Google Scholar] [CrossRef]
- Mathers, A.J.; Peirano, G.; Pitout, J.D. Escherichia coli ST131: The quintessential example of an international multiresistant high-risk clone. Adv. Appl. Microbiol. 2015, 90, 109–154. [Google Scholar] [CrossRef]
- Ma, L.; Xie, M.; Yang, Y.; Ding, X.; Li, Y.; Yan, Z.; Chan, E.W.; Chen, S.; Chen, G.; Zhang, R. Prevalence and genomic characterization of clinical Escherichia coli strains that harbor the plasmid-borne tet(X4) gene in China. Microbiol. Res. 2024, 285, 127730. [Google Scholar] [CrossRef]
- Raivio, T.L.; Leblanc, S.K.; Price, N.L. The Escherichia coli Cpx envelope stress response regulates genes of diverse function that impact antibiotic resistance and membrane integrity. J. Bacteriol. 2013, 195, 2755–2767. [Google Scholar] [CrossRef]
- Kurabayashi, K.; Hirakawa, Y.; Tanimoto, K.; Tomita, H.; Hirakawa, H. Role of the CpxAR two-component signal transduction system in control of fosfomycin resistance and carbon substrate uptake. J. Bacteriol. 2014, 196, 248. [Google Scholar] [CrossRef] [PubMed]
- Hazan, R.; Sat, B.; Engelberg-Kulka, H. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 2004, 186, 3663. [Google Scholar] [CrossRef] [PubMed]
- Erental, A.; Sharon, I.; Engelberg-Kulka, H. Two programmed cell death systems in Escherichia coli: An apoptotic-like death is inhibited by the mazEF-mediated death pathway. PLoS Biol. 2012, 10, e1001281. [Google Scholar] [CrossRef]
- Lee, D.C.; Zheng, J.; She, Y.M.; Jia, Z. Structure of Escherichia coli tyrosine kinase Etk reveals a novel activation mechanism. EMBO J. 2008, 27, 1758–1766. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, A.W.P.; Llabrés, S.; Neuberger, A.; Blaza, J.N.; Bai, X.C.; Okada, U.; Murakami, S.; van Veen, H.W.; Zachariae, U.; Scheres, S.H.W.; et al. Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump. Nat. Microbiol. 2017, 2, 17070. [Google Scholar] [CrossRef]
- Cannatelli, A.; D’Andrea, M.M.; Giani, T.; Di Pilato, V.; Arena, F.; Ambretti, S.; Gaibani, P.; Rossolini, G.M. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 2013, 57, 5521–5526. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef]
- Wand, M.E.; Sutton, J.M. Mutations in the two component regulator systems PmrAB and PhoPQ give rise to increased colistin resistance in Citrobacter and Enterobacter spp. J. Med. Microbiol. 2020, 69, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Ko, K.S. Mutations and expression of PmrAB and PhoPQ related with colistin resistance in Pseudomonas aeruginosa clinical isolates. Diagn. Microbiol. Infect. Dis. 2014, 78, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Wang, N.; Li, X.; Shi, K.; Zhou, Z.; Yu, Y.; Hua, X. The Effect of Colistin Resistance-Associated Mutations on the Fitness of Acinetobacter baumannii. Front. Microbiol. 2016, 7, 1715. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.D.; Nickel, G.C.; Bajaksouzian, S.; Lavender, H.; Murthy, A.R.; Jacobs, M.R.; Bonomo, R.A. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 2009, 53, 3628–3634. [Google Scholar] [CrossRef]
- Huang, J.; Li, C.; Song, J.; Velkov, T.; Wang, L.; Zhu, Y.; Li, J. Regulating polymyxin resistance in Gram-negative bacteria: Roles of two-component systems PhoPQ and PmrAB. Future Microbiol. 2020, 15, 445–459. [Google Scholar] [CrossRef]
- Nirwan, P.K.; Chatterjee, N.; Panwar, R.; Dudeja, M.; Jaggi, N. Mutations in two component system (PhoPQ and PmrAB) in colistin resistant Klebsiella pneumoniae from North Indian tertiary care hospital. J. Antibiot. 2021, 74, 450–457. [Google Scholar] [CrossRef]
- Lin, J.C.; Kristopher Siu, L.K.; Chang, F.Y.; Wang, C.H. Mutations in the pmrB gene constitute the major mechanism underlying chromosomally encoded colistin resistance in clinical Escherichia coli. J. Glob. Antimicrob. Resist. 2024, 38, 275–280. [Google Scholar] [CrossRef]
- Alsahlani, F.; Haeili, M. Genetic Alterations Associated with Colistin Resistance Development in Escherichia coli. Microb. Drug Resist. 2024, 30, 325–331. [Google Scholar] [CrossRef]
- Kim, S.; Woo, J.H.; Kim, N.; Kim, M.H.; Kim, S.Y.; Son, J.H.; Moon, D.C.; Lim, S.K.; Shin, M.; Lee, J.C. Characterization of Chromosome-Mediated Colistin Resistance in Escherichia coli Isolates from Livestock in Korea. Infect. Drug Resist. 2019, 12, 3291–3299. [Google Scholar] [CrossRef]
- Li, B.; Ke, B.; Zhao, X.; Guo, Y.; Wang, W.; Wang, X.; Zhu, H. Antimicrobial Resistance Profile of mcr-1 Positive Clinical Isolates of Escherichia coli in China From 2013 to 2016. Front. Microbiol. 2018, 9, 2514. [Google Scholar] [CrossRef]
- Zakaria, A.S.; Edward, E.A.; Mohamed, N.M. Genomic Insights into a Colistin-Resistant Uropathogenic Escherichia coli Strain of O23:H4-ST641 Lineage Harboring mcr-1.1 on a Conjugative IncHI2 Plasmid from Egypt. Microorganisms 2021, 9, 799. [Google Scholar] [CrossRef]
- Hirakawa, H.; Nishino, K.; Yamada, J.; Hirata, T.; Yamaguchi, A. Beta-lactam resistance modulated by the overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Antimicrob. Chemother. 2003, 52, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Jing, W.; Liu, J.; Wu, S.; Li, X.; Liu, Y. Role of cpxA Mutations in the Resistance to Aminoglycosides and β-Lactams in Salmonella enterica serovar Typhimurium. Front. Microbiol. 2021, 12, 604079. [Google Scholar] [CrossRef]
- Raivio, T.L. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol. Microbiol. 2005, 56, 1119–1128. [Google Scholar] [CrossRef]
- Sato, T.; Suzuki, Y.; Shiraishi, T.; Honda, H.; Shinagawa, M.; Yamamoto, S.; Ogasawara, N.; Takahashi, H.; Takahashi, S.; Tamura, Y.; et al. Tigecycline Nonsusceptibility Occurs Exclusively in Fluoroquinolone-Resistant Escherichia coli Clinical Isolates, Including the Major Multidrug-Resistant Lineages O25b:H4-ST131-H30R and O1-ST648. Antimicrob. Agents Chemother. 2017, 61, e01654-16. [Google Scholar] [CrossRef]
- Du, D.; Wang-Kan, X.; Neuberger, A.; van Veen, H.W.; Pos, K.M.; Piddock, L.J.V.; Luisi, B.F. Multidrug efflux pumps: Structure, function, and regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. [Google Scholar] [CrossRef]
- Zhao, Z.; Xu, Y.; Jiang, B.; Qi, Q.; Tang, Y.J.; Xian, M.; Wang, J.; Zhao, G. Systematic Identification of CpxRA-Regulated Genes and Their Roles in Escherichia coli Stress Response. mSystems 2022, 7, e0041922. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- GBD 2019 Risk Factors Collaborators. Global burden of 87 risk factors in 204 countries and territories, 1990-2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1223–1249. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Shaik, S.; Ranjan, A.; Suresh, A.; Sarker, N.; Semmler, T.; Wieler, L.H.; Alam, M.; Watanabe, H.; Chakravortty, D.; et al. Genomic and Functional Characterization of Poultry Escherichia coli From India Revealed Diverse Extended-Spectrum β-Lactamase-Producing Lineages with Shared Virulence Profiles. Front. Microbiol. 2019, 10, 2766. [Google Scholar] [CrossRef]
- Suresh, A.; Ranjan, A.; Jadhav, S.; Hussain, A.; Shaik, S.; Alam, M.; Baddam, R.; Wieler, L.H.; Ahmed, N. Molecular Genetic and Functional Analysis of pks-Harboring, Extra-Intestinal Pathogenic Escherichia coli From India. Front. Microbiol. 2018, 9, 2631. [Google Scholar] [CrossRef] [PubMed]
- Zanditenas, E.; Ankri, S. Unraveling the interplay between unicellular parasites and bacterial biofilms: Implications for disease persistence and antibiotic resistance. Virulence 2024, 15, 2289775. [Google Scholar] [CrossRef]
- Helfand, M.S.; Bonomo, R.A. Extended-spectrum beta-lactamases in multidrug-resistant Escherichia coli: Changing the therapy for hospital-acquired and community-acquired infections. Clin. Infect. Dis. 2006, 43, 1415–1416. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Galperin, M.Y. Diversity of structure and function of response regulator output domains. Curr. Opin. Microbiol. 2010, 13, 150–159. [Google Scholar] [CrossRef]
- Masi, M.; Pinet, E.; Pagès, J.M. Complex Response of the CpxAR Two-Component System to β-Lactams on Antibiotic Resistance and Envelope Homeostasis in Enterobacteriaceae. Antimicrob. Agents Chemother. 2020, 64, e00291-20. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, V.B.; Rajamohan, G. Genome analysis of urease positive Serratia marcescens, co-producing SRT-2 and AAC (6′)-Ic with multidrug efflux pumps for antimicrobial resistance. Genomics 2019, 111, 653–660. [Google Scholar] [CrossRef]
- Feng, Y.; Zou, S.; Chen, H.; Yu, Y.; Ruan, Z. BacWGSTdb 2.0: A one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 2021, 49, D644–D650. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Dereeper, A.; Summo, M.; Meyer, D.F. PanExplorer: A web-based tool for exploratory analysis and visualization of bacterial pan-genomes. Bioinformatics 2022, 18, 4412–4414. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed]
- David, S.W.; Scott, H.; Sukanta, S.; Eponine, O.; Harrison, P.; Jason, R.G.; Paul, S.; Vasuk, G. Phastest: Faster than PHASTER, better than PHAST. Nucleic Acids Res. 2023, 51, W443–W450. [Google Scholar] [CrossRef]
- Sidorczuk, K.; Burdukiewicz, M.; Cerk, K.; Fritscher, J.; Kingsley, R.A.; Schierack, P.; Hildebrand, F.; Kolenda, R. adhesiomeR: A tool for Escherichia coli adhesin classification and analysis. BMC Genom. 2024, 25, 609. [Google Scholar] [CrossRef]
- Ferrer Florensa, A.; Almagro Armenteros, J.J.; Kaas, R.S.; Clausen, P.T.; Nielsen, H.; Rost, B.; Aarestrup, F.M. Whole-genome prediction of bacterial pathogenic capacity on novel bacteria using protein language models, with PathogenFinder2. bioRxiv 2025, 2025-04. [Google Scholar] [CrossRef]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7, S10. [Google Scholar] [CrossRef]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher sequence analysis tools framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. Swiss-Model: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Labbé, C.M.; Rey, J.; Lagorce, D.; Vavruša, M.; Becot, J.; Sperandio, O.; Villoutreix, B.O.; Tufféry, P.; Miteva, M.A. MTiOpenScreen: A web server for structure-based virtual screening. Nucleic Acids Res. 2015, 43, W448–W454. [Google Scholar] [CrossRef]
- Wang, Z.; Pan, H.; Sun, H.; Kang, Y.; Liu, H.; Cao, D.; Hou, T. fastDRH: A webserver to predict and analyze protein-ligand complexes based on molecular docking and MM/PB(GB)SA computation. Brief. Bioinform. 2022, 23, bbac201. [Google Scholar] [CrossRef]
- Fährrolfes, R.; Bietz, S.; Flachsenberg, F.; Meyder, A.; Nittinger, E.; Otto, T.; Volkamer, A.; Rarey, M. ProteinsPlus: A web portal for structure analysis of macromolecules. Nucleic Acids Res. 2017, 45, W337–W343. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Tao, H.; He, J.; Huang, S.Y. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef] [PubMed]
- Honorato, R.V.; Trellet, M.E.; Jiménez-García, B.; Schaarschmidt, J.J.; Giulini, M.; Reys, V.; Koukos, P.I.; Rodrigues, J.P.G.L.M.; Karaca, E.; van Zundert, G.C.P.; et al. The HADDOCK2.4 web server for integrative modelling of biomolecular complexes. Nat. Protoc. 2024, 19, 3219–3241. [Google Scholar] [CrossRef]
- Del Conte, A.; Camagni, G.F.; Clementel, D.; Minervini, G.; Monzon, A.M.; Ferrari, C.; Piovesan, D.; Tosatto, S.C.E. RING 4.0: Faster residue interaction networks with novel interaction types across over 35,000 different chemical structures. Nucleic Acids Res. 2024, 52, W306–W312. [Google Scholar] [CrossRef]
- Audrain, B.; Ferrières, L.; Zairi, A.; Soubigou, G.; Dobson, C.; Coppée, J.Y.; Beloin, C.; Ghigo, J.M. Induction of the Cpx envelope stress pathway contributes to Escherichia coli tolerance to antimicrobial peptides. Appl. Environ. Microbiol. 2013, 79, 7770–7779. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Z.; Feng, Y. BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 2016, 44, D682–D687. [Google Scholar] [CrossRef]
- Ruan, Z.; Yu, Y.; Feng, Y. The global dissemination of bacterial infections necessitates the study of reverse genomic epidemiology. Brief. Bioinform. 2020, 21, 741–750. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
| Name | Volume (Å) | Surface (Å) | Drug Score | Simple Score |
|---|---|---|---|---|
| P_0 | 1970.39 | 2486.25 | 0.809533 | 0.61 |
| P_1 | 898.95 | 1030.4 | 0.830871 | 0.58 |
| P_2 | 519.03 | 784.57 | 0.815272 | 0.32 |
| P_3 | 472.18 | 689.34 | 0.76657 | 0.24 |
| P_4 | 396.54 | 782.71 | 0.745362 | 0.33 |
| P_5 | 374.19 | 711.25 | 0.619852 | 0.21 |
| P_6 | 363.16 | 596.45 | 0.556378 | 0.17 |
| Name | Volume (Å) | Surface (Å) | Drug Score | Simple Score |
|---|---|---|---|---|
| P_0 | 1117.44 | 1388.63 | 0.8 | 0.6 |
| P_1 | 209.28 | 287.32 | 0.45 | 0.0 |
| P_2 | 191.36 | 488.81 | 0.46 | 0.15 |
| P_3 | 185.15 | 253.78 | 0.27 | 0.0 |
| P_4 | 138.24 | 265.49 | 0.35 | 0.05 |
| P_5 | 133.95 | 314.54 | 0.23 | 0.0 |
| P_6 | 121.41 | 199.14 | 0.15 | 0.0 |
| Name | Docking Score | Confidence Score | Ligand Rmsd (A) | LG Score | MaxSub |
|---|---|---|---|---|---|
| acrA1-CpxR | −215.11 | 0.7862 | 34.10 | 6.100 | 0.516 |
| acrA2-CpxR | −222.27 | 0.8093 | 25.12 | 6.100 | 0.516 |
| AcrD-CpxR | −209.86 | 0.7680 | 65.49 | 6.100 | 0.516 |
| SrkA-CpxR | −208.93 | 0.7647 | 67.59 | 6.100 | 0.516 |
| Wzc-CpxR | −219.20 | 0.7996 | 34.56 | 6.100 | 0.516 |
| sugE-CpxR | −221.04 | 0.8055 | 39.87 | 6.100 | 0.516 |
| mdtJ1-CpxR | −212.14 | 0.7761 | 46.48 | 6.100 | 0.516 |
| mdtJ2-CpxR | −221.18 | 0.8059 | 350.99 | 6.100 | 0.516 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srinivasan, V.B.; Rajasekar, N.; Krishnan, K.; Kumar, M.; Giri, C.; Singh, B.; Rajamohan, G. Genomic and Functional Characterization of Multidrug-Resistant E. coli: Insights into Resistome, Virulome, and Signaling Systems. Antibiotics 2025, 14, 667. https://doi.org/10.3390/antibiotics14070667
Srinivasan VB, Rajasekar N, Krishnan K, Kumar M, Giri C, Singh B, Rajamohan G. Genomic and Functional Characterization of Multidrug-Resistant E. coli: Insights into Resistome, Virulome, and Signaling Systems. Antibiotics. 2025; 14(7):667. https://doi.org/10.3390/antibiotics14070667
Chicago/Turabian StyleSrinivasan, Vijaya Bharathi, Naveenraj Rajasekar, Karthikeyan Krishnan, Mahesh Kumar, Chankit Giri, Balvinder Singh, and Govindan Rajamohan. 2025. "Genomic and Functional Characterization of Multidrug-Resistant E. coli: Insights into Resistome, Virulome, and Signaling Systems" Antibiotics 14, no. 7: 667. https://doi.org/10.3390/antibiotics14070667
APA StyleSrinivasan, V. B., Rajasekar, N., Krishnan, K., Kumar, M., Giri, C., Singh, B., & Rajamohan, G. (2025). Genomic and Functional Characterization of Multidrug-Resistant E. coli: Insights into Resistome, Virulome, and Signaling Systems. Antibiotics, 14(7), 667. https://doi.org/10.3390/antibiotics14070667






