Variable In Vitro Efficacy of Delafloxacin on Multidrug-Resistant Pseudomonas aeruginosa and the Detection of Delafloxacin Resistance Determinants
Abstract
1. Introduction
2. Results
2.1. Antimicrobial Susceptibility Testing
2.2. Genome Sequencing
3. Discussion
4. Materials and Methods
4.1. Strains
4.2. Investigation of Minimum Inhibitory Concentration (MIC) Values
4.3. Whole-Genome Sequencing (WGS)
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oliver, A.; Rojo-Molinero, E.; Arca-Suarez, J.; Beşli, Y.; Bogaerts, P.; Cantón, R.; Cimen, C.; Croughs, P.D.; Denis, O.; Giske, C.G.; et al. Pseudomonas aeruginosa antimicrobial susceptibility profiles, resistance mechanisms and international clonal lineages: Update from ESGARS-ESCMID/ISARPAE Group. Clin. Microbiol. Infect. 2024, 30, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Raman, G.; Avendano, E.E.; Chan, J.; Merchant, S.; Puzniak, L. Risk factors for hospitalized patients with resistant or multidrug-resistant Pseudomonas aeruginosa infections: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 2018, 7, 79. [Google Scholar] [CrossRef]
- Palavutitotai, N.; Jitmuang, A.; Tongsai, S.; Kiratisin, P.; Angkasekwinai, N. Epidemiology and risk factors of extensively drug-resistant Pseudomonas aeruginosa infections. PLoS ONE 2018, 13, e0193431. [Google Scholar] [CrossRef]
- Camus, L.; Vandenesch, F.; Moreau, K. From genotype to phenotype: Adaptations of Pseudomonas aeruginosa to the cystic fibrosis environment. Microb. Genom. 2021, 7, mgen000513. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Baigenzhin, A.; Bissenova, N.; Yergaliyeva, A.; Marassulov, S.; Tuleubayeva, E.; Aitysheva, U. ESKAPE pathogens in pediatric cardiac surgery patients: 5-year microbiological monitoring in a tertiary hospital in Kazakhstan. Acta Microbiol. Immunol. Hung. 2024, 71, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef]
- Lorusso, A.B.; Carrara, J.A.; Barroso, C.D.N.; Tuon, F.F.; Faoro, H. Role of Efflux Pumps on Antimicrobial Resistance in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2022, 23, 15779. [Google Scholar] [CrossRef]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Karruli, A.; Catalini, C.; D’amore, C.; Foglia, F.; Mari, F.; Harxhi, A.; Galdiero, M.; Durante-Mangoni, E. Evidence-Based Treatment of Pseudomonas aeruginosa Infections: A Critical Reappraisal. Antibiotics 2023, 12, 399. [Google Scholar] [CrossRef]
- Reid, E.; Walters, R.W.; Destache, C.J. Beta-Lactam vs. Fluoroquinolone Monotherapy for Pseudomonas aeruginosa Infection: A Systematic Review and Meta-Analysis. Antibiotics 2021, 10, 1483. [Google Scholar] [CrossRef]
- Mataracı Kara, E.; Çakmak, S.M.; Er, S. Assessment of in vitro interactions between delafloxacin and other antimicrobials against multi-drug resistant Pseudomonas aeruginosa strains. J. Chemother. 2024, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kunz Coyne, A.J.; El Ghali, A.; Holger, D.; Rebold, N.; Rybak, M.J. Therapeutic Strategies for Emerging Multidrug-Resistant Pseudomonas aeruginosa. Infect. Dis. Ther. 2022, 11, 661–682. [Google Scholar] [CrossRef] [PubMed]
- Jones, F.; Hu, Y.; Coates, A. The Efficacy of Using Combination Therapy against Multi-Drug and Extensively Drug-Resistant Pseudomonas aeruginosa in Clinical Settings. Antibiotics 2022, 11, 323. [Google Scholar] [CrossRef] [PubMed]
- Venkataramana, G.P.; Lalitha, A.K.V.; Mariappan, S.; Sekar, U. Plasmid-Mediated Fluoroquinolone Resistance in Pseudomonas aeruginosa and Acinetobacter baumannii. J. Lab. Physicians 2022, 14, 271–277. [Google Scholar] [CrossRef]
- Pazhani, G.P.; Chakraborty, S.; Fujihara, K.; Yamasaki, S.; Ghosh, A.; Nair, G.; Ramamurthy, T. QRDR mutations, efflux system & antimicrobial resistance genes in enterotoxigenic Escherichia coli isolated from an outbreak of diarrhoea in Ahmedabad, India. Indian. J. Med. Res. 2011, 134, 214–223. [Google Scholar]
- Juan, C.; Peña, C.; Oliver, A. Host and Pathogen Biomarkers for Severe Pseudomonas aeruginosa Infections. J. Infect. Dis. 2017, 215 (Suppl. 1), S44–S51. [Google Scholar] [CrossRef]
- Strateva, T.; Keuleyan, E.; Peykov, S. Genomic insights into NDM-1-producing Pseudomonas aeruginosa: Current status in a Bulgarian tertiary hospital and on the Balkans. Acta Microbiol. Immunol. Hung. 2024, 71, 99–109. [Google Scholar] [CrossRef]
- Tenover, F.C.; Nicolau, D.P.; Gill, C.M. Carbapenemase-producing Pseudomonas aeruginosa—An emerging challenge. Emerg. Microbes Infect. 2022, 11, 811–814. [Google Scholar] [CrossRef]
- WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2024.
- Butler, M.S.; Paterson, D.L. Antibiotics in the clinical pipeline in October 2019. J. Antibiot. 2020, 73, 329–364. [Google Scholar] [CrossRef] [PubMed]
- Mogle, B.T.; Steele, J.M.; Thomas, S.J.; Bohan, K.H.; Kufel, W.D. Clinical review of delafloxacin: A novel anionic fluoroquinolone. J. Antimicrob. Chemother. 2018, 73, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- Kocsis, B.; Gulyás, D.; Szabó, D. Delafloxacin, Finafloxacin, and Zabofloxacin: Novel Fluoroquinolones in the Antibiotic Pipeline. Antibiotics 2021, 10, 1506. [Google Scholar] [CrossRef]
- Craddock, V.D.; Steere, E.L.; Harman, H.; Britt, N.S. Activity of Delafloxacin and Comparator Fluoroquinolones against Multidrug-Resistant Pseudomonas aeruginosa in an In Vitro Cystic Fibrosis Sputum Model. Antibiotics 2023, 12, 1078. [Google Scholar] [CrossRef]
- Ocheretyaner, E.R.; Park, T.E. Delafloxacin: A novel fluoroquinolone with activity against methicillin-resistant Staphylococcus aureus (MRSA) and Pseudomonas aeruginosa. Expert Rev. Anti-Infect. Ther. 2018, 16, 523–530. [Google Scholar] [CrossRef]
- Nascimento-Carvalho, C.M. Delafloxacin as a treatment option for community-acquired pneumonia infection. Expert. Opin. Pharmacother. 2021, 22, 1975–1982. [Google Scholar] [CrossRef] [PubMed]
- Jordán-Chaves, J.D.D.; Lobato-Cano, R.; Casas-Ciria, J.; Freyre-Carillo, C.; Santotoribio, J.D.; de-la-Rubia-Martin, M.F. In vitro susceptibility to delafloxacin of Pseudomonas aeruginosa with resistance to other quinolones (ciprofloxacin and levofloxacin). Clin. Microbiol. Infect. 2024, 30, 405–406. [Google Scholar] [CrossRef]
- Del Barrio-Tofiño, E.; López-Causapé, C.; Oliver, A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int. J. Antimicrob. Agents 2020, 56, 106196. [Google Scholar] [CrossRef]
- Vatansever, C.; Menekse, S.; Dogan, O.; Gucer, L.S.; Ozer, B.; Ergonul, O.; Can, F. Co-existence of OXA-48 and NDM-1 in colistin resistant Pseudomonas aeruginosa ST235. Emerg. Microbes Infect. 2020, 9, 152–154. [Google Scholar] [CrossRef]
- Lee, A.C.; Jones, A.L. Multi-resistant Pseudomonas aeruginosa ST235 in cystic fibrosis. Paediatr. Respir. Rev. 2018, 27, 18–20. [Google Scholar] [CrossRef]
- Treepong, P.; Kos, V.; Guyeux, C.; Blanc, D.; Bertrand, X.; Valot, B.; Hocquet, D. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin. Microbiol. Infect. 2018, 24, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Saharman, Y.R.; Pelegrin, A.C.; Karuniawati, A.; Sedono, R.; Aditianingsih, D.; Goessens, W.H.; Klaassen, C.H.; van Belkum, A.; Mirande, C.; Verbrugh, H.A.; et al. Epidemiology and characterisation of carbapenem-non-susceptible Pseudomonas aeruginosa in a large intensive care unit in Jakarta, Indonesia. Int. J. Antimicrob. Agents 2019, 54, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Yang, F.; Ji, J.; Cao, W.; Liu, C.; Ding, B.; Xu, X. Dissecting the genotypic features of a fluoroquinolone-resistant Pseudomonas aeruginosa ST316 sublineage causing ear infections in Shanghai, China. Microb. Genom. 2023, 9, mgen000989. [Google Scholar] [CrossRef] [PubMed]
- Zowawi, H.M.; Syrmis, M.W.; Kidd, T.J.; Balkhy, H.H.; Walsh, T.R.; Al Johani, S.M.; Al Jindan, R.Y.; Alfaresi, M.; Ibrahim, E.; Al-Jardani, A.; et al. Identification of carbapenem-resistant Pseudomonas aeruginosa in selected hospitals of the Gulf Cooperation Council States: Dominance of high-risk clones in the region. J. Med. Microbiol. 2018, 67, 846–853. [Google Scholar] [CrossRef]
- Cholley, P.; Thouverez, M.; Hocquet, D.; van der Mee-Marquet, N.; Talon, D.; Bertrand, X. Most multidrug-resistant Pseudomonas aeruginosa isolates from hospitals in eastern France belong to a few clonal types. J. Clin. Microbiol. 2011, 49, 2578–2583. [Google Scholar] [CrossRef]
- Quick, J.; Cumley, N.; Wearn, C.M.; Niebel, M.; Constantinidou, C.; Thomas, C.M.; Pallen, M.J.; Moiemen, N.S.; Bamford, A.; Oppenheim, B.; et al. Seeking the source of Pseudomonas aeruginosa infections in a recently opened hospital: An observational study using whole-genome sequencing. BMJ Open 2014, 4, e006278. [Google Scholar] [CrossRef]
- Petitjean, M.; Martak, D.; Silvant, A.; Bertrand, X.; Valot, B.; Hocquet, D. Genomic characterization of a local epidemic Pseudomonas aeruginosa reveals specific features of the widespread clone ST395. Microb. Genom. 2017, 3, e000129. [Google Scholar] [CrossRef]
- Bolanos, S.; Acebes, C.; Martinez-Exposito, Ó.; Boga, J.A.; Fernandez, J.; Rodriguez-Lucas, C. Role of parC Mutations at Position 84 on High-Level Delafloxacin Resistance in Methicillin-Resistant Staphylococcus aureus. Antibiotics 2024, 13, 641. [Google Scholar] [CrossRef]
- Luzarraga, V.; Cremniter, J.; Plouzeau, C.; Michaud, A.; Broutin, L.; Burucoa, C.; Pichon, M. In vitro activity of delafloxacin against clinical levofloxacin-resistant Helicobacter pylori isolates. J. Antimicrob. Chemother. 2024, 79, 2633–2639. [Google Scholar] [CrossRef]
- Gulyás, D.; Kamotsay, K.; Szabó, D.; Kocsis, B. Investigation of Delafloxacin Resistance in Multidrug-Resistant Escherichia coli Strains and the Detection of E. coli ST43 International High-Risk Clone. Microorganisms 2023, 11, 1602. [Google Scholar] [CrossRef]
- Kubicskó, A.; Kamotsay, K.; Szabó, D.; Kocsis, B. Analysis of molecular mechanisms of delafloxacin resistance in Escherichia coli. Sci. Rep. 2024, 14, 26423. [Google Scholar] [CrossRef] [PubMed]
- Kubicskó, A.; Juhász, J.; Kamotsay, K.; Szabo, D.; Kocsis, B. Detection of Delafloxacin Resistance Mechanisms in Multidrug-Resistant Klebsiella pneumoniae. Antibiotics 2025, 14, 62. [Google Scholar] [CrossRef] [PubMed]
- Fuzi, M.; Szabo, D.; Csercsik, R. Double-Serine Fluoroquinolone Resistance Mutations Advance Major International Clones and Lineages of Various Multi-Drug Resistant Bacteria. Front. Microbiol. 2017, 8, 2261. [Google Scholar] [CrossRef] [PubMed]
- Holland, M.; Bjanes, E.; Nizet, V.; Dillon, N. Bicarbonate modulates delafloxacin activity against MDR Staphylococcus aureus and Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2022, 77, 433–442. [Google Scholar] [CrossRef]
- Millar, B.C.; McCaughan, J.; Rendall, J.C.; Moore, J.E. Delafloxacin--A novel fluoroquinolone for the treatment of ciprofloxacin-resistant Pseudomonas aeruginosa in patients with cystic fibrosis. Clin. Respir. J. 2021, 15, 116–120. [Google Scholar] [CrossRef]
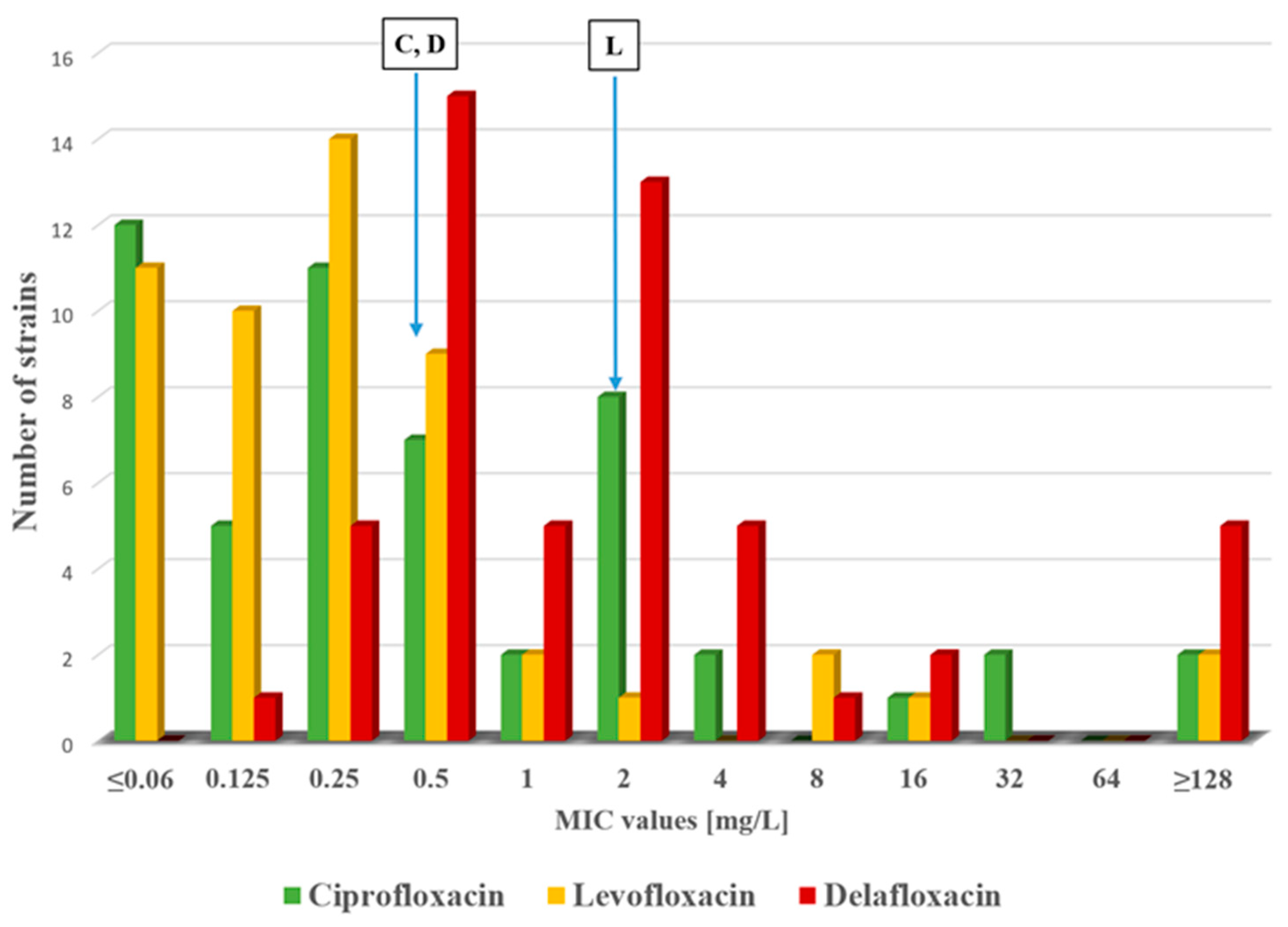
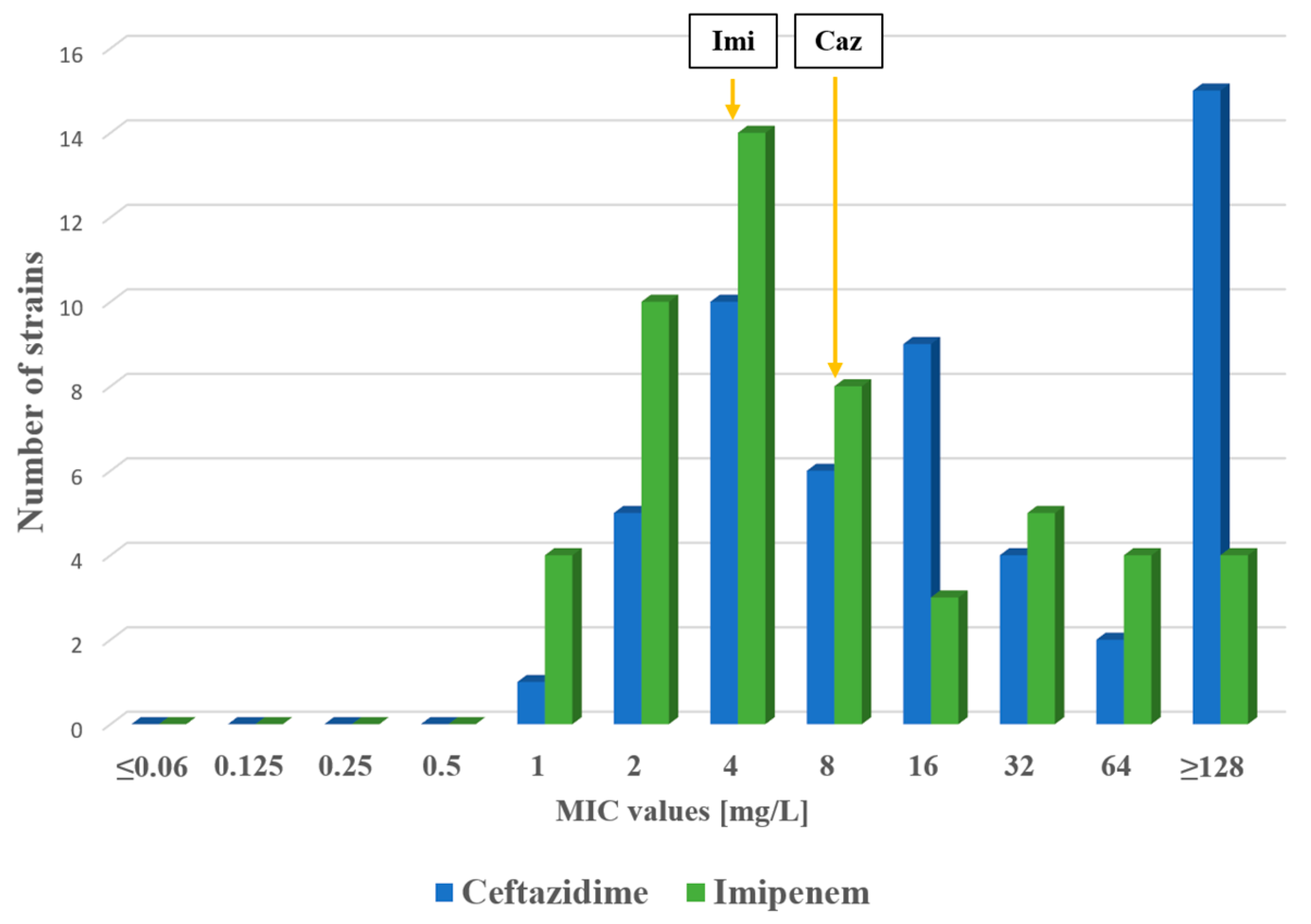
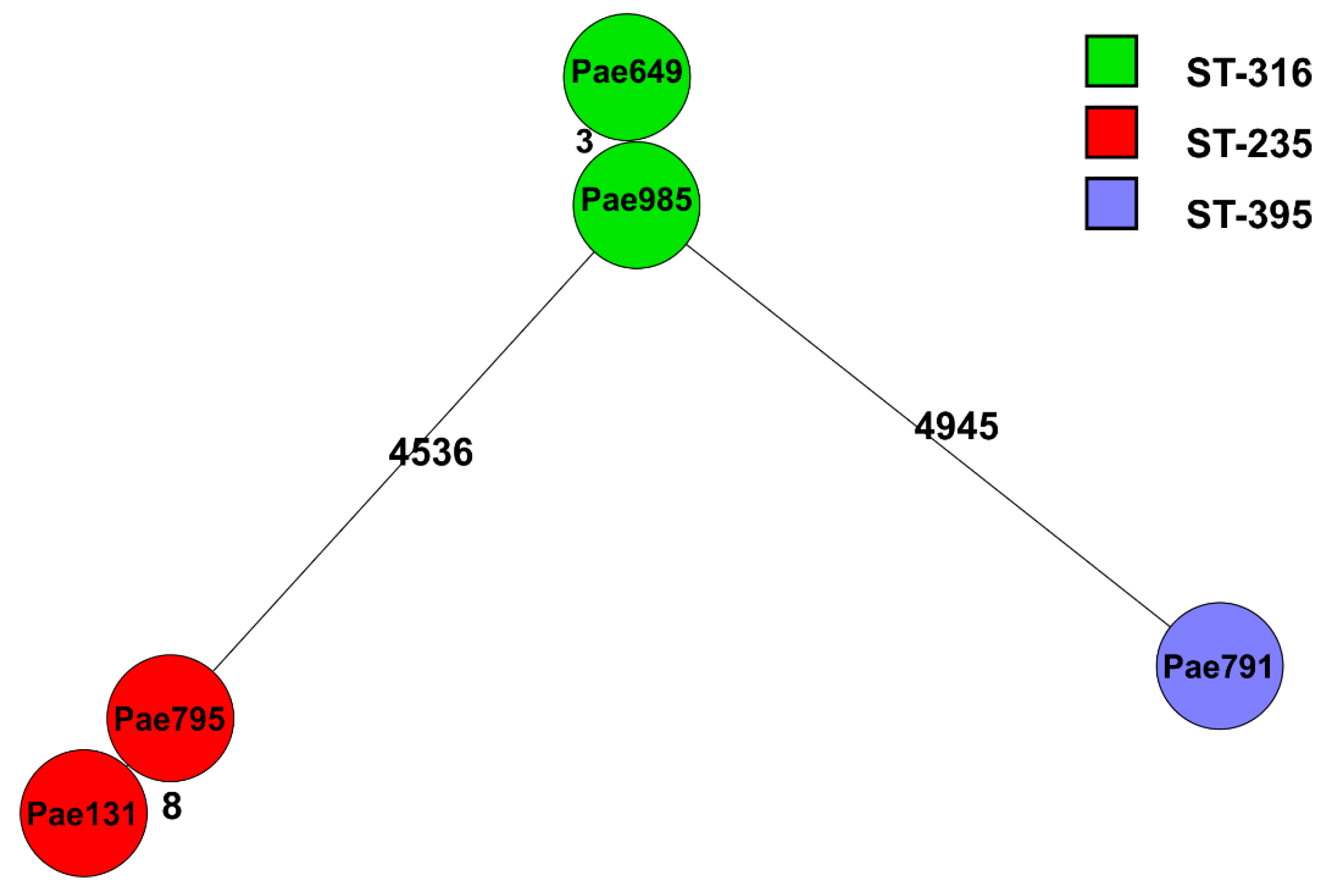
| MLST | Beta-Lactamases | Other Resistance Genes | cip | lev | del | caz | imi | |
|---|---|---|---|---|---|---|---|---|
| P. aeruginosa 131 | ST235 | blaNDM-1, blaOXA-488, blaPDC-35 | sul1, catB7, fosA, aph(3′)-IIb | 32 | 8 | 128 | 128 | 32 |
| P. aeruginosa 649 | ST316 | blaNDM-1, blaOXA-395, blaPDC-36, blaPME-1 | sul1, catB7, fosA, aph(3′)-IIb, aph(3′)-Ib, aph(6)-Id, ant(4′)-IIb, ble, aac(3)-II | 128 | 128 | 128 | 128 | 128 |
| P. aeruginosa 791 | ST395 | blaOXA-905, blaPDC-8 | catB7, fosA, aph(3′)-IIb | 32 | 16 | 128 | 16 | 8 |
| P. aeruginosa 795 | ST235 | blaNDM-1, blaOXA-488, blaPDC-35 | catB7, fosA, aph(3′)-IIb | 16 | 8 | 128 | 128 | 64 |
| P. aeruginosa 985 | ST316 | blaNDM-1, blaOXA-395, blaPDC-36, blaPME-1 | sul1, catB7, fosA, aph(3′)-IIb, aph(3′)-Ib, aph(6)-Id, ant(4′)-IIb, ble, aac(3)-II | 128 | 128 | 128 | 128 | 128 |
| (a) | |||||
| ST | ST235 P. aeruginosa 131 | ST316 P. aeruginosa 649 | ST395 P. aeruginosa 791 | ST235 P. aeruginosa 795 | ST316 P. aeruginosa 985 |
| QRDR | gyrA: Thr83Ile parC: Ser87Leu parE: Thr223Ala | gyrA: Thr83Ile parC: Ser87Leu parE: Glu459Val | gyrA: Thr83Ile parC: Ser87Leu parE: Val200Met | gyrA: Thr83Ile parC: Ser87Leu parE: Thr223Ala | gyrA: Thr83Ile parC: Ser87Leu parE: Glu459Val |
| (b) | |||||
| ST | ST235 P. aeruginosa 131 | ST316 P. aeruginosa 649 | ST395 P. aeruginosa 791 | ST235 P. aeruginosa 795 | ST316 P. aeruginosa 985 |
| Fluoroquinolone antibiotic efflux | MexA, MexB, MexC, MexD, MexF, MexG, MexH, MexI, MexR, MexS, MexT MexV, MexW, MexY, MexZ, OprJ, OprM, OprN, rsmA, soxR, CpxR, YajC, PmpM, OpmD, adeF, ParS, ParR, Typa A NfxB, nalC, nalD | MexA, MexB, MexC, MexD, MexE, MexG, MexI, MexR, MexS, MexT, MexY, MexV, MexW, OprJ, OprM, OprN CpxR, rsmA, PmpM, adeF, OpmD, YajC, ParS, ParR, soxR, Type A NfxB, nalC, nalD | MexA, MexB, MexC, MexD MexE, MexF, MexG, MexH, MexI, MexR, MexS, MexT, MexV, MexW, MexY, OprJ, OprN, OprM, OpmD, PmpM, rsmA, soxR, ParS, YajC, ParR, Type A NfxB, nalC, nalD | MexA, MexB, MexC, MexD, MexE, MexF, MexG, MexH, MexI, MexS, MexT, MexV, MexW, OprJ, OprN, OprM, OpmD, YajC, ParS, ParR, Type A NfxB, rsmA, adeF, PmpM, soxR, CpxR, nalC, nalD, MexR | MexA, MexB, MexC, MexD, MexE, MexG, MexH, MexI, MexR, MexS, MexT, MexV, MexW, MexY, OprJ, OprM OprN, YajC, ParS, PmpM, adeF, ParR, CpxR, OpmD, rsmA, soxR, Type A, NfxB, nalC, nalD |
| Sample Name | Average Denovo Coverage | Number of Contigs | N50 Value |
|---|---|---|---|
| P. aeruginosa 131 | 98 | 77 | 251 514 |
| P. aeruginosa 649 | 97 | 92 | 260 809 |
| P. aeruginosa 791 | 96 | 61 | 447 543 |
| P. aeruginosa 795 | 96 | 84 | 226 439 |
| P. aeruginosa 985 | 95 | 89 | 217 956 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kubicskó, A.; Kamotsay, K.; Banczerowski, P.; Sipos, L.; Szabó, D.; Kocsis, B. Variable In Vitro Efficacy of Delafloxacin on Multidrug-Resistant Pseudomonas aeruginosa and the Detection of Delafloxacin Resistance Determinants. Antibiotics 2025, 14, 542. https://doi.org/10.3390/antibiotics14060542
Kubicskó A, Kamotsay K, Banczerowski P, Sipos L, Szabó D, Kocsis B. Variable In Vitro Efficacy of Delafloxacin on Multidrug-Resistant Pseudomonas aeruginosa and the Detection of Delafloxacin Resistance Determinants. Antibiotics. 2025; 14(6):542. https://doi.org/10.3390/antibiotics14060542
Chicago/Turabian StyleKubicskó, András, Katalin Kamotsay, Péter Banczerowski, László Sipos, Dóra Szabó, and Béla Kocsis. 2025. "Variable In Vitro Efficacy of Delafloxacin on Multidrug-Resistant Pseudomonas aeruginosa and the Detection of Delafloxacin Resistance Determinants" Antibiotics 14, no. 6: 542. https://doi.org/10.3390/antibiotics14060542
APA StyleKubicskó, A., Kamotsay, K., Banczerowski, P., Sipos, L., Szabó, D., & Kocsis, B. (2025). Variable In Vitro Efficacy of Delafloxacin on Multidrug-Resistant Pseudomonas aeruginosa and the Detection of Delafloxacin Resistance Determinants. Antibiotics, 14(6), 542. https://doi.org/10.3390/antibiotics14060542





