Blood Culture Contamination Creep Independent of COVID-19 Pandemics: An Interrupted Time-Series Analysis
Abstract
:1. Introduction
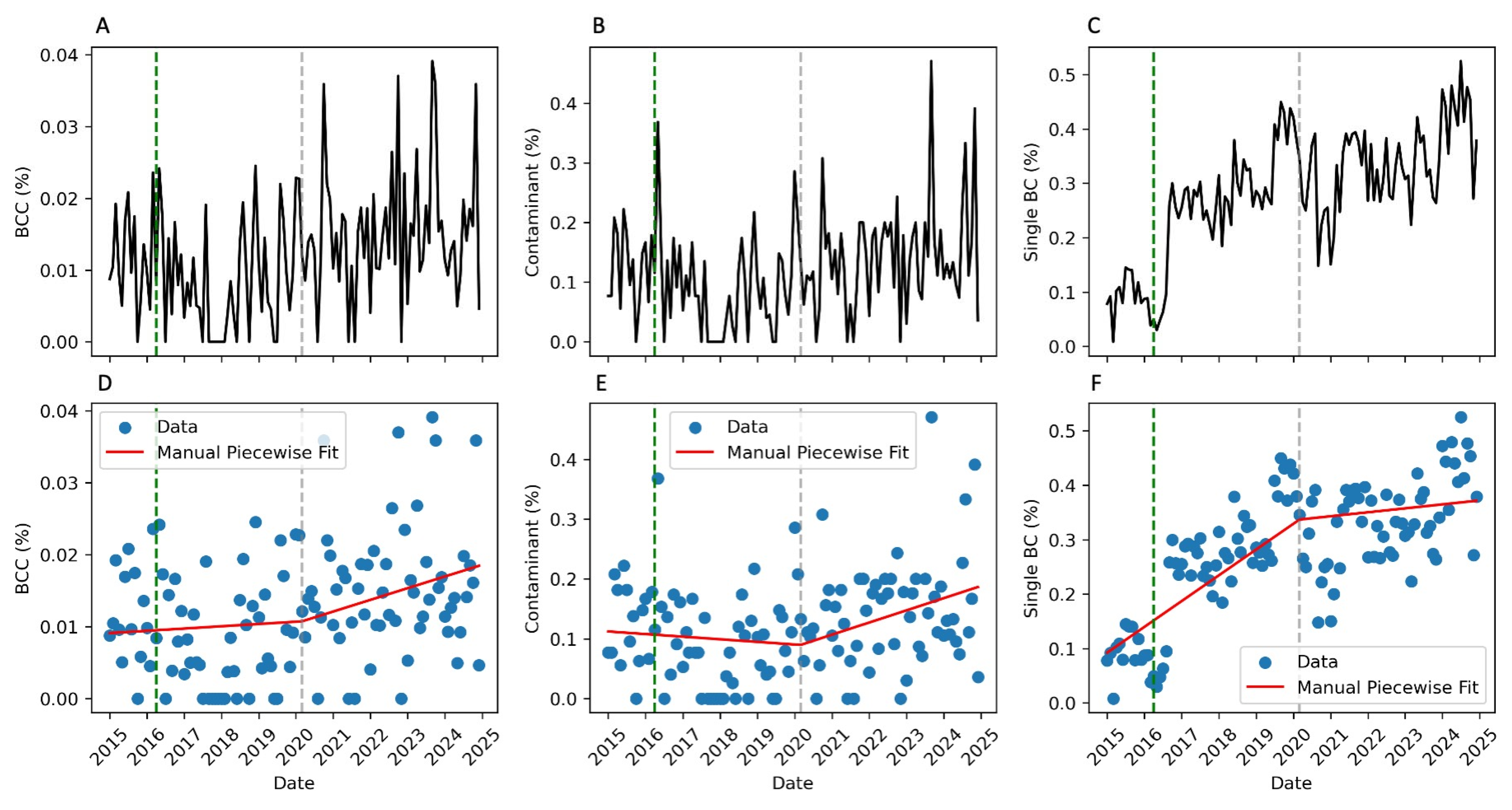
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Design and Clinical Setting
4.2. Blood Culture Collection
4.3. Microbiological Methods, Definitions, and Indicators
4.4. Statistical Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IGH | Izola Genera Hospital |
| BC | Blood culture |
| BCC | Blood culture contamination |
| ICU | Intensive care unit |
| IQR | Interquartile range |
| PD | Patient-day |
References
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, Regional, and National Sepsis Incidence and Mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef]
- Doern, G.V.; Carroll, K.C.; Diekema, D.J.; Garey, K.W.; Rupp, M.E.; Weinstein, M.P.; Sexton, D.J. Practical Guidance for Clinical Microbiology Laboratories: A Comprehensive Update on the Problem of Blood Culture Contamination and a Discussion of Methods for Addressing the Problem. Clin. Microbiol. Rev. 2019, 33, e00009-19. [Google Scholar] [CrossRef] [PubMed]
- Hall, K.K.; Lyman, J.A. Updated Review of Blood Culture Contamination. Clin. Microbiol. Rev. 2006, 19, 788–802. [Google Scholar] [CrossRef] [PubMed]
- Fabre, V.; Carroll, K.C.; Cosgrove, S.E. Blood Culture Utilization in the Hospital Setting: A Call for Diagnostic Stewardship. J. Clin. Microbiol. 2022, 60, e01005-21. [Google Scholar] [CrossRef] [PubMed]
- Schoffelen, T.; Papan, C.; Carrara, E.; Eljaaly, K.; Paul, M.; Keuleyan, E.; Martin Quirós, A.; Peiffer-Smadja, N.; Palos, C.; May, L.; et al. European Society of Clinical Microbiology and Infectious Diseases Guidelines for Antimicrobial Stewardship in Emergency Departments (Endorsed by European Association of Hospital Pharmacists). Clin. Microbiol. Infect. 2024, 30, 1384–1407. [Google Scholar] [CrossRef] [PubMed]
- Theophanous, R.; Ramos, J.; Calland, A.R.; Krcmar, R.; Shah, P.; Da Matta, L.T.; Shaheen, S.; Wrenn, R.H.; Seidelman, J. Blood Culture Algorithm Implementation in Emergency Department Patients as a Diagnostic Stewardship Intervention. Am. J. Infect. Control 2024, 52, 985–991. [Google Scholar] [CrossRef]
- Lamy, B.; Dargère, S.; Arendrup, M.C.; Parienti, J.-J.; Tattevin, P. How to Optimize the Use of Blood Cultures for the Diagnosis of Bloodstream Infections? A State-of-the Art. Front. Microbiol. 2016, 7, 697. [Google Scholar] [CrossRef]
- Wilson, M.L. Principles and Procedures for Blood Cultures (CLSI Guideline M47), 2nd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022. [Google Scholar]
- US Centers for Disease Control and Prevention. Blood Culture Contamination: An Overview for Infection Control and Antibiotic Stewardship Programs Working with the Clinical Laboratory. Available online: https://www.cdc.gov/antibiotic-use/core-elements/pdfs/fs-bloodculture-508.pdf (accessed on 23 April 2025).
- Sautter, R.L.; Parrott, J.S.; Nachamkin, I.; Diel, C.; Tom, R.J.; Bobenchik, A.M.; Bradford, J.Y.; Gilligan, P.; Halstead, D.C.; LaSala, P.R.; et al. American Society for Microbiology Evidence-Based Laboratory Medicine Practice Guidelines to Reduce Blood Culture Contamination Rates: A Systematic Review and Meta-Analysis. Clin. Microbiol. Rev. 2024, 37, e00087-24. [Google Scholar] [CrossRef]
- Msemburi, W.; Karlinsky, A.; Knutson, V.; Aleshin-Guendel, S.; Chatterji, S.; Wakefield, J. The WHO Estimates of Excess Mortality Associated with the COVID-19 Pandemic. Nature 2023, 613, 130–137. [Google Scholar] [CrossRef]
- Janezic, S.; Mahnic, A.; Kuhar, U.; Kovač, J.; Jenko Bizjan, B.; Koritnik, T.; Tesovnik, T.; Šket, R.; Krapež, U.; Slavec, B.; et al. SARS-CoV-2 Molecular Epidemiology in Slovenia, January to September 2021. Eurosurveillance 2023, 28, 2200451. [Google Scholar] [CrossRef]
- Jeverica, S.; Maganja, D.B.; Dernič, J.; Golob, P.; Stepišnik, A.; Novak, B.; Papst, L.; Dodič, A.J.; Gasparini, M. The Influence of COVID-19 on Antimicrobial Resistance Trends at a Secondary Care Hospital in Slovenia: An Interrupted Time Series Analysis. Antibiotics 2024, 13, 1033. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Kim, T.S.; Lee, C.M.; Kang, C.K.; Park, W.B.; Kim, N.J.; Choe, P.G.; Oh, M. Effect of Wearing Personal Protective Equipment (PPE) for COVID-19 Treatment on Blood Culture Contamination: Implication for Optimal PPE Strategies. J. Korean Med. Sci. 2023, 38, e180. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Kim, T.S.; Jo, H.J.; Lee, C.M.; Lee, M.; Kang, C.K.; Choe, P.G.; Park, W.B.; Kim, N.J. Reduction of Blood Culture Contamination Rates through Simplified Personal Protective Equipment in COVID-19 Patient Care Setting. J. Hosp. Infect. 2024, 147, 219–220. [Google Scholar] [CrossRef] [PubMed]
- Fabre, V.; Hsu, Y.-J.; Carroll, K.C.; Salinas, A.B.; Gadala, A.; Bower, C.; Boyd, S.; Degnan, K.O.; Dhaubhadel, P.; Diekema, D.J.; et al. Blood Culture Use in Medical and Surgical Intensive Care Units and Wards. JAMA Netw. Open 2025, 8, e2454738. [Google Scholar] [CrossRef]
- Tolle, H.; Nguyen, A.; MacPhail, A.; Hassoun-Kheir, N.; Chraiti, M.-N.; Boroli, F.; Zanella, M.-C.; Harbarth, S.; Catho, G.; Buetti, N. Increased Incidence of Blood Culture Contaminations during and after the COVID-19 Pandemic. Infection 2025, 53, 711–716. [Google Scholar] [CrossRef]
- Farfour, E.; Clichet, V.; Péan De Ponfilly, G.; Carbonnelle, E.; Vasse, M. Impact of COVID-19 Pandemic on Blood Culture Practices and Bacteremia Epidemiology. Diagn. Microbiol. Infect. Dis. 2023, 107, 116002. [Google Scholar] [CrossRef]
- Sacchetti, B.; Travis, J.; Steed, L.L.; Webb, G. Identification of the Main Contributors to Blood Culture Contamination at a Tertiary Care Academic Medical Center. Infect. Prev. Pract. 2022, 4, 100219. [Google Scholar] [CrossRef]
- Andrei, A.-I.; Popescu, G.-A.; Popoiu, M.A.; Mihai, A.; Tălăpan, D. Changes in Use of Blood Cultures in a COVID-19-Dedicated Tertiary Hospital. Antibiotics 2022, 11, 1694. [Google Scholar] [CrossRef]
- Saleh, L.; Chamieh, A.; El Basst, R.; Azar, E. The Trends of Blood Culture Contamination and Utilization Rates in an LMIC Tertiary Care Center from 2010 to 2022: A Call for Diagnostic Stewardship? Antimicrob. Steward. Healthc. Epidemiol. 2025, 5, e27. [Google Scholar] [CrossRef]
- Rupp, M.E.; Cavalieri, R.J.; Marolf, C.; Lyden, E. Reduction in Blood Culture Contamination Through Use of Initial Specimen Diversion Device. Clin. Infect. Dis. 2017, 65, 201–205. [Google Scholar] [CrossRef]
- Callado, G.Y.; Lin, V.; Thottacherry, E.; Marins, T.A.; Martino, M.D.V.; Salinas, J.L.; Marra, A.R. Diagnostic Stewardship: A Systematic Review and Meta-Analysis of Blood Collection Diversion Devices Used to Reduce Blood Culture Contamination and Improve the Accuracy of Diagnosis in Clinical Settings. Open Forum Infect. Dis. 2023, 10, ofad433. [Google Scholar] [CrossRef]
- Arenas, M.; Boseman, G.M.; Coppin, J.D.; Lukey, J.; Jinadatha, C.; Navarathna, D.H. Asynchronous Testing of 2 Specimen-Diversion Devices to Reduce Blood Culture Contamination: A Single-Site Product Supply Quality Improvement Project. J. Emerg. Nurs. 2021, 47, 256–264.e6. [Google Scholar] [CrossRef]
- Patton, R.G.; Schmitt, T. Innovation for Reducing Blood Culture Contamination: Initial Specimen Diversion Technique. J. Clin. Microbiol. 2010, 48, 4501–4503. [Google Scholar] [CrossRef]
- Lalezari, A.; Cohen, M.J.; Svinik, O.; Tel-Zur, O.; Sinvani, S.; Al-Dayem, Y.A.; Block, C.; Moses, A.E.; Oster, Y.; Salameh, S.; et al. A Simplified Blood Culture Sampling Protocol for Reducing Contamination and Costs: A Randomized Controlled Trial. Clin. Microbiol. Infect. 2020, 26, 470–474. [Google Scholar] [CrossRef]
- US Centers for Disease Control and Prevention. National Healthcare Safety Network (NHSN) Master Organism Common Commensals List. Available online: https://www.cdc.gov/nhsn/xls/master-organism-com-commensals-lists.xlsx (accessed on 22 April 2025).
- Jekel, C.F.; Venter, G. Pwlf: A Python Library for Fitting 1D Continuous Piecewise Linear Functions. Available online: https://github.com/cjekel/piecewise_linear_fit_py (accessed on 20 May 2025).
| Characteristic | Pre-COVID-19 | Post-COVID-19 | All | p-Value | |||
|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | n | (%) | ||
| Hospital | |||||||
| Admissions sum | 79,608 | (53.9) | 68,125 | (46.1) | 147,733 | (100) | |
| Patient-day sum | 347,504 | (54.8) | 286,654 | (45.2) | 634,158 | (100) | |
| Patient-day median (IQR) a | 5447 | (5115–5881) | 5013 | (4610–5441) | 5267 | (4874–5653) | <0.001 |
| Blood culture (BC) | |||||||
| BC sum | 13,968 | (55.7) | 11,100 | (44.3) | 25,068 | (100) | |
| BC/PD b median (IQR) a | 39.6 | (35.6–44.8) | 37.8 | (34.8–43.2) | 38.9 | (35.1–44.1) | 0.161 |
| Adult BC/PD b median (IQR) a | 35.0 | (31.3–40.3) | 34.1 | (30.1–38.7) | 34.7 | (30.8–39.9) | 0.825 |
| Pediatric BC/PD b median (IQR) a | 4.3 | (3.45–5.2) | 4.3 | (3.4–5.4) | 4.3 | (3.4–5.3) | 0.173 |
| Characteristic | Pre-COVID-19 | Post-COVID-19 | All | p-Value | |||
|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | n | (%) | ||
| Demographic | |||||||
| BCC $ | 131 | (43.2) | 172 | (56.8) | 303 | (100) | |
| Male | 80 | (61.1) | 116 | (67.4) | 196 | (64.7) | 0.222 |
| Female | 51 | (38.9) | 55 | (32.6) | 106 | (35.3) | |
| Age median (IQR) a | 66 | (45–77) | 66 | (49–76) | 66 | (49–76) | 0.820 |
| Patient group | |||||||
| Adult | 112 | (85.5) | 147 | (85.5) | 259 | (85.5) | 0.994 |
| Pediatric | 19 | (14.5) | 25 | (14.5) | 44 | (14.5) | |
| Department | |||||||
| Internal medicine | 82 | (62.6) | 95 | (55.2) | 177 | (58.4) | 0.001 |
| Surgery | 26 | (19.8) | 31 | (18.0) | 57 | (18.8) | |
| Pediatrics | 16 | (12.2) | 11 | (6.4) | 27 | (8.9) | |
| Emergency | 4 | (3.1) | 33 | (19.2) | 37 | (13.2) | |
| Gynecology | 3 | (2.3) | 2 | (1.2) | 5 | (1.7) | |
| Microorganism groups | |||||||
| CoNS * | 76 | (58.0) | 119 | (69.2) | 195 | (64.4) | 0.193 |
| Streptococcus | 23 | (17.6) | 17 | (9.9) | 40 | (13.2) | |
| Cutibacterium | 8 | (6.1) | 11 | (6.4) | 19 | (6.3) | |
| Corynebacterium | 7 | (5.3) | 4 | (2.3) | 11 | (3.6) | |
| Anaerobes | 4 | (3.1) | 4 | (2.3) | 8 | (2.6) | |
| Bacillus | 3 | (2.3) | 1 | (0.6) | 4 | (1.3) | |
| Other | 10 | (7.6) | 16 | (9.3) | 26 | (8.6) | |
| Polymicrobial BCC $ | |||||||
| Yes | 6 | (4.6) | 14 | (8.1) | 20 | (6.6) | 0.216 |
| No | 125 | (95.4) | 158 | (91.9) | 283 | (93.4) | |
| Collection type | |||||||
| Percutaneous | 82 | (62.6) | 83 | (48.3) | 165 | (54.5) | 0.013 |
| Catheter | 49 | (37.4) | 89 | (51.7) | 138 | (45.5) | |
| BCC # Indicators | Pre-COVID-19 | Post-COVID-19 | Total | p-Value b | |||
|---|---|---|---|---|---|---|---|
| % | (95% CI) | % | (95% CI) | % | (95% CI) | ||
| BCC # rate | 0.9 | (0.8–1.1) | 1.5 | (1.3–1.8) | 1.2 | (1.1–1.4) | 0.001 |
| Contaminant proportion | 9.8 | (7.8–11.8) | 14.2 | (11.8–16.6) | 11.9 | (10.3–13.5) | 0.016 |
| Single BC * rate | 23.1 | (20.2–26.1) | 33.6 | (31.5–35.8) | 28.1 | (26.0–30.2) | <0.001 |
| First-to-second bottle ratio (n1, n2, all) a | 0.886 | (31, 35, 66) | 1.917 | (115, 60, 175) | 1.537 | (146, 95, 241) | 0.024 |
| Interrupted Time Series Parameters | Pre-COVID-19 | Post-COVID-19 | p-Value |
|---|---|---|---|
| Slope [×10−3] | Slope [×10−3] | ||
| BCC # rate | 0.350 | 1.620 | 0.700 |
| Contaminant proportion | −3.960 | 20.591 | 0.065 |
| Single BC * rate | 46.187 | 7.474 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeverica, S.; Dernič, J.; Golob, P.; Stepišnik, A.; Novak, B.; Gantar, T.; Papst, L.; Dodič, A.J.; Maganja, D.B.; Zmazek, J.; et al. Blood Culture Contamination Creep Independent of COVID-19 Pandemics: An Interrupted Time-Series Analysis. Antibiotics 2025, 14, 533. https://doi.org/10.3390/antibiotics14060533
Jeverica S, Dernič J, Golob P, Stepišnik A, Novak B, Gantar T, Papst L, Dodič AJ, Maganja DB, Zmazek J, et al. Blood Culture Contamination Creep Independent of COVID-19 Pandemics: An Interrupted Time-Series Analysis. Antibiotics. 2025; 14(6):533. https://doi.org/10.3390/antibiotics14060533
Chicago/Turabian StyleJeverica, Samo, Jani Dernič, Peter Golob, Alenka Stepišnik, Bojan Novak, Tomaž Gantar, Lea Papst, Anamarija Juriševič Dodič, Darja Barlič Maganja, Jan Zmazek, and et al. 2025. "Blood Culture Contamination Creep Independent of COVID-19 Pandemics: An Interrupted Time-Series Analysis" Antibiotics 14, no. 6: 533. https://doi.org/10.3390/antibiotics14060533
APA StyleJeverica, S., Dernič, J., Golob, P., Stepišnik, A., Novak, B., Gantar, T., Papst, L., Dodič, A. J., Maganja, D. B., Zmazek, J., & Gasparini, M. (2025). Blood Culture Contamination Creep Independent of COVID-19 Pandemics: An Interrupted Time-Series Analysis. Antibiotics, 14(6), 533. https://doi.org/10.3390/antibiotics14060533






