Antibiotics Resistance Profile of Clinical Isolates of Pseudomonas aeruginosa Obtained from Farwaniya Hospital in Kuwait Using Phenotypic and Molecular Methods
Abstract
1. Introduction
2. Results
2.1. Sample Collection
2.2. Isolation and Identification
2.3. Susceptibility Test by Broth Microdilution Method
2.4. Molecular Detection of Resistance Genes
2.5. Sequencing of gyrA and parC Genes in MDR Isolates of P. aeruginosa
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Isolation and Identification
4.3. Antibiotic Susceptibility Testing by Broth Microdilution Method
4.4. Molecular Detection of Resistance Genes
4.5. Sequencing of Resistance Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAC | Aminoglycoside acetyltransferase |
| AMEs | Aminoglycoside modifying enzymes |
| AMI | Amikacin |
| AMR | Antimicrobial resistance |
| ANT | Aminoglycoside nucleotidyl transferase |
| APH | Aminoglycoside phosphoryl transferase |
| Asn | Asparagine |
| Asp | Aspartic acid or aspartate |
| AUG | Amoxicillin-clavulanic acid |
| AZT | Aztreonam |
| C/T | Ceftolozane-tazobactam |
| CDC | Centers for Disease Control and Prevention |
| CIP | Ciprofloxacin |
| CLSI | Clinical Laboratory Standards Institute |
| COL | Colistin |
| CZA | Ceftazidime-avibactam |
| ESBL | Extended-Spectrum Beta-Lactamase |
| ETP | Ertapenem |
| FOT | Cefotaxime |
| GEN | Gentamicin |
| Gly | Glycine |
| ICUs | Intensive Care Units |
| IMI | Imipenem |
| MDR | Multidrug-resistant |
| MERO | Meropenem |
| MIC | Minimum inhibitory concentration |
| P. aeruginosa | Pseudomonas aeruginosa |
| P/T4 | Piperacillin-tazobactam |
| PBP3 | Penicillin-binding protein |
| PCR | Polymerase chain reaction |
| QRDR | Quinolone-resistant determinative region |
| RND | Resistance-nodulation-division |
| SXT | Trimethoprim-sulfamethoxazole |
| TAZ | Ceftazidime |
| TGC | Tigecycline |
| TOB | Tobramycin |
| WHO | World Health Organization |
References
- Wilson, M.G.; Pandey, S. Pseudomonas Aeruginosa. StatPearls-NCBI Bookshelf. Available online: https://www.ncbi.nlm.nih.gov/books/NBK557831/ (accessed on 10 December 2024).
- Mulcahy, L.R.; Isabella, V.M.; Lewis, K. Pseudomonas aeruginosa biofilms in disease. Microb. Ecol. 2013, 68, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Resistance Collaborators Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [CrossRef]
- WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development, and Strategies to Prevent and Control Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2024.
- Pai, H.; Kim, J.-W.; Kim, J.; Lee, J.H.; Choe, K.W.; Gotoh, N. Carbapenem resistance Mechanisms in Pseudomonas aeruginosa Clinical Isolates. Antimicrob. Agents Chemother. 2001, 45, 480–484. [Google Scholar] [CrossRef]
- Glen, K.A.; Lamont, I.L. Characterization of acquired β-lactamases in Pseudomonas aeruginosa and quantification of their contributions to resistance. Microbiol. Spectr. 2024, 12, e00694-24. [Google Scholar] [CrossRef]
- Michaelis, C.; Grohmann, E. Horizontal gene transfer of antibiotic resistance genes in biofilms. Antibiotics 2023, 12, 328. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside modifying enzymes. Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer. Chemother. 2010, 13, 151–171. [Google Scholar] [CrossRef]
- Collins, J.A.; Osheroff, N. Gyrase and Topoisomerase IV: Recycling old targets for New Antibacterials to Combat Fluoroquinolone Resistance. ACS Infect. Dis. 2024, 10, 1097–1115. [Google Scholar] [CrossRef]
- Pachori, P.; Gothalwal, R.; Gandhi, P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes. Dis. 2019, 6, 109–119. [Google Scholar] [CrossRef]
- Li, Y.; Roberts, J.A.; Walker, M.M.; Aslan, A.T.; Harris, P.N.A.; Sime, F.B. The global epidemiology of ventilator-associated pneumonia caused by multi-drug resistant Pseudomonas aeruginosa: A systematic review and meta-analysis. J. Infect. Dis. 2024, 139, 78–85. [Google Scholar] [CrossRef]
- Ranke, T.D.; Strassle, P.; Harris, A.D.; Zhu, J.; Johnson, J.K. Recovery of Gram-negative bacilli in stored endotracheal aspirates. J. Clin. Microbiol. 2012, 50, 2791–2792. [Google Scholar] [CrossRef]
- Al-Orphaly, M.; Hadi, H.A.; Eltayeb, F.K.; Al-Hail, H.; Samuel, B.G.; Sultan, A.A.; Skariah, S. Epidemiology of Multidrug-Resistant Pseudomonas aeruginosa in the Middle East and North Africa Region. mSphere 2021, 6, e00202-21. [Google Scholar] [CrossRef] [PubMed]
- Hays, J.P.; Safain, K.S.; Almogbel, M.S.; Habib, I.; Khan, M.A. Extended Spectrum- and Carbapenemase-Based Β-Lactam resistance in the Arabian Peninsula—A descriptive review of recent years. Antibiotics 2022, 11, 1354. [Google Scholar] [CrossRef] [PubMed]
- Boushra, M.R.; Gad, G.F.M.; Hassuna, N.A.; Waly, N.G.F.; Ibrahem, R.A. Phenotypic and genotypic assessment of fluoroquinolones and aminoglycosides resistance in Pseudomonas aeruginosa collected from Minia hospitals, Egypt during COVID-19 pandemic. BMC Infect. Dis. 2024, 24, 763. [Google Scholar] [CrossRef]
- Arefin, M.S.; Mitu, M.J.; Mitu, S.Y.; Nurjahan, A.; Mobin, M.; Nahar, S.; Rahman, M.H. Mutational alterations in the QRDR regions associated with fluoroquinolone resistance in Pseudomonas aeruginosa of clinical origin from Savar, Dhaka. PLoS ONE 2025, 20, e0302352. [Google Scholar] [CrossRef]
- Hafiz, T.A.; Essa, E.A.B.; Alharbi, S.R.; Alyami, A.S.; Alkudmani, Z.S.; Mubaraki, M.A.; Alturki, N.A.; Alotaibi, F. Epidemiological, microbiological, and clinical characteristics of multi-resistant Pseudomonas aeruginosa isolates in King Fahad Medical City, Riyadh, Saudi Arabia. Trop. Med. Infect. Dis. 2023, 8, 205. [Google Scholar] [CrossRef]
- Alali, W.Q.; AlFouzan, W.; Dhar, R. Prevalence of antimicrobial resistance in Gram-negative clinical isolates from a major secondary hospital in Kuwait: A retrospective descriptive study. GERMS 2021, 11, 498–511. [Google Scholar] [CrossRef]
- Yoon, E.J.; Jeong, S.H. Mobile carbapenemase genes in Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 614058. [Google Scholar] [CrossRef]
- Maharjan, N. Pseudomonas aeruginosa isolates among clinical samples showing growth in a tertiary care centre: A descriptive cross-sectional study. J. Nep. Med. Assoc. 2022, 60, 676–680. [Google Scholar] [CrossRef]
- Alshammari, H.O.; Somily, A.; Qattan, M.Y.; Alsubki, R.A.; Moussa, I.M. Susceptibility pattern of multi-drug resistance Pseudomonas aeruginosa isolates from tertiary care hospital in Riyadh, KSA. J. King Saud Univ. Sci. 2023, 35, 102702. [Google Scholar] [CrossRef]
- Patel, J.; Javiya, V.; Ghatak, S.; Patel, K. Antibiotic susceptibility patterns of Pseudomonas aeruginosa at a tertiary care hospital in Gujarat, India. Indian. J. Pharmacol. 2008, 40, 230. [Google Scholar] [CrossRef]
- Alqahtani, M.; Tickler, I.A.; Deesi, Z.A.; AlFouzan, W.; Jabri, A.A.; Jindan, R.A.; Johani, S.A.; Alkahtani, S.A.; Kharusi, A.A.; Mokaddas, E.; et al. Molecular detection of carbapenem resistance genes in rectal swabs from patients in Gulf Cooperation Council hospitals. J. Hosp. Infect. 2021, 112, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Khater, E. Detection of carbapenem-resistant Pseudomonas aeruginosa in tertiary care hospital in Saudi Arabia. Microb. Infect. Dis. 2022, 3, 693–702. [Google Scholar] [CrossRef]
- Gondal, A.J.; Choudhry, N.; Niaz, A.; Yasmin, N. Molecular analysis of carbapenem and aminoglycoside resistance genes in carbapenem-resistant Pseudomonas aeruginosa clinical strains: A Challenge for Tertiary Care Hospitals. Antibiotics 2024, 13, 191. [Google Scholar] [CrossRef]
- Hosu, M.C.; Vasaikar, S.D.; Okuthe, G.E.; Apalata, T. Detection of extended spectrum beta- lactamase genes in Pseudomonas aeruginosa isolated from patients in rural Eastern Cape Province, South Africa. Sci. Rep. 2021, 11, 7110. [Google Scholar] [CrossRef]
- Haghighi, S.; Goli, H.R. High prevalence of blaVEB, blaGES, and blaPER genes in beta- lactam resistant clinical isolates of Pseudomonas aeruginosa. AIMS Microbiol. 2022, 8, 153. [Google Scholar] [CrossRef]
- Jalal, N.A.; Hariri, S.H.; Momenah, A.M.; Khan, S.; Bantun, F. Molecular detection of blaPER-1, blaVEB-1, and blaPSE-1 βlactamase genes from P. aeruginosa Severe Urogenital UTI Infection. Fish. Sci. 2023, 10, 1628–1635. [Google Scholar] [CrossRef][Green Version]
- Mohammedkheir, M.I.A.; Gaafar, E.M.; AbdAlla, E.G.E. Assessment of Bla TEM, Bla SHV, and Bla CTX-M genes of antibiotic resistance in Gram-negative bacilli causing urinary tract infections in Khartoum State: A cross-sectional study. BMC Infect. Dis. 2024, 24, 141. [Google Scholar] [CrossRef]
- Bahrami, M.; Mmohammadi-Sichani, M.; Karbasizadeh, V. Prevalence of SHV, TEM, CTX-M and OXA-48 β-Lactamase Genes in Clinical Isolates of Pseudomonas aeruginosa in Bandar-Abbas, Iran. Avicenna J. Clin. Microbiol. Infect. 2018, 5, 86–90. [Google Scholar] [CrossRef]
- Marwa, M.A.; Atef, S.; Mai, M. OXA-10 and GES-1 extended-spectrum beta-lactamases play a major role in causing antibiotic resistance of Pseudomonas aeruginosa isolated from nosocomial infections in Ismailia, Egypt. Egypt. J. Med. Microbiol. 2015, 24, 81–88. [Google Scholar]
- Hashemi, A.B.; Moghaddam, M.N.; Forghanifard, M.M.; Yousefi, E. Detection of blaOXA-10 and blaOXA-48 Genes in Pseudomonas aeruginosa Clinical Isolates by Multiplex PCR. J. Med. Microbiol. Infect. Dis. 2021, 9, 142–147. [Google Scholar] [CrossRef]
- Thabit, A.K.; Alghamdi, A.M.; Miaji, M.Y.; Alharbi, F.S.; Jawah, A.F.; Alturki, F.; Hosin, N.; Bazuqamah, M.; Almutairi, M.S.; Alhamed, H.; et al. Antibiotic susceptibility of Pseudomonas aeruginosa in Saudi Arabia: A national antimicrobial resistance surveillance study. Front. Public. Health 2024, 12, 1436648. [Google Scholar] [CrossRef] [PubMed]
- Atassi, G.; Medernach, R.; Scheetz, M.; Nozick, S.; Rhodes, N.J.; Murphy-Belcaster, M.; Hauser, A.R. Genomics of aminoglycoside resistance in Pseudomonas aeruginosa bloodstream infections at a United States Academic Hospital. Microbiol. Spectr. 2023, 11, e05087-22. [Google Scholar] [CrossRef]
- Azimi, L.; Armin, S.; Kafil, H.S.; Abdollahi, N.; Ghazvini, K.; Hasanzadeh, S.; Zahedani, S.S.; Tabatabaei, S.R.; Fallah, F. Evaluation of phenotypic and genotypic patterns of aminoglycoside resistance in the Gram-negative bacteria isolates collected from pediatric and general hospitals. Mol. Cell Pediatr. 2022, 9, 2. [Google Scholar] [CrossRef]
- Ahmadian, L.; Bazgir, Z.N.; Ahanjan, M.; Valadan, R.; Goli, H.R. Role of Aminoglycoside- modifying enzymes (AMEs) in resistance to aminoglycosides among clinical isolates of Pseudomonas aeruginosa in the north of Iran. BioMed Res. Int. 2021, 2024, 7077344. [Google Scholar] [CrossRef]
- Abdelrahim, S.S.; Hassuna, N.A.; Waly, N.G.; Kotb, D.N.; Abdelhamid, H.; Zaki, S. Coexistence of plasmid-mediated quinolone resistance (PMQR) and extended-spectrum beta-lactamase (ESBL) genes among clinical Pseudomonas aeruginosa isolates in Egypt. BMC Microbiol. 2024, 24, 175. [Google Scholar] [CrossRef]
- Alfouzan, W.; Dhar, R.; Nicolau, D.P. In vitro activity of newer and conventional antimicrobial agents, including fosfomycin and colistin, against selected Gram-negative bacilli in Kuwait. Pathogens 2018, 7, 75. [Google Scholar] [CrossRef]
- Benwan, K.A.; Jamal, W. Etiology and antibiotic susceptibility Patterns of urinary tract infections in children in a general hospital in Kuwait: A 5-ear retrospective study. Med. Princ. Pract. 2022, 31, 562–569. [Google Scholar] [CrossRef]
- Saki, M.; Sheikh, A.F.; Seyed-Mohammadi, S.; Dezfuli, A.A.Z.; Shahin, M.; Tabasi, M.; Veisi, H.; Keshavarzi, R.; Khani, P. Occurrence of plasmid-mediated quinolone resistance genes in Pseudomonas aeruginosa strains isolated from clinical specimens in southwest Iran: A multicentral study. Sci. Rep. 2022, 12, 2296. [Google Scholar] [CrossRef]
- Shariati, A.; Arshadi, M.; Khosrojerdi, M.A.; Abedinzadeh, M.; Ganjalishahi, M.; Maleki, A.; Heidary, M.; Khoshnood, S. The resistance mechanisms of bacteria against ciprofloxacin and new approaches for enhancing the efficacy of this antibiotic. Front. Public. Health 2022, 10, 1025633. [Google Scholar] [CrossRef]
- El-Badawy, M.F.; Alrobaian, M.M.; Shohayeb, M.M.; Abdelwahab, S.F. Investigation of six plasmid-mediated quinolone resistance genes among clinical isolates of pseudomonas: A genotypic study in Saudi Arabia. Infect. Drug Resist. 2019, 12, 915–923. [Google Scholar] [CrossRef]
- Shanan, R.; Yousef, N.; Balid, M.E.; Tahan, Z.S. Prevalence of Plasmid-Mediated Fluoroquinolone Resistance Genes in Pseudomonas aeruginosa Among Patients at Aleppo University Hospital, Syria. J. Clin. Lab. Anal. 2025, 39, e25153. [Google Scholar] [CrossRef] [PubMed]
- Al-Marjani, M.F. Presence of qnr gene in environmental and clinical Pseudomonas aeruginosa isolates in Baghdad. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 853–857. [Google Scholar]
- Akhlaghi, F.; Nikokar, I.; Mojtahedi, A.; Mobin, M.; Atrkar Roshan, Z.; Karampour, M. Molecular detection of mutations in gyrA, gyrB, parC, and parE genes in the quinolone resistance determining region among Pseudomonas aeruginosa isolated from burn wound infection. Iran. J. Med. Microbiol. 2024, 18, 89–97. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.-J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2018, 37, 177–192. [Google Scholar] [CrossRef]
- Alajmi, R.Z.; Alfouzan, W.A.; Mustafa, A.S. The prevalence of multidrug-resistant Enterobacteriaceae among neonates in Kuwait. Diagnostics 2023, 13, 1505. [Google Scholar] [CrossRef]
- Neyestanaki, D.K.; Mirsalehian, A.; Rezagholizadeh, F.; Jabalameli, F.; Taherikalani, M.; Emaneini, M. Determination of extended spectrum beta-lactamases, metallo-beta-lactamases and AmpC-beta- lactamases among carbapenem resistant Pseudomonas aeruginosa isolated from burn patients. Burns 2014, 40, 1556–1561. [Google Scholar] [CrossRef]
- Bert, F. Identification of PSE and OXA beta-lactamase genes in Pseudomonas aeruginosa using PCR- restriction fragment length polymorphism. J. Antimicrob. Chemother. 2002, 50, 11–18. [Google Scholar] [CrossRef]
- Cho, H.H.; Kwon, G.C.; Kim, S.; Koo, S.H. Distribution of pseudomonas-derived cephalosporinase and metallo- β -lactamases in carbapenem-resistant Pseudomonas aeruginosa isolates from Korea. J. Microbiol. Biotechnol. 2015, 25, 1154–1162. [Google Scholar] [CrossRef]
- Khosravi, A.D.; Mihani, F. Detection of metallo-β-lactamase–producing Pseudomonas aeruginosa strains isolated from burn patients in Ahwaz, Iran. Diagn. Microbiol. Infect. Dis. 2007, 60, 125–128. [Google Scholar] [CrossRef]
- Michalska, A.D.; Sacha, P.T.; Ojdana, D.; Wieczorek, A.; Tryniszewska, E. Prevalence of resistance to aminoglycosides and fluoroquinolones among Pseudomonas aeruginosa strains in a University Hospital in Northeastern Poland. Braz. J. Microbiol. 2014, 45, 1455–1458. [Google Scholar] [CrossRef]
- Farahi, R.M.; Ali, A.A.; Gharavi, S. Characterization of gyrA and parC mutations in ciprofloxacin-resistant Pseudomonas aeruginosa isolates from Tehran hospitals in Iran. Iran. J. Microbiol. 2018, 10, 242–249. [Google Scholar]
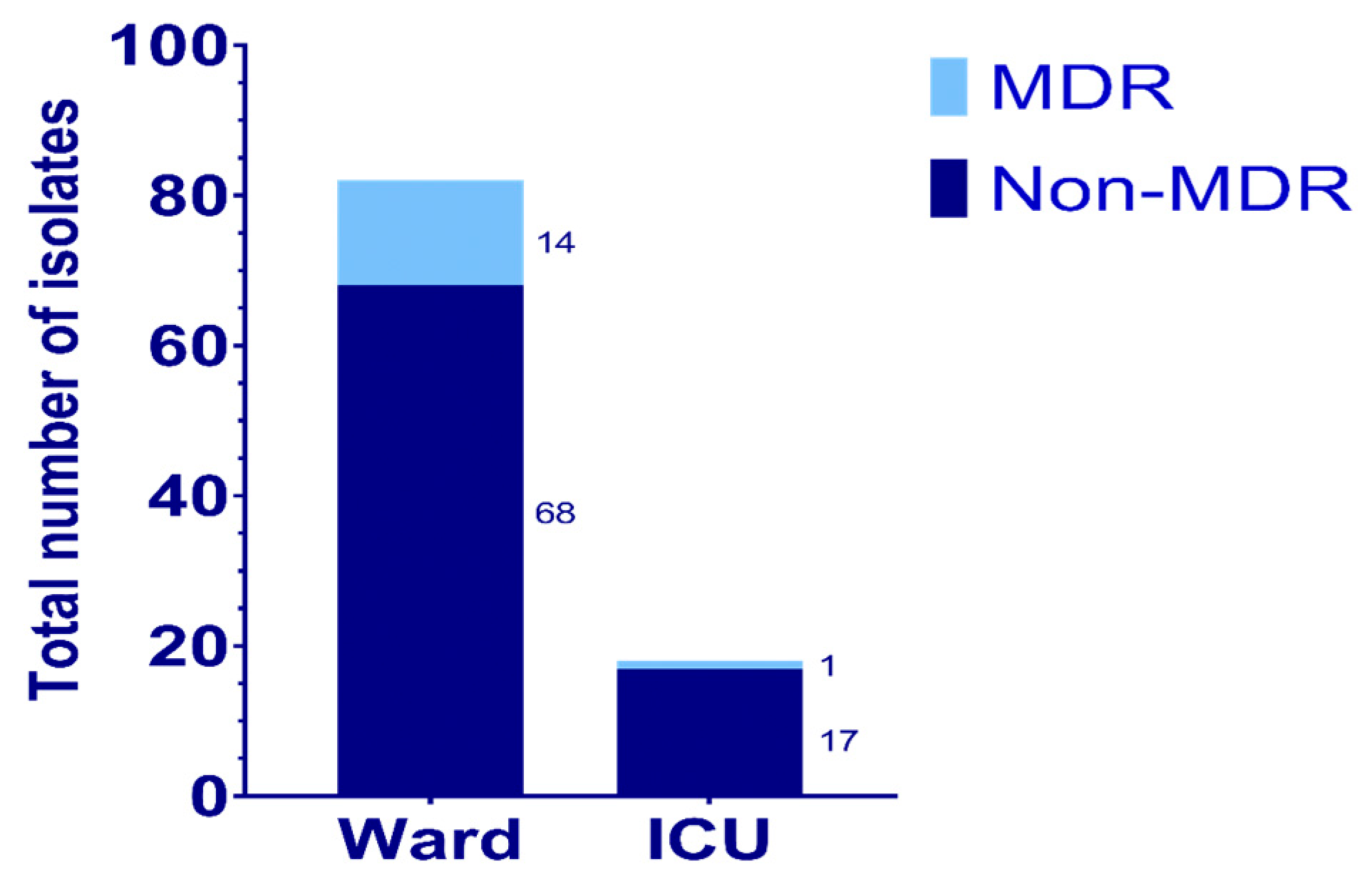
| Organism | Respiratory | Blood | Urine | Others |
|---|---|---|---|---|
| Klebsiella pneumonia | 46 | 3 | 3 | 5 |
| Acinetobacter baumannii | 52 | 0 | 0 | 0 |
| Pseudomonas aeruginosa | 46 | 4 | 22 | 36 |
| Escherichia coli | 0 | 0 | 0 | 1 |
| Organism | Wards Samples | ICUs Samples |
|---|---|---|
| Klebsiella pneumonia | 27 | 30 |
| Acinetobacter baumannii | 15 | 37 |
| Pseudomonas aeruginosa | 90 | 18 |
| Escherichia coli | 1 | 0 |
| Antibiotic Name | Number of Resistances Isolates from Wards, n(%) | Number of Resistances Isolates from ICUs’, n (%) |
|---|---|---|
| Piperacillin-tazobactam | 6 (7.3%) | 1 (5.5%) |
| Ceftazidime-avibactam | 23 (28%) | 3 (16.6%) |
| Ceftolozane-tazobactam | 21 (25.6%) | 1 (5.5%) |
| Aztreonam | 19 (23.1%) | 2 (11.1%) |
| Colistin | 9 (10.9%) | 1 (5.5%) |
| Tobramycin | 17 (20.7%) | 1 (5.5%) |
| Amikacin | 14 (17%) | 1 (5.5%) |
| Gentamicin | 19 (23.1%) | 1 (5.5%) |
| Ciprofloxacin | 25 (30.4%) | 3 (16.6%) |
| Meropenem | 26 (31.7%) | 2 (11.1%) |
| Imipenem | 27 (32.9%) | 3 (16.6%) |
| Ceftazidime | 22 (26.8%) | 3 (16.6%) |
| Isolate # | gyrA Position | parC Position | ||
|---|---|---|---|---|
| Point Mutation | Silent Mutation | Point Mutation | Silent Mutation | |
| 69 | Thr>Ile ACC83ATC | - | Ser>Leu TCG87TTG | Ala>Ala GCG115GCT |
| 71 | Thr>Ile ACC83ATC | - | Ser>Leu TCG87TTG | Ala>Ala GCG115GCT |
| 72 | Thr>Ile ACC83ATC | - | Ser>Leu TCG87TTG | Ala>Ala GCG115GCT |
| 77 | Thr>Ile ACC83ATC | - | Ser>Leu TCG87TTG | Ala>Ala GCG115GCT |
| 94 | Thr>Ile ACC83ATC | - | Ser>Leu TCG87TTG | Ala>Ala GCG115GCT |
| 95 | Thr>Ile ACC83ATC | Arg>Arg CGT68CGA His>His CAC132CAT | Ser>Leu TCG87TTG | Ala>Ala GCG115GCT |
| 99 | Thr>Ile ACC83ATC | His>His CAC132CAT | - | Ala>Ala GCG115GCT |
| 123 | No mutation | - | Ala>Ala GCG115GCT | |
| 160 | Thr>Ile ACC83ATC | Arg>Arg CGT68CGA His>His CAC132CAT | Ser>Leu TCG87TTG | Ala>Ala GCG115GCT |
| 182 | Thr>Ile ACC83ATC | - | - | Ala>Ala GCG115GCT |
| 194 | Thr>Ile ACC83ATC | - | Ser>Leu TCG87TTG | Ala>Ala GCG115GCT |
| 196 | Thr>Ile ACC83ATC | Arg>Arg CGT68CGA | Ser>Leu TCG87TTG | Ala>Ala GCG115GCT |
| 201 | Thr>Ile ACC83ATC | - | Ser>Leu TCG87TTG | Ala>Ala GCG115GCT |
| 203 | Thr>Ile ACC83ATC | His>His CAC132CAT | Ser>Leu TCG87TTG | Ala>Ala GCG115GCT |
| 204 | Thr>Ile ACC83ATC | - | Ser>Leu TCG87TTG | Ala>Ala GCG115GCT |
| Antibiotic Group | Gene | Denaturation | Annealing | Extension | References |
|---|---|---|---|---|---|
| β-lactam-β-lactamase inhibitor combinations (amoxicillin-clavulanic acid) | bla-TEM, bla-SHV, and bla-OXA-10 | 12 min at 95 °C/30 cycles | 50 °C for 30 s | 72 °C for 1 min | [48,49] |
| ESBL genes | bla-CTX, bla-TEM, bla-SHV, and bla-VEB | 12 min at 95 °C/30 cycles | 50 °C for 30 s | 72 °C for 1 min | [48,50] |
| Carbapenems | bla-IMP, bla-VIM, bla-OXA-48, bla-OXA-23, bla-NDM | 12 min at 95 °C/30 cycles | 50 °C for 30 s | 72 °C for 1 min | [50,51] |
| Aminoglycosides | aac(6′)-Ib, ant(2″)-Ia, ant(3″)-Ia, aph(3′)-Ib, and aac(3)-Ia | 12 min at 95 °C/30 cycles | 50 °C for 30 s | 72 °C for 1 min | [35,36,40] |
| Fluoroquinolones | qnrA, qnrB, qnrC, qnrD, and qnrS | 12 min at 95 °C/30 cycles | 50 °C for 30 s | 72 °C for 1 min | [40] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Althaferi, R.S.; Alfouzan, W.A.; Mustafa, A.S. Antibiotics Resistance Profile of Clinical Isolates of Pseudomonas aeruginosa Obtained from Farwaniya Hospital in Kuwait Using Phenotypic and Molecular Methods. Antibiotics 2025, 14, 539. https://doi.org/10.3390/antibiotics14060539
Althaferi RS, Alfouzan WA, Mustafa AS. Antibiotics Resistance Profile of Clinical Isolates of Pseudomonas aeruginosa Obtained from Farwaniya Hospital in Kuwait Using Phenotypic and Molecular Methods. Antibiotics. 2025; 14(6):539. https://doi.org/10.3390/antibiotics14060539
Chicago/Turabian StyleAlthaferi, Rawan Saad, Wadha Ahmed Alfouzan, and Abu Salim Mustafa. 2025. "Antibiotics Resistance Profile of Clinical Isolates of Pseudomonas aeruginosa Obtained from Farwaniya Hospital in Kuwait Using Phenotypic and Molecular Methods" Antibiotics 14, no. 6: 539. https://doi.org/10.3390/antibiotics14060539
APA StyleAlthaferi, R. S., Alfouzan, W. A., & Mustafa, A. S. (2025). Antibiotics Resistance Profile of Clinical Isolates of Pseudomonas aeruginosa Obtained from Farwaniya Hospital in Kuwait Using Phenotypic and Molecular Methods. Antibiotics, 14(6), 539. https://doi.org/10.3390/antibiotics14060539





