Abstract
Background/Objectives: Non-typhoidal Salmonella (NTS) is one of the most common causative agents of food poisoning in Vietnam, and contaminated livestock meat poses a major risk to human health. The present study aims to provide the genetic characteristics of NTS with a particular focus on antimicrobial resistance and determine phylogenetic relationships between isolates from different sources in Southern Vietnam based on whole-genome sequencing (WGS) data. Methods: A total of 49 NTS isolates from pork/broiler meat, pigs, chickens, and humans were collected in Ho Chi Minh City and four provinces of Southern Vietnam. Phenotypic antimicrobial susceptibility testing (AST) and WGS for all isolates were performed. Results: As a result, 14 different serotypes were identified, among which S. Typhimurium and its monophasic variant were the dominant serotypes for human and pig sources. All chicken samples belonged to S. Indiana, whereas S. Infantis predominated in broiler meat. AST results revealed that 98% of isolates were multidrug resistant. NTS strains isolated from poultry and pigs exhibited resistance to the highest priority antimicrobials—quinolones and polymyxin, as well as to β-lactams, aminoglycosides, tetracycline, and sulfonamide, which are considered to be critical for the treatment of severe diseases. Conclusions: The results highlight the utmost importance of issues related to the selection, spreading, and transmission of multi-resistant strains from animals to humans.
1. Introduction
Non-typhoidal Salmonella (NTS) is a food-borne pathogen that causes a significant health and economic global burden worldwide, accounting for 4.38 million DALYs (Disability-Adjusted Life Years) (2007–2015) [1,2,3]. Being zoonotic, NTS is hosted by livestock and, due to further contamination of the environment, water, and food, manifests as a leading causative agent in food poisoning worldwide [1,2,4]. Among different food sources, contaminated poultry and pork products pose a main risk to human health as they participate in different food chain stages [2,3].
The use of antibiotics in the food animal sector plays a key role in the emergence of multidrug-resistant (MDR) bacteria: so far, approximately 80% of farm animals are treated with broad-spectrum antibiotics, which, given the increasing globalization of the livestock industry, exerts strong selective pressure on bacterial populations. Through adaptive evolution, Salmonella have developed diverse and complex resistance mechanisms that facilitate the rapid acquisition and spread of antibiotic resistance across diverse host species. Horizontal gene transfer, point mutations, the presence of heavy metal resistance genes and virulence genes, compensatory mechanisms, epistatic interactions, formation of biofilms, and the activation of efflux pumps, – all contribute to the MDR evolution in Salmonella. Moreover, it is suggested that Salmonella can exchange resistance genes with other bacteria of the Enterobacteriaceae family during passage through the intestine [3,4,5].
The exposure of human populations to antibiotic-resistant bacteria is facilitated by the consumption of animal products or direct contact with contaminated sources, both of which resulted from inadequate hygiene practices throughout the food chain production. This situation gives rise to global human health issues, including treatment failure, limited antimicrobial choices, increased morbidity, heightened disease burdens, outbreaks, and a higher risk of mortality.
As a global health problem, key international documents and guidelines were developed to prevent and reduce the use of antibiotics in animal agriculture: the World Health Organization (WHO) Guidelines on the Use of Medically Important Antimicrobials in Food-Producing Animals presented recommendations on the use of medically important antimicrobials in food-producing animals to preserve their effectiveness in human and veterinary medicine; the Food and Agriculture Organization (FAO) and the World Organization for Animal Health (OIE) provide guidelines for monitoring the use of antimicrobials across the human, animal and plant sectors; the Global Repository of Available Guidelines for Prudent Use of Antimicrobials compiled by the World Veterinary Association created a global repository of available guidelines for responsible use of antimicrobials in animal health; the ASEAN Guidelines for Prudent Use of Antimicrobials in Livestock are intended for consideration by ASEAN Member States for developing national strategies to combat antimicrobial resistance through the prudent use of antimicrobials in livestock sectors; and the 79th United Nations General Assembly Declaration committing to a significant reduction in the global quantity of antimicrobials use in the agrifood system by 2030 [4,5,6,7,8].
Although the prevalence of multidrug-resistant Salmonella varies geographically, the incidence of MDR NTS worldwide is rising steadily. The situation is further exacerbated by growing resistance to critical classes of antibiotics that are essential for severe infection treatment [7]. Globally, the highest MDR rates have been reported in South and Southeast Asia (Thailand—69%, Taiwan—53.8%, India—68%, Bangladesh—94%) and sub-Saharan Africa (77.2–87.4%), which are considered super regions for the global burden of multidrug-resistant Salmonella. In contrast, the lowest levels of MDR-NTS were recorded in the United States and Europe (9.6–22.6%) [7,8,9,10].
Vietnam, one of the leading countries in Southeast Asia in meat product manufacturing, ranks first in the prevalence of NTS, which is 2–5% in humans and 30–41% in retail meats and the farm environment [1]. Moreover, high rates of MDR NTS have emerged both in pigs (86%) and chickens (95%), pork and poultry meat production (40–75%), and in human sources (12.5–50%) [1,11,12,13]. Interestingly, the highest NTS prevalence in Vietnam was reported for duck and pig (94.3% and 91.3%) compared to chicken (64.7%) and exhibited a high resistance rate for tetracycline (58.5%), trimethoprim/sulfamethoxazole (58.1%), streptomycin (47.3%), ampicillin (39.8%), and chloramphenicol (37.3%) [12,13]. According to a study conducted by Coyne et al. (2019), four antibiotics (chlortetracycline, neomycin, colistin, and amoxicillin) considered crucial by the World Health Organization (WHO) for human infection treatment were found in pig and chicken feeds [14]. All this underlines the importance of localized antimicrobial resistance surveillance as well as careful monitoring of changes and distribution of drug-resistant microbial populations in animals and humans across this territory.
Based on whole-genome sequencing data, this study aims to provide genetic characteristics of multidrug-resistant non-typhoidal Salmonella from different sources with a focus on both antimicrobial resistance patterns and phylogenetic analysis. This research addresses the lack of data on multidrug-resistant salmonellosis in Southern Vietnam, a region underrepresented in the scientific literature compared to more extensively studied areas. Through a One Health approach, we provide a comprehensive examination of phenotypic and genotypic multidrug resistance patterns, with an emphasis on WGS-based serotype prediction and phylogenetic analysis of MDR Salmonella from livestock animals (pigs and chickens), retail meat (pork and broiler meat), and humans with acute diarrhea. This study elucidates the background of multidrug resistance, transmission pathways, population structure, and within-host diversity, potentially revealing local factors contributing to the selection, evolution, and spreading of multi-resistant Salmonella strains.
2. Results
2.1. NTS Serotype Identification
According to WGS analysis of 49 Salmonella enterica isolates, 14 different serotypes were identified: S. Typhimurium, S. Typhimurium monophasic, S. Indiana, S. London, S. Infantis, S. Rissen, S. Newport, S. Derby, S. Give, S. Enteritidis, S. Meleagridis, S. Kentucky, S. Panama, and S. Agona (Table 1). Five S. Typhimurium isolates were divided into three sequence types (STs), among which only ST34 belonged to a human source. All S. Typhimurium monophasic isolates belonged to the type ST34 without differences between host origin. Both S. Typhimurium and its monophasic variant were the dominant serotypes among human and pig sources and were identified in 72.7 and 44.4% of cases, respectively. All chicken samples belonged to S. Indiana ST17, whereas S. Infantis predominated in broiler meat samples (63.6%) and included two ST: one belonged to ST10, while ST32 represented the highest number of broiler meat isolates. All S. London ST155 isolates were detected in either pig or pork samples. The remaining serotypes (S. Newport, S. Derby, S. Give, S. Enteritidis, S. Meleagridis, S. Kentucky, S. Panama, and S. Agona) included one to two isolates predominantly from pig samples.

Table 1.
Serotyping results of 49 NTS isolated from different sources.
2.2. Antimicrobial Susceptibility Testing (AST) of NTS
The phenotypic AST results showed that among 49 Salmonella isolates, 48 (97.9%) were resistant to more than three antibiotic classes and were assigned as multidrug resistant (MDR), while four isolates (8.2%) were resistant to eight different antibiotic classes and were therefore defined as extensively drug resistant (XDR). All isolates (100%) were susceptible to only one of 15 antibiotics—MEM, whereas the highest percentage of resistance was observed against AMP and TET (45/49, 91.8%), followed by CIP/NAL and CHL (40/49, 81.6%, and 38/49, 77.6%, respectively).
The resistance rate for human isolates was highest for TET (11/11, 100%), followed by AMP, NAL/CIP, and CHL (10/11, 90.9% each) (Table 2). Similarly, Salmonella isolated from livestock and pork/broiler raw meat were most resistant to AMP, TET, CTX, NAL/CIP, and CHL. However, it is worth noting that isolates obtained from pigs have demonstrated a significantly higher resistance rate to CT (12/18, 66.7%) than those from other sources (χ2 = 17.479; df = 4; p = 0.002): pairwise comparison of the proportion of CT-resistant strains revealed differences between pig and chicken isolates (p = 0.01373) as well as pig and broiler meat isolates (p = 0.00039). A similar pattern was observed for broiler meat isolates, which showed the highest resistance to CTX (81.8%) compared to isolates from other sources (χ2 = 27.246; df = 4; p < 0.001): pairwise comparison of the proportion of CTX-resistant strains revealed differences between broiler meat and chicken isolates (p = 0.00481), broiler meat and humans (p = 0.00463), and broiler meat and pig isolates (p = 0.00001). All chicken isolates (100%) showed resistance to AMP and GEN/TOB, while 4/5 (80.0%) isolates were resistant to TET. Interestingly, none of the chicken and broiler isolates had similar resistance profiles.

Table 2.
Antimicrobial resistance of 49 NTS isolates from different sources. Non-wild-type isolates were considered as resistant to NAL, TET, CHL, and AZM.
We also analyzed antimicrobial resistance profiles of particular Salmonella serotypes combined with more than three isolates. For S. Typhimurium isolates and its monophasic variant, AMP and TET resistance rates were highest (94.1%), followed by NAL/CIP and CHL (88.2%). All S. Infantis isolates were resistant to β-lactams (except meropenem), NAL/CIP, TET, and GEN/TOB, whereas all S. Indiana and S. London isolates showed resistance to AMP, NAL/CIP, CHL, GEN/TOB, and AMP, NAL/CIP, CHL, and TMP/SMX, respectively. Among all Salmonella serotypes, only one S. Indiana isolate was resistant to AMI, while S. Infantis showed resistance to the third-generation cephalosporin CTX compared to other serotypes.
2.3. Correlation Between Phenotypic and Genotypic Antimicrobial Resistance of Salmonella Isolates
To investigate the relationship between phenotypic and genotypic antimicrobial resistance, we analyzed acquired resistance genes and/or resistance mutations for eight antibiotic classes using the ResFinder tool and assessed their correlation with the MIC pro-files of 49 Salmonella isolates (Table 3). Among all NTS isolates, 42 antimicrobial resistance genes (ARGs) have been identified. Overall, Salmonella isolates cultured from pig, chicken, and meat samples harbored more resistance genes compared to those from humans with acute diarrhea.

Table 3.
Correlation between genotypic and phenotypic AST and comparison of the resistance profile of NTS isolates from different sources.
2.3.1. Beta-Lactam Antibiotics
Phenotypic AST results demonstrated that among five β-lactam antibiotics, the majority (45/49, 91.8%) of NTS isolates were resistant to aminopenicillins (AMP: MIC ≥ 512 µg/mL), while 75% (37/49) showed susceptibility to third- and fourth-generation cephalosporins. All isolates were susceptible to carbapenems (MEM, MIC 0.008–0.047 mg/L) (not represented in Table 3). Twelve Salmonella isolates resistant to third- and fourth-generation cephalosporins (CTX with/or not CTZ and FEP) were isolated from all sources (human, pig, chicken, and broiler samples) and belonged to 4 serotypes—S. Infantis, S. Newport, S. Typhimurium monophasic, and S. London (Table 3).
All resistant phenotypes were confirmed by the presence of at least one gene from the beta-lactamases (BLs) group in the genome of Salmonella isolates. Among all gene types, blaTEM-1B was predominant (35/49, 71.4%), followed by blaCTX-Ms (12/45, 26.7%), blaOXA-1 (7/49, 14.3%), blaLAP-2 (3/49, 6.1%), and blaSHV-25 (1/49, 2.0%). Interestingly, the majority of resistant isolates (34/45, 75.6%) carried a single type of BL, among which the blaTEM-1B gene was predominantly found in isolates resistant only to AMP (24/33, 72.7%), whereas blaCTX-Ms prevailed in isolates resistant to cephalosporins. Two isolates from broiler and pork meat samples with the highest MIC values were positive for three blaCTX-M-55 + blaTEM-1B + blaLAP-2 BLs genes. Four isolates susceptible to β-lactams did not possess any BLs resistance genes.
2.3.2. Aminoglycosides
Of 49 NTS isolates, 48 (97.9%) showed phenotypic susceptibility to AMI, while only 16 (32.7%) were susceptible to GEN and TOB (Table 3). All isolates from chicken samples and the vast majority of broiler isolates (10/11, 90.9%) were resistant to GEN/TOB. Isolates from pig and pork samples have demonstrated the lowest resistance to aminoglycosides.
Genotypically, this class of antimicrobials demonstrated the largest resistance diversity. In total, we identified 11 aminoglycoside-modifying enzyme (AME) genes: aac(6′)-Iaa, aac(3)-IV, aac3IId, aac(6′)-lb-cr, aadA, aadB, aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib, aph(4)-Ia, and rmtB. One of the acetyl-transferase genes is aac(6′)-Iaa. Moreover, different combinations of genes, such as aac(6′)-Iaa, aph(6)-Id, aph(3″)-Ib, aadA, and aadB, were found in susceptible NTS isolates and were not associated with phenotypic resistance to GEN, TOB, and AMI in our study. The genetic profile in Salmonella isolates phenotypically resistant to GEN and TOB was marked by the presence aac3IId gene (which was predominantly detected in human and pig isolates) and aac(3)-IV and aph(4)-Ia (which prevailed in chicken and broiler meat isolates).
One isolate (S. Indiana) phenotypically resistant to all aminoglycosides with MIC > 256 µg/mL possessed seven resistance genes (aac(6′)-Iaa + aac(3)-IV + aph(4)-Ia + aph(3″)-Ib + aph(6)-Id + aac(6′)-lb-cr + rmtB) in its genome. Furthermore, the gene rmtB, which encodes for 16S rRNA methyltransferase, was detected specifically in this isolate and was absent in the other 48 Salmonella isolates.
2.3.3. Quinolones
According to AST results, only 3 (6.1%) Salmonella isolates from human and pig samples were fully susceptible to quinolones—CIP and NAL (Table 3). Only one susceptible isolate did not have resistance genes. However, discrepancy between phenotypic and genotypic results was found for two isolates, assigned as quinolone-susceptible but carrying aac(6′)-Ib-cr, parC(T57S), and qnrA1 resistance patterns. The qnrS1 gene was the most frequent, present in 65.3% of resistant isolates. NAL- and CIP-resistant isolates with MIC > 256 µg/mL and MIC > 32 µg/mL, respectively, were characterized by the presence of double mutations D87Y/S83Y in the gyrA gene and T57S/S80R in the parC gene. It might also be noted that multiple resistance gene carriage in Salmonella was associated with resistance level to quinolones: the higher the MIC value, the more resistance genes were detected in Salmonella isolates. Both NAL- and CIP-resistant isolates with the highest MIC values had the most diverse genetic profiles and were associated with the following resistance patterns: gyrA(D87Y)/gyrA(S83Y), parC(T57S)/parC(S80R/I), aac(6′)-Ib-cr, and qnrS1.
2.3.4. Other Antibiotics
We found that 16 out of 17 TMP/SMX-susceptible isolates carried sul1, sul2, and sul3 resistance genes, whereas dfrA12, dfrA14, and dfrA5 genes, involved in trimethoprim resistance development, were observed in only TMP/SMX-resistant isolate genomes.
Due to the lack of clinical breakpoints for NAL, CHL, AZM, and TET antimicrobials, ECOFF values were used to distinguish WT isolates (without phenotypically detectable resistance mechanisms) from NWT isolates with acquired resistance.
Of 49 Salmonella isolates, five (10.2%) were WT to CHL and did not harbor resistance genes in their genome. Genes cmlA1, floR, and catA2/catA3/catB3 were associated with the NWT phenotype. All NWT isolates with MIC > 256 carried either the floR gene (encodes chloramphenicol acetyltransferase) or a combination of floR with catA/B genes. All chicken isolates were NWT to CHL and carried combinations of three resistance genes: floR + cmlA1 + catB3 and floR + catA1+ catB3 (Table 3).
Overall, the majority of NTS isolates (38/49, 77.6%) exhibited a WT phenotype to AZM, and no resistance genes were found in their genome. In contrast, NWT isolates predominantly carried the phosphotransferase mph(A) gene (10/11, 90.9%); one isolate contained the macrolide efflux protein mef(B) gene.
Among 49 NTS, 33 (67.3%) isolates were phenotypically susceptible to colistin, and no resistance genes were found in their genome. Resistance to CT was associated with three variants of the mobile colistin resistance mcr gene (1.1, 3.1, 3.5). Interestingly, all isolates from chicken and broiler meat samples were susceptible to CT, whereas the majority of pig isolates (12/18, 66.7%) showed resistance to CT.
Four isolates (8.2%) had a WT phenotype to TET; two of them contained the tetM gene. NWT isolates harbored either tetA, tetB, or combinations of tetA + tetB and tetA + tetM genes.
2.4. Plasmid Analysis
Using PlasmidFinder 2.1 software, we identified 35 AMR plasmids among 49 NTS isolates: 25 different incompatibility (Inc) family plasmids were detected in 48/49 (97.9%) isolates, while the colicin (Col) group plasmids were contained within 28/49 (57.1%) isolates (Figure 1).
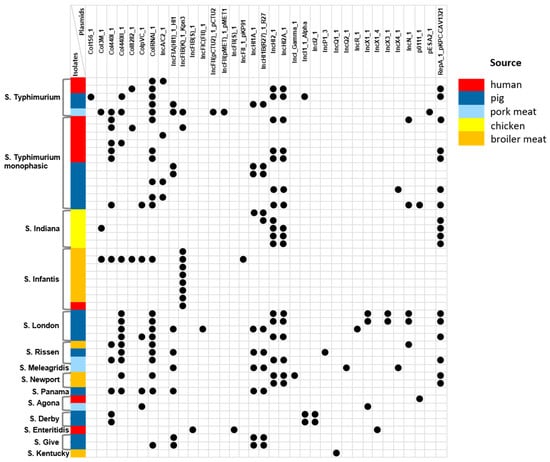
Figure 1.
Plasmid profiling in the Vietnamese NTS isolates from different sources.
The most frequently detected incompatibility groups were IncHI2_1 and IncHI2A_1, found in 17 (34.7%) NTS isolates from various serovars and sources. Both plasmids were associated with the plasmid RepA_1_pKPC-CAV1321. Other replicon types, IncHI1A_1 and IncHI1B(R27)_1_R27, were detected in 10 (20.4%) isolates, mostly from pigs and pork meat (9/10, 90.0%). Interestingly, the IncFIA(HI1)_1_HI1 replicon was carried by 11 Salmonella strains, of which 10 (90.9%) were also isolated from either pig or pork meat samples. The plasmid replicon IncFIB(K)_1_Kpn3, on the contrary, was predominant among isolates from broiler meat (70.0%) and was harbored by all S. Infantis isolates. Notably, many previous studies reported that the IncFII(S) plasmid was one of the most widely represented types and was present in all clinical Salmonella isolates [15,16]. Our results are inconsistent with these findings and showed that only one human S. Enteritidis isolate carries the IncFII(S)_1 replicon. Interestingly, only two S. London isolates from pigs presented the IncX3_1 plasmid (primarily responsible for the dissemination of antibiotic resistance genes) alongside the IncX1_1 and IncN_1 plasmid types.
Among the colicin family plasmids, ColRNAI_1 was the most frequently detected across all S. London, S. Rissen, and S. Typhimurium isolates, irrespective of source. Another Col440I_1 plasmid was detected in 28.6% of NTS isolates regardless of serotype and source, whereas Col440II_1 and ColpVC_1 plasmids were the most frequently observed in pig and pork meat isolates (9/13, 69.2%, and 4/5, 80.0%, respectively). All S. London and S. Rissen isolates contained ColRNAI_1 along with the Col440II_1 plasmid, while one S. Infantis broiler meat isolate presented five types of Col plasmids (Col3M_1, Col440I_1, Col440II_1, Col8282_1, ColpVC_1, and ColRNAI_1). Notably, six types of Col plasmids were detected in broiler isolates, whereas only one Col440I_1 plasmid was detected among chicken isolates.
2.5. Phylogenetic Analysis
Given that MLST (Multi Locus Sequence Typing) can not sufficiently differentiate Salmonella serotypes, for phylogenetic analysis we used core genome SNPs as having the highest resolution compared to MLST, core genome MLST, and whole-genome MLST [17]. Based on core SNPs of 49 Salmonella genomes, we constructed the maximum likelihood phylogenetic tree with respect to the phenotypic antibiotic resistance profile (Figure 2).

Figure 2.
Maximum likelihood phylogenetic tree of 49 Vietnamese NTS isolates from different sources based on core SNVs with respect to phenotypic resistance profile. A, B, C and d, e, f, g, h stand for clusters and subclusters, respectively. AMP—ampicillin, CTZ—ceftazidime, CTX—cefotaxime, FEP—cefepime, MEM—meropenem, NAL—nalidixic acid, CIP—ciprofloxacin, TET—tetracycline, CHL—chloramphenicol, TMP/SMX—trimethoprim/sulfamethoxazole, CT—colistin, GEN—gentamycin, TOB—tobramycin, AMI—amikacin, AZM—azithromycin.
As a result, all 49 NTS isolates clustered according to the ST serotypes and formed three distinct phylogenetic clusters: A, B, and C. The most predominant cluster A was divided into two subclusters, d and e. Subcluster d comprised S. Typhimurium isolates and its monophasic variant; however, within this subcluster, ST34 and ST19 isolates (including reference ST19 strain) formed a separate group adjacent to the ST36 isolate. Subcluster e included S. Newport ST4157 isolates. S. London ST155 isolates grouped together and formed Cluster B. Cluster C combined 10 different Salmonella serotypes, among which a single S. Enteritidis ST11 from a human formed a distant subcluster. Subclusters f and g were formed by S. Infantis ST32/ST10 and S. Indiana ST17 strains, isolated predominantly from chicken and broiler meat samples. The remaining groups of subclusters contained one to three Salmonella isolates.
Notably, each Salmonella serotype in this analysis formed a distinct cluster or subcluster, whereas the phylogenetic tree showed close genetic relatedness between strains isolated from different sources. These observations indicate that the clustering of NTS isolates is significantly affected by serotypes rather than isolation source. Moreover, since Salmonella strains exhibit different mechanisms of genomic diversity and transmission pathways, obtained results suggest the possibility of NTS transmission across different hosts and locations, ultimately leading to the foodborne illness in humans.
To assess the genetic diversity and relationships between Vietnamese isolates from different sources, we generated cladograms based on core-genome SNPs for two serotypes that predominated in our investigation: S. Infantis and S. Typhimurium (and its monophasic variant) isolates (Figure 3). In total, for this analysis, we used 292 S. Infantis and 283 S. Typhimurium genomes of the corresponding serotypes from different sources, regions, and years, randomly selected from the international GenBank database.
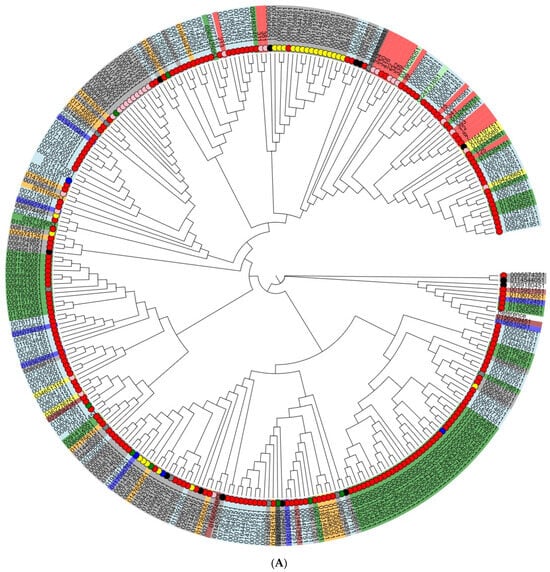
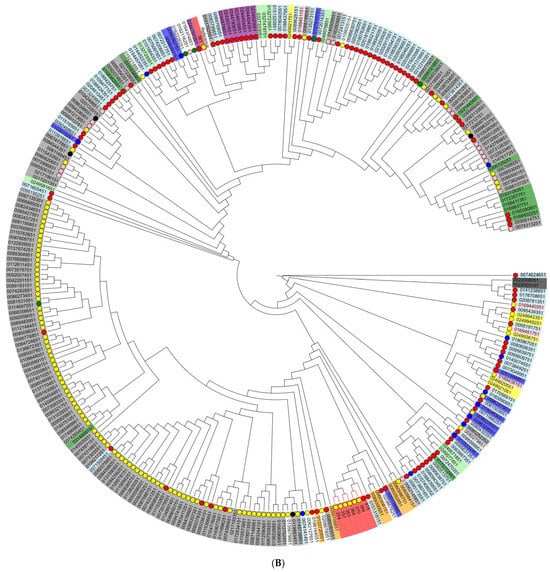
Figure 3.
Phylogenetic (cladogram) tree based on core-genome single nucleotide polymorphisms (SNPs) of (A) S. Typhimurium (and its monophasic variant) isolates and (B) S. Infantis isolates. The cladogram was created using the maximum-likelihood algorithm. Small colored circles innermost stand for the sources of NTS isolates: red—human, pink—porcine, yellow—poultry, black—bovine, and green—food; the absence of a circle indicates isolates with unknown origin. The countries of origin of Salmonella isolates are marked by color in rectangles: (A) green: Australia; orange: Canada; yellow: China; light green: Denmark; dark red: Japan; light blue: United Kingdom; grey: USA; red: Vietnam; blue: others. (B) purple: South Africa; light green: Australia; white: Brazil; green: Canada; blue: Germany; pink: Hungary; yellow: Slovenia; orange: Peru; light blue: United Kingdom; grey: USA; red: Vietnam; white: others.
As a result, seven of eight Vietnamese S. Infantis isolates from both broiler meat and humans were clustered together in one subcluster; interestingly, these isolates showed a close genetic relationship with human isolates from the United Kingdom. The majority of Vietnamese S. Typhimurium isolates did not completely cluster together; however, irrespective of pig or human source, they were included in one subcluster with those isolated predominantly from humans. Two S. Typhimurium isolates from pork meat had a close relationship with isolates from poultry samples obtained in the USA.
3. Discussion
Since WGS has been proven to be the most reliable, accurate, and effective method for serovar and sequence type identification, as well as antimicrobial resistance patterns and phylogenetic relationships estimation between different isolates, we used this approach for the comprehensive characterization of Salmonella isolates from different sources in southern regions of Vietnam.
According to our study, monophasic S. Typhimurium ST34 [4: i: -] (one of the most common in Vietnam) was the predominant serotype found among pigs and patients with diarrhea [13,18]. Moreover, a high prevalence of monophasic S. Typhimurium in pig farms was detected in northern and central Vietnam. Recent studies have shown that the highest similarity is observed between human and pig serovars [19]. Our phylogenetic analysis showed that monophasic S. Typhimurium isolates from humans and pigs clustered together into the same clades, which implies the transmission of this pathogen. S. London was typical for both pig and pork samples, while S. Derby and S. Give were only typical for pig samples. Taking into account that pork is the most consumed meat in Vietnam and has the highest (91.3%) NTS prevalence among farm livestock products, the pig production sector poses one of the main reservoirs for human infection [13,20]. All chicken samples in our study belonged to the S. Infantis serotype, while S. Indiana was mostly found in broiler meat samples. It has been shown that S. Indiana is a relevant serotype associated with chicken meat in Vietnam and exhibited resistance to four to six classes of antimicrobials [21]. However, according to the study by Ta et al. (2014), S. Indiana was the fifth most frequently identified serotype in retail raw poultry meat in Vietnam after Albany, Agona, Dabou, and Hadar [22]. The variety of Salmonella serotypes may reflect the geographical, collection time, and identification methods diversity.
In the current study, we deliberately included NTS isolates that were resistant to more than three antibiotic classes and assigned as MDR. The spread of MDR bacteria in humans, the livestock sector, veterinary medicine, and ready-to-eat products is an alarming public health concern and affects not only humans but also animals, causing emergency situations worldwide. Overall, data from all regions of Vietnam show an increase in AMR and MDR over time. A study from 2000–2020 reported an increasing trend in the pooled AMR prevalence in NTS for quinolones (15.6%), 3rd-, 4th-, and 5th-generation cephalosporins (23.7%), tetracyclines (12.9%), amphenicol (17.8%), and multidrug resistance (11.4%) [1].
Our results demonstrated similarity in phenotypic resistance profiles between different serotypes, except that S. Infantis showed the highest resistance to third- and fourth-generation cephalosporins compared to others. Despite the MDR status, all NTS isolates were susceptible to meropenem—a beta-lactam antibiotic of the carbapenem class—which has been proven efficacious as monotherapy in the treatment of a broad spectrum of infections as well as those caused by organisms resistant to other antimicrobials. On the contrary, the highest resistance was observed against fluoroquinolones—CIP and NAL—which are among the highest priority critically important antimicrobials (HPCIAs) and the drugs of choice for salmonellosis treatment. Notably, among all NTS isolates, the highest MIC values showed chicken isolates S. Indiana ST17, which was recognized as an epidemiologically important type due to its high resistance level to most widely used classes of antimicrobials [23]. Moreover, the majority of pig isolates have demonstrated significantly higher MICs to another class of HPCIAs—polymyxin, represented by CT in our study. This finding is in line with other studies from Northern Vietnam: an NTS survey (n = 138) in pigs (2011–2012) revealed phenotypic resistance to CT in only four S. Typhimurium isolates (2.9%), while subsequent surveillance data (2013–2019) showed a CT resistance rate of 9–26%, predominantly in isolates from pigs, retail chickens, and pork [1,13]. The resistance rate for Salmonella isolates from humans as well as those from animals and pork/broiler raw meat was highest for tetracycline, beta-lactams, phenicol, and quinolones. This important observation is consistent with the previous study by Nhung et al. (2017), which found that sulfonamides, tetracyclines, and macrolides were the most frequently detected residues in pig and poultry production in Vietnam [24]. This finding indicates not only poor sanitation in the small-scale livestock farms but also limited surveillance under inadequate usage of antibiotic agents in veterinary or medicated feed, boosting the risk of selecting multi-resistant strains and transferring them from animals to humans, which, after all, leads to a negative impact on the population’s health in general. However, so far Vietnam does not have a nationwide surveillance system to monitor NTS and resistance level to AMR in human and livestock populations [1].
It is well known that the presence of AMR genes is responsible for Salmonella resistance development. In our study, all NTS-resistant and NWT had at least one resistance gene in their genome. Moreover, the number of resistance genes was associated with resistance level: the higher the MIC value, the broader the genetic profile detected in Salmonella isolates. This observation suggests that high antibiotic concentrations promote selective pressure on cooperative genes, implying that multiple genes or sequential mutations in AMR genes are required for antibiotic resistance to develop. The highest correlation between phenotypic and genotypic resistance was found for azithromycin, chloramphenicol, and ampicillin (100%) and above 90 and 95% for tetracycline and colistin, respectively. However, it is worth noting that for four antimicrobial classes (quinolones, aminoglycosides, sulfonamides, and tetracycline), AMR gene content did not match the phenotype: the presence of one to four resistance genes was common for both susceptible/WT and resistant/NWT isolates. This discrepancy may indicate either a lack of expression in the absence of antibiotic exposure conditions or a low level of resistance genes for high-level clinical resistance development, since Salmonella resistance is polygenic [23].
The blaTEM-1B resistance gene has predominated among β-lactam-resistant isolates, while blaCTX-M-55 and blaCTX-M-65 genes (or co-harboring blaCTX-M-55/blaCTX-M-65 with blaTEM-1B) were associated with resistance to cephalosporins. There are many β-lactamase variants disseminated in Salmonella isolates globally; however, among Vietnamese isolates, we revealed quite a narrow spectrum of BL genes, and the majority of resistant isolates (75.6%) carried only one gene, which is in line with a prior study from Vietnam [25]. Moreover, blaCTX-M-producing isolates were mostly identified from broiler meat samples and showed the highest resistance to cephalosporins. Previous studies have demonstrated the high prevalence of Salmonella in broiler meat in Vietnam, ranging from 69 to 77%, depending on the region [26]. Since blaCTX-Ms are often cross-resistant to other various antibiotics and mobilize to all major plasmids approximately 10 times more frequently than other class A β-lactamases, blaCTX-Ms-harboring Salmonella in broiler meat represent a troubling and intractable issue for human and food safety [27].
Fluoroquinolone resistance in NTS isolates from Vietnam was attributed not only to point mutations in the quinolone resistance-determining regions (QRDR), involving chromosomal gyr and par topoisomerase genes, but also to plasmid-mediated quinolone resistance (PMQR) genes qnr and aac(6′)-lb-cr, encoding pentapeptide topoisomerase-binding proteins and quinolone-modifying enzymes, respectively. The low-level resistance to CIP was mediated by the presence of qnrS1 and aac(6′)-lb-cr PMQR genes, whilst double mutations S83F/D87N in gyrA and T57S/S80I in parC QRDR genes were related to high resistance levels to both NAL and CIP. Our results are in agreement with other studies suggesting that high-level quinolone resistance results from the gradual accumulation of QRDR mutations [28,29,30,31]. Compared with the highest cephalosporin resistance in broiler meat isolates, the highest resistance to quinolones was demonstrated by S. Indiana isolates from chicken, which had the most diverse genetic resistance profile and harbored both QRDR and PMQR genes.
Salmonella isolates resistant to aminoglycosides exhibited the most complex and diverse genetic profile compared to those resistant to other classes of antimicrobials. Four families of aminoglycoside-modifying enzyme (AME) genes were identified in this study: aac, aph, aad, and rmt, among which aminoglycoside acetyltransferase aac(6′)-Iaa was predominant and possessed by all 49 NTS isolates regardless of phenotypic resistance-susceptibility status. Notably, nucleotidyltransferase ant genes associated with enzymatic inactivation of aminoglycosides were not detected in our sample. Among the entire spectrum of AME genes, only aac3IId, aac(3)-IV, aph(4)-Ia, and aac(6′)-lb-cr genes were associated with resistance to GEN and TOB. Since the aac(6′)-Ib-cr gene is known to be involved in cross-resistance to aminoglycosides and fluoroquinolones, five chicken isolates harboring this gene were resistant to CIP/NAL and GEN/TOB antibiotics. Moreover, among 49 NTS isolates, only one chicken isolate was resistant to all aminoglycosides (GEN, TOB, and AMI) and was characterized by the presence of the 16S rRNA methylase rmtB gene in its genetic profile, which has been shown to have a low prevalence in Salmonella and confers resistance to multiple aminoglycosides [32,33].
The recent emergence of colistin resistance and the rapid transmission of the mobile colistin resistance gene (mcr) among different bacterial species worldwide raise concerns about the uncontrolled transmission of colistin-resistant bacteria across all ecological niches [34]. Our study demonstrated that all isolates from chicken and broiler meat were susceptible to colistin, whilst NTS from pig isolates showed the highest resistance (66.7%) compared to human (27.3%) and pork meat (25%) isolates. Among all mcr gene variants (mcr-1 to mcr-10), only mcr-1 and mcr-3 variants were detected in CT-resistant isolates. Similar results were obtained in another study from Vietnam that showed the widespread use of colistin or colistin-containing feeds in livestock breeding: more than 70% of pig and chicken fecal samples contained CT-resistant E. coli with mcr-1 or mcr-3 genes [1]. Interestingly, one human and three pig isolates showed resistance to CT; however, without association with the plasmid-borne mcr genes. This suggests the presence of other underlying resistance mechanisms for Salmonella to develop colistin resistance, such as efflux pumps, chromosomal mutations, and/or alterations in the O-antigen polymerase gene [35]. Thus, increased attention must be directed towards monitoring CT-resistant bacteria and excessive antimicrobial utilization in the agricultural sector.
In this study, high rates of NWT-phenotype to chloramphenicol and tetracycline (~90 and 92%) were observed for NTS serovars from different sources. The genetic profile for CHL-NWT isolates was related to chloramphenicol acetyltransferases cat genes types A/B, as well as chloramphenicol exporters cmlA and floR efflux pump genes. Despite the general observation that the presence of more than one phenicol resistance gene in the same Salmonella isolate is rare, in our study, 48% of the Vietnamese CHL-resistant isolates contained two and three phenicol resistance genes each [36]. So far, among the wide variety of tetracycline resistance genes, only five—tetA, tetB, tetC, tetD, and tetG—have been reported for Salmonella [32]. Intriguingly, we found another ribosomal protection gene, tetM, co-carried with the tetA gene in 20% of TET-NWT isolates, predominantly from pigs. It is well known that the tetM gene is usually carried by Enterococcus sp., is atypical for Gram-negative bacteria, and was first described in E. coli strains from chickens and pigs [36,37]. Subsequently, Jurado-Rabadán S. et al. (2014) identified the tetM gene in doxycycline-resistant E. coli isolates from pigs [38]. Taken together with our results, these data suggest that the presence of the tetM gene in porcine Salmonella isolates may be the result of possible horizontal transfer of this gene from intestinal enterococci or E. coli [37,38].
Vietnamese NTS isolates exhibited a relatively high prevalence of resistance (65%) to trimethoprim/sulfamethoxazole, with the highest rates observed in isolates from pigs and pork meat. However, we found that sulfonamide resistance sul1/sul2/sul3 genes by themselves were present only in TMP/SMX-susceptible isolates, while the combination of sul genes with transferable dihydrofolate reductase dfrA was typical exclusively for TMP/SMX-resistant isolates. Thus, according to our findings, the presence of dfrA genes is responsible for the development of TMP/SMX resistance. On the other hand, this is contrary to previous studies, which have suggested that TMP/SMX resistance in Salmonella is typically linked to the presence of either sul1 or sul2 genes [39]. Notably, for phenotypic testing we used a synergistic TMP and SMX combination, which does not allow us to determine whether the dfrA gene alone is sufficient to confer resistance to TMP/SMX.
One of the most effective antimicrobials in our study was azithromycin, to which Salmonella isolates showed a low NWT-phenotype rate (25%) with a strong association of phenotypic resistance with the macrolide 2′-phosphotransferase mph(A) and efflux pump mef(B) genes. Given that macrolides remain one of the mainstays of treatment for MDR in countries with a high burden of fluoroquinolone resistance, the spread of azithromycin resistance among circulating strains could have a major impact on the dwindling options for treatment strategies.
The present study has several limitations. First, the sample size is rather small and may not reflect the overall resistance patterns in Vietnam. Second, serotyping analysis classified the 49 NTS isolates into 14 different serotypes, resulting in an imbalance of Salmonella serotypes and source categories that tend to be statistically fragile. Third, due to the limitations of short-read sequencing technology, we were unable to precisely determine the location of resistance genes on the chromosome or plasmids. However, despite these limitations, our data provided relevant information on the genetic patterns of multidrug-resistant Salmonella isolates from different sources and highlighted the need for a controlled approach to antibiotic overuse in animal husbandry.
4. Materials and Methods
4.1. Study Design, Sampling Collection, and Bacterial Isolation
This study was performed within a framework of scientific cooperation between the St. Petersburg Pasteur Institute, Russia, and the Pasteur Institute in Ho Chi Minh City, Vietnam. A total of 49 isolates from pigs (n = 18) and chickens (n = 5) from family-run livestock farms, pork (n = 4) and broiler (n = 11) meat from retail outlets, and humans (n = 11) with acute diarrhea were collected during 2012–2021 from Ho Chi Minh City (n = 20), Dong Nai (n = 24), Long An (n = 1), Ben Tre (n = 1), and An Giang (n = 3) provinces, Vietnam.
Salmonella strains were isolated according to the International Standard ISO 6579-1:2017, using buffered peptone water (pre-enrichment), tetrathionate broth, and Rappaport Vassiliadis soy peptone broth (selective enrichment), followed by subcultivation onto XLD agar plates and Hektoen Enteric agar plates (Condalab, Madrid, Spain) [40]. The isolated bacteria were identified to a species level with MALDI-TOF MS (Autof MS1000, Zhengzhou, China). The isolated strains were stored in cryotubes with beads for microorganisms (DELTALAB, S.L., Barcelona, Spain) at −70 °C. Salmonella serotypes and appropriate sequence types (STs) were determined by whole-genome sequencing data.
4.2. Phenotypic Antimicrobial Susceptibility Testing (AST)
Phenotypic antimicrobial susceptibility testing (AST) was performed according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommendations by the serial broth microdilution testing using “MPC-MICRO PS” strips (St. Petersburg Pasteur Institute, Russia) with Muller Hinton broth (Condalab, Madrid, Spain) and gradient diffusion method using Muller Hinton agar (Condalab, Madrid, Spain) with the following antibiotics: β-lactams (ampicillin (AMP)); cephalosporins (ceftazidime (CTZ), cefotaxime (CTX), cefepime (FEP)); carbapenem (meropenem (MEM)); quinolones (nalidixic acid (NAL), ciprofloxacin (CIP)); tetracycline (TET); phenicol (chloramphenicol (CHL)); trimethoprim/sulfamethoxazole (TMP/SMX); polymyxins (colistin (CT)); aminoglycosides (gentamycin (GEN), tobramycin (TOB), amikacin (AMI)); and macrolides (azithromycin (AZM)). The results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical breakpoint interpretation (EUCAST, 2024). In some cases where clinical breakpoints were not available, epidemiological cut-off values (ECOFF) were used to distinguish wild-type (WT) isolates from isolates with acquired resistance (non-wild-type, NWT): NAL (8 mg/mL), TET (8 mg/mL), CHL (16 mg/mL), AZM (16 mg/mL). Salmonella isolates showed resistance to three or more different classes of antimicrobials and were defined as multidrug resistant (MDR).
4.3. DNA Extraction and Sequencing
Genomic DNA of all 49 Salmonella isolates was isolated by the QIAamp DNA Mini Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s guidelines. The DNA concentration was quantified on a Qubit 4.0 fluorometer. Whole-genome shotgun DNA libraries were prepared using the MGIEasy FS DNA Library Prep Set and then sequenced on a DNBSEQ-G50 sequencer (MGI Tech Co. Ltd., Beijing, China) in a 2 × 100 paired-end (PE) mode.
4.4. Bioinformatics Analysis
The raw paired-end reads were analyzed using FastQC software (v.0.12.1; Babraham Institute, Cambridge, UK), then trimmed and filtered by Trim Galore! (version 0.6.7). Bacterial genomes were assembled de novo by SPAdes assembler software (version 3.13.1), and the results were evaluated by QUAST (version 5.2.0). The assembled genome sequences for all isolates were deposited to the NCBI GenBank under the project number PRJNA933104.
Core-genome SNPs were called using Snippy pipeline v.4.6.0 (https://github.com/tseemann/snippy, accessed on 16 June 2022). Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 was used as a reference genome (GenBank accession number AE006468.2). Mutation clustering analysis, maximum likelihood tree, and cladograms were constructed using the RAxML-NG inference tool based on the core genome SNPs obtained using the Snippy core. To analyze the phylogeographic distribution of Salmonella isolates from Vietnam, 34,291 assembled Salmonella genomes were downloaded from the NCBI Genome database (Nucleic Acids Research; accessed 1 January 2013; http://www.ncbi.nlm.nih.gov/sites/genome) as available in February 2024. By metadata filtering, 575 Salmonella Infantis and Typhimurium genomes were randomly selected for further investigation and added to the phylogenetic analysis. The reliability of the phylogenetic tree branches was evaluated by the bootstrapping method (1000 replications). Phylogenetic trees and cladograms were visualized by Evolview v3 (https://www.evolgenius.info/evolview/, accessed on 22 May 2019).
Additionally, the ResFinder 4.0 database with default parameters was used for antimicrobial resistance gene (ARG) detection. The SeqSero 1.2 service was used for Salmonella serotype prediction based on WGS information (Center for Genomic Epidemiology, https://www.genomicepidemiology.org/, accessed on 11 August 2020). For plasmid detection, PlasmidFinder 2.1 was used (https://github.com/genomicepidemiology/plasmidfinder, accessed on 31 January 2021).
4.5. Statistical Analysis
All statistics were performed in the R programming language, environment version 4.3.1. A p-value < 0.05 was defined as statistical significance. The data were tested by the chi-square (χ2) test and Fisher’s exact test.
5. Conclusions
Using a WGS-based approach, we conducted a comprehensive investigation of non-typhoidal Salmonella serovars from different sources in Southern Vietnam with a particular focus on multidrug resistance patterns. This study showed that both livestock animals and meat products are a major source of Salmonella with high levels of resistance to multiple antimicrobials and an expanded spectrum of resistance genes and plasmids. Despite the limited number of samples, we were able to determine that NTS strains isolated from poultry and pigs exhibit resistance to the highest priority antimicrobials—quinolones and β-lactams—which are essential for severe salmonellosis treatment. Additionally, these strains showed resistance to aminoglycosides, tetracyclines, and sulfonamides. The obtained results highlight the utmost importance of issues related to the selection, spreading, and transmission of multi-resistant strains along the entire meat production chain from animals to humans. As the level of multidrug-resistant NTS in Vietnam is steadily increasing, this should serve as an incentive to develop strict preventive control strategies and food safety regulations, along with more stringent and enhanced surveillance under sanitation practices and antibiotic overuse in the animal husbandry sector. Thus, ongoing monitoring of antimicrobial resistance patterns in different Salmonella serovars among humans, animals, and meat products is a prerequisite for efficient control of genome dynamics, NTS diversity, prevalence, and relationships between different hosts.
Author Contributions
Software, formal analysis, visualization, writing—original draft preparation, D.S.; conceptualization, validation, data curation, writing—review and editing, S.E.; formal analysis, investigation—L.S.; project administration, supervision—L.K.; project administration—M.M.; investigation—S.Z.; software, formal analysis—D.P.; software, formal analysis—A.S.; data curation, resources, formal analysis—T.Q.N.; project administration, data curation, resources—V.H.N.; resources—T.K.V.; investigation—L.T.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Ethics Committee in Biomedical Research–Pasteur Institute in Ho Chi Minh City (certificate 12/CN_HDDD, 11 September 2020) and the Independent Ethics Committee of St. Petersburg Pasteur Institute (No. 73, 30 November 2021).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All data of this study are presented in the article. The assembled genome sequences for all isolates were uploaded to the NCBI Sequence Read Archive under the project number PRJNA933104 at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA933104 (accessed on 1 March 2024).
Acknowledgments
The authors thank Nikita Gladyshev for providing expert support with phylogenetic tree construction, contributions to data analysis, and for his overall valuable assistance during this work.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; and in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| NTS | Non-typhoidal Salmonella |
| WGS | Whole-genome sequencing |
| MDR | Multidrug resistance |
| ARG | Antimicrobial resistance genes |
| AST | Antimicrobial susceptibility testing |
| WT | Wild-type |
| NWT | Non-wild-type |
| BLs | Beta-lactamases |
| AMP | Ampicillin |
| CTZ | Ceftazidime |
| CTX | Cefotaxime |
| FEP | Cefepime |
| MEM | Meropenem |
| NAL | Nalidixic acid |
| CIP | Ciprofloxacin |
| TET | Tetracycline |
| CHL | Chloramphenicol |
| TMP/SMX | Trimethoprim/Sulfamethoxazole |
| CT | Colistin |
| GEN | Gentamycin |
| TOB | Tobramycin |
| AMI | Amikacin |
| AZM | Azithromycin |
References
- Nhung, N.T.; Phu, D.H.; Carrique-Mas, J.J.; Padungtod, P. A review and meta-analysis of non-typhoidal Salmonella in Vietnam: Challenges to the control and antimicrobial resistance traits of a neglected zoonotic pathogen. One Health 2024, 18, 100698. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kang, H.; Excler, J.-L.; Kim, J.H.; Lee, J.-S. The Economic Burden of Non-Typhoidal Salmonella and Invasive Non-Typhoidal Salmonella Infection: A Systematic Literature Review. Vaccines 2024, 12, 758. [Google Scholar] [CrossRef] [PubMed]
- Billah, M.M.; Rahman, M.S. Salmonella in the environment: A review on ecology, antimicrobial resistance, seafood contaminations, and human health implications. J. Hazard. Mater. Adv. 2024, 13, 100407. [Google Scholar] [CrossRef]
- Caneschi, A.; Bardhi, A.; Barbarossa, A.; Zaghini, A. The Use of Antibiotics and Antimicrobial Resistance in Veterinary Medicine, a Complex Phenomenon: A Narrative Review. Antibiotics 2023, 12, 487. [Google Scholar] [CrossRef] [PubMed]
- Acosta, A.; Tirkaso, W.; Nicolli, F.; Van Boeckel, T.P.; Cinardi, G.; Song, J. The future of antibiotic use in livestock. NaNat. Commun. 2025, 16, 2469. [Google Scholar] [CrossRef]
- Founou, L.L.; Founou, R.C.; Essack, S.Y. Antibiotic Resistance in the Food Chain: A Developing Country-Perspective. Front. Microbiol. 2016, 7, 1881. [Google Scholar] [CrossRef]
- Sima, C.M.; Buzilă, E.R.; Trofin, F.; Păduraru, D.; Luncă, C.; Duhaniuc, A.; Dorneanu, O.S.; Nastase, E.V. Emerging Strategies against Non-Typhoidal Salmonella: From Pathogenesis to Treatment. Curr. Issues Mol. Biol. 2024, 46, 7447–7472. [Google Scholar] [CrossRef]
- Kumar, G.; Kumar, S.; Jangid, H.; Dutta, J.; Shidiki, A. The rise of non-typhoidal Salmonella: An emerging global public health concern. Front. Microbiol. 2025, 16, 1524287. [Google Scholar] [CrossRef]
- Hengkrawit, K.; Tangjade, C. Prevalence and Trends in Antimicrobial Susceptibility Patterns of Multi-Drug-Resistance Non-Typhoidal Salmonella in Central Thailand, 2012–2019. Infect. Drug Resist. 2022, 15, 1305–1315. [Google Scholar] [CrossRef]
- Sahu, A.A.; Sephalika, S.; Mohakud, N.K.; Sahu, B.R. Prevalence and Multidrug Resistance in Non-Typhoidal Salmonella in India: A 20-Year Outlook. Acta Microbiol. Hell. 2025, 70, 6. [Google Scholar] [CrossRef]
- Tuat, C.V.; Hue, P.T.; Loan, N.T.P.; Thuy, N.T.; Hue, L.T.; Giang, V.N.; Erickson, V.I.; Padungtod, P. Antimicrobial Resistance Pilot Surveillance of Pigs and Chickens in Vietnam, 2017–2019. Front. Vet. Sci. 2021, 8, 618497. [Google Scholar] [CrossRef] [PubMed]
- Bloomfield, S.; Duong, V.T.; Tuyen, H.T.; Campbell, J.I.; Thomson, N.R.; Parkhill, J.; Le Phuc, H.; Chau, T.T.H.; Maskell, D.J.; Perron, G.G.; et al. Mobility of antimicrobial resistance across serovars and disease presentations in non-typhoidal Salmonella from animals and humans in Vietnam. Microb. Genom. 2022, 8, mgen000798. [Google Scholar] [CrossRef]
- Holohan, N.; Wallat, M.; Luu, T.H.Y.; Clark, E.; Truong, D.T.Q.; Xuan, S.D.; Vu, H.T.K.; Van Truong, D.; Huy, H.T.; Nguyen-Viet, H.; et al. Analysis of Antimicrobial Resistance in Non-typhoidal Salmonella Collected From Pork Retail Outlets and Slaughterhouses in Vietnam Using Whole Genome Sequencing. Front. Vet. Sci. 2022, 9, 816279. [Google Scholar] [CrossRef] [PubMed]
- Coyne, L.; Arief, R.; Benigno, C.; Giang, V.N.; Huong, L.Q.; Jeamsripong, S.; Kalpravidh, W.; McGrane, J.; Padungtod, P.; Patrick, I.; et al. Characterizing Antimicrobial Use in the Livestock Sector in Three South East Asian Countries (Indonesia, Thailand, and Vietnam). Antibiotics 2019, 8, 33. [Google Scholar] [CrossRef]
- Redondo-Salvo, S.; Fernández-López, R.; Ruiz, R.; Vielva, L.; DE Toro, M.; Rocha, E.P.C.; Garcillán-Barcia, M.P.; De La Cruz, F. Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat. Commun. 2020, 11, 3602. [Google Scholar] [CrossRef]
- Commichaux, S.; Rand, H.; Javkar, K.; Molloy, E.K.; Pettengill, J.B.; Pightling, A.; Hoffmann, M.; Pop, M.; Jayeola, V.; Foley, S.; et al. Assessment of plasmids for relating the 2020 Salmonella enterica serovar Newport onion outbreak to farms implicated by the outbreak investigation. BMC Genom. 2023, 24, 165. [Google Scholar] [CrossRef]
- Yan, S.; Jiang, Z.; Zhang, W.; Liu, Z.; Dong, X.; Li, D.; Liu, Z.; Li, C.; Liu, X.; Zhu, L. Genomes-based MLST, cgMLST, wgMLST and SNP analysis of Salmonella Typhimurium from animals and humans. Comp. Immunol. Microbiol. Infect. Dis. 2023, 96, 101973. [Google Scholar] [CrossRef] [PubMed]
- Mather, A.E.; Phuong, T.L.T.; Gao, Y.; Clare, S.; Mukhopadhyay, S.; Goulding, D.A.; Hoang, N.T.D.; Tuyen, H.T.; Lan, N.P.H.; Thompson, C.N.; et al. New Variant of Multidrug-Resistant Salmonella enterica Serovar Typhimurium Associated with Invasive Disease in Immunocompromised Patients in Vietnam. mBio 2018, 9, e01056-18. [Google Scholar] [CrossRef]
- Soliani, L.; Rugna, G.; Prosperi, A.; Chiapponi, C.; Luppi, A. Salmonella Infection in Pigs: Disease, Prevalence, and a Link between Swine and Human Health. Pathogens 2023, 12, 1267. [Google Scholar] [CrossRef]
- Tu, L.T.P.; Hoang, N.V.M.; Cuong, N.V.; Campbell, J.; Bryant, J.E.; Hoa, N.T.; Kiet, B.T.; Thompson, C.; Duy, D.T.; Phat, V.V.; et al. High levels of contamination and antimicrobial-resistant non-typhoidal Salmonella serovars on pig and poultry farms in the Mekong Delta of Vietnam. Epidemiol. Infect. 2015, 143, 3074–3086. [Google Scholar] [CrossRef]
- Nguyen, D.T.A.; Kanki, M.; Nguyen, P.D.; Le, H.T.; Ngo, P.T.; Tran, D.N.M.; Le, N.H.; Van Dang, C.; Kawai, T.; Kawahara, R.; et al. Prevalence, antibiotic resistance, and extended-spectrum and AmpC beta-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int. J. Food Microbiol. 2016, 236, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Ta, Y.T.; Nguyen, T.T.; To, P.B.; Pham, D.X.; Le, H.T.H.; Thi, G.N.; Alali, W.Q.; Walls, I.; Doyle, M.P. Quantification, serovars, and antibiotic resistance of salmonella isolated from retail raw chicken meat in Vietnam. J. Food Prot. 2014, 77, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Leinyuy, J.F.; Ali, I.M.; Ousenu, K.; Tume, C.B. Molecular characterization of antimicrobial resistance related genes in E. coli, Salmonella and Klebsiella isolates from broilers in the West Region of Cameroon. PLoS ONE 2023, 18, e0280150. [Google Scholar] [CrossRef] [PubMed]
- Nhung, N.T.; Van, N.T.B.; Van Cuong, N.; Duong, T.T.Q.; Nhat, T.T.; Hang, T.T.T.; Nhi, N.T.H.; Kiet, B.T.; Hien, V.B.; Ngoc, P.T.; et al. Antimicrobial residues and resistance against critically important antimicrobials in non-typhoidal Salmonella from meat sold at wet markets and supermarkets in Vietnam. Int. J. Food Microbiol. 2017, 266, 301–309. [Google Scholar] [CrossRef]
- Da Pham, X.; Hong, H.L.T.; Thanh, H.T.T.; Le, L.T.; Le, H.V.; Thi, N.H.; Le Tran, M.; Trung, N.T. Strains and virulence genes of salmonella with multidrug resistance isolated from chicken carcasses (Ha-noi, Vietnam). Health Risk Analysis 2023, 1, 115–123. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Le, H.V.; Hai, H.V.T.; Tuan, T.N.; Nguyen, H.M.; Xuan, D.P.; Thanh, H.T.T.; Le Thi, H.H. Whole-Genome Analysis of Antimicrobial-Resistant Salmonella enterica Isolated from Duck Carcasses in Hanoi, Vietnam. Curr. Issues Mol. Biol. 2023, 45, 2213–2229. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby, G.A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef]
- Song, Q.; Xu, Z.; Gao, H.; Zhang, D. Overview of the development of quinolone resistance in Salmonella species in China, 2005-2016. Infect. Drug Resist. 2018, 11, 267–274. [Google Scholar] [CrossRef]
- Pribul, B.R.; Festivo, M.L.; Rodrigues, M.S.; Costa, R.G.; Rodrigues, E.C.d.P.; de Souza, M.M.S.; Rodrigues, D.d.P. Characteristics of Quinolone Resistance in Salmonella spp. Isolates from the Food Chain in Brazil. Front. Microbiol. 2017, 8, 299. [Google Scholar] [CrossRef]
- Weng, R.; Gu, Y.; Zhang, W.; Hou, X.; Wang, H.; Tao, J.; Deng, M.; Zhou, M.; Zhao, Y. Corrigendum: Whole-genome sequencing provides insight into antimicrobial resistance and molecular characteristics of Salmonella from livestock meat and diarrhea patient in Hanzhong, China. Front. Microbiol. 2022, 13, 981414. [Google Scholar] [CrossRef]
- Kotb, D.N.; Mahdy, W.K.; Mahmoud, M.S.; Khairy, R.M.M. Impact of co-existence of PMQR genes and QRDR mutations on fluoroquinolones resistance in Enterobacteriaceae strains isolated from community and hospital acquired UTIs. BMC Infect. Dis. 2019, 19, 979. [Google Scholar] [CrossRef] [PubMed]
- Navickaite, I.; Holmes, H.; Dondi, L.; Randall, L.; Fearnley, C.; Taylor, E.; Fullick, E.; Horton, R.; Williamson, S.; AbuOun, M.; et al. Occurrence and characterization of rmtB-harbouring Salmonella and Escherichia coli isolates from a pig farm in the UK. J. Antimicrob. Chemother. 2024, 79, 1329–1336. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Z.-Y.; Wang, Y.; Sun, F.; Li, W.; Wu, H.; Shen, P.-C.; Pan, Z.-M.; Jiao, X. Emergence of 16S rRNA Methylase Gene rmtB in Salmonella Enterica Serovar London and Evolution of RmtB-Producing Plasmid Mediated by IS26. Front. Microbiol. 2021, 11, 604278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, J.; Yang, X.; Wu, Y.; Wang, Z.; Xu, Y.; Zhou, L.; Wang, J.; Jiao, X.; Sun, L. Emerging Mobile Colistin Resistance Gene Mcr-1 and Mcr-10 in Enterobacteriaceae Isolates From Urban Sewage in China. Infect. Drug Resist. 2025, 18, 1035–1048. [Google Scholar] [CrossRef]
- Fortini, D.; Owczarek, S.; Dionisi, A.M.; Lucarelli, C.; Arena, S.; Carattoli, A.; Enter-Net Italia Colistin Resistance Study Group; Villa, L.; García-Fernández, A. Colistin Resistance Mechanisms in Human Salmonella enterica Strains Isolated by the National Surveillance Enter-Net Italia (2016–2018). Antibiotics 2022, 11, 102. [Google Scholar] [CrossRef]
- Pavelquesi, S.L.S.; Ferreira, A.C.A.d.O.; Rodrigues, A.R.M.; Silva, C.M.d.S.; Orsi, D.C.; da Silva, I.C.R. Presence of Tetracycline and Sulfonamide Resistance Genes in Salmonella spp.: Literature Review. Antibiotics 2021, 10, 1314. [Google Scholar] [CrossRef] [PubMed]
- Bryan, A.; Shapir, N.; Sadowsky, M.J. Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 2004, 70, 2503–2507. [Google Scholar] [CrossRef]
- Jurado-Rabadán, S.; de la Fuente, R.; A Ruiz-Santa-Quiteria, J.; Orden, J.A.; de Vries, L.E.; Agersø, Y. Detection and linkage to mobile genetic elements of tetracycline resistance gene tet(M) in Escherichia coli isolates from pigs. BMC Vet. Res. 2014, 10, 155. [Google Scholar] [CrossRef]
- Chia-Wei, L.; Cheng, J.-F.; Tung, K.-C.; Hong, Y.-K.; Lin, J.-H.; Lin, Y.-H.; Tsai, C.-A.; Lin, S.-P.; Chen, Y.-C.; Shi, Z.-Y.; et al. Evolution of trimethoprim/sulfamethoxazole resistance in Shewanella algae from the perspective of comparative genomics and global phylogenic analysis. J. Microbiol. Immunol. Infect. 2022, 55, 1195–1202. [Google Scholar] [CrossRef]
- ISO 6579-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of salmonella spp. International Organization for Standardization: Geneva, Switzerland, 2017.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).