Outcome of Bloodstream Infections Caused by Antibiotic-Resistant Bacteria: A 7-Year Retrospective Study at the University Hospital of Palermo, Italy
Abstract
1. Introduction
2. Results
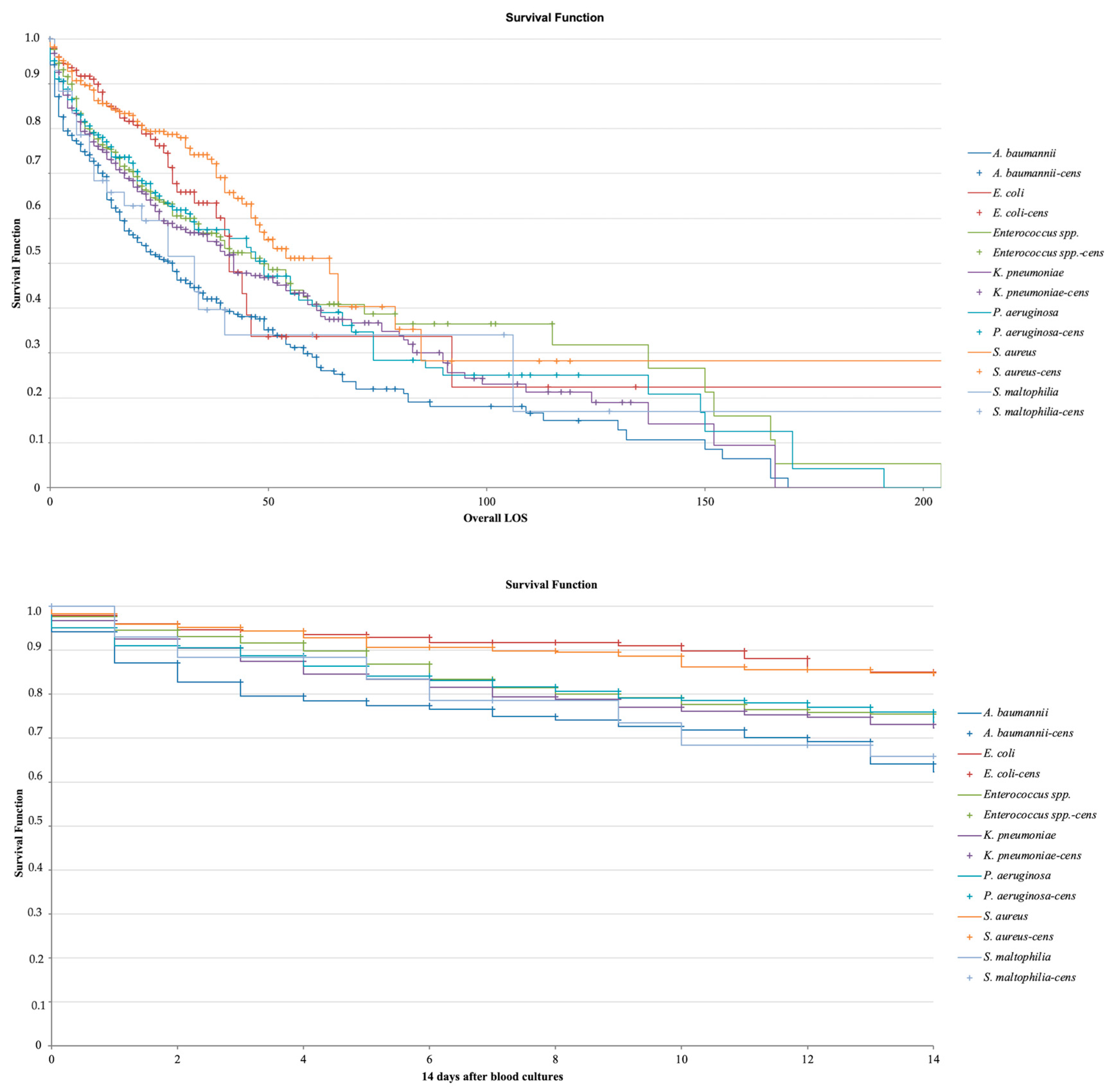
2.1. Study Population and Mortality
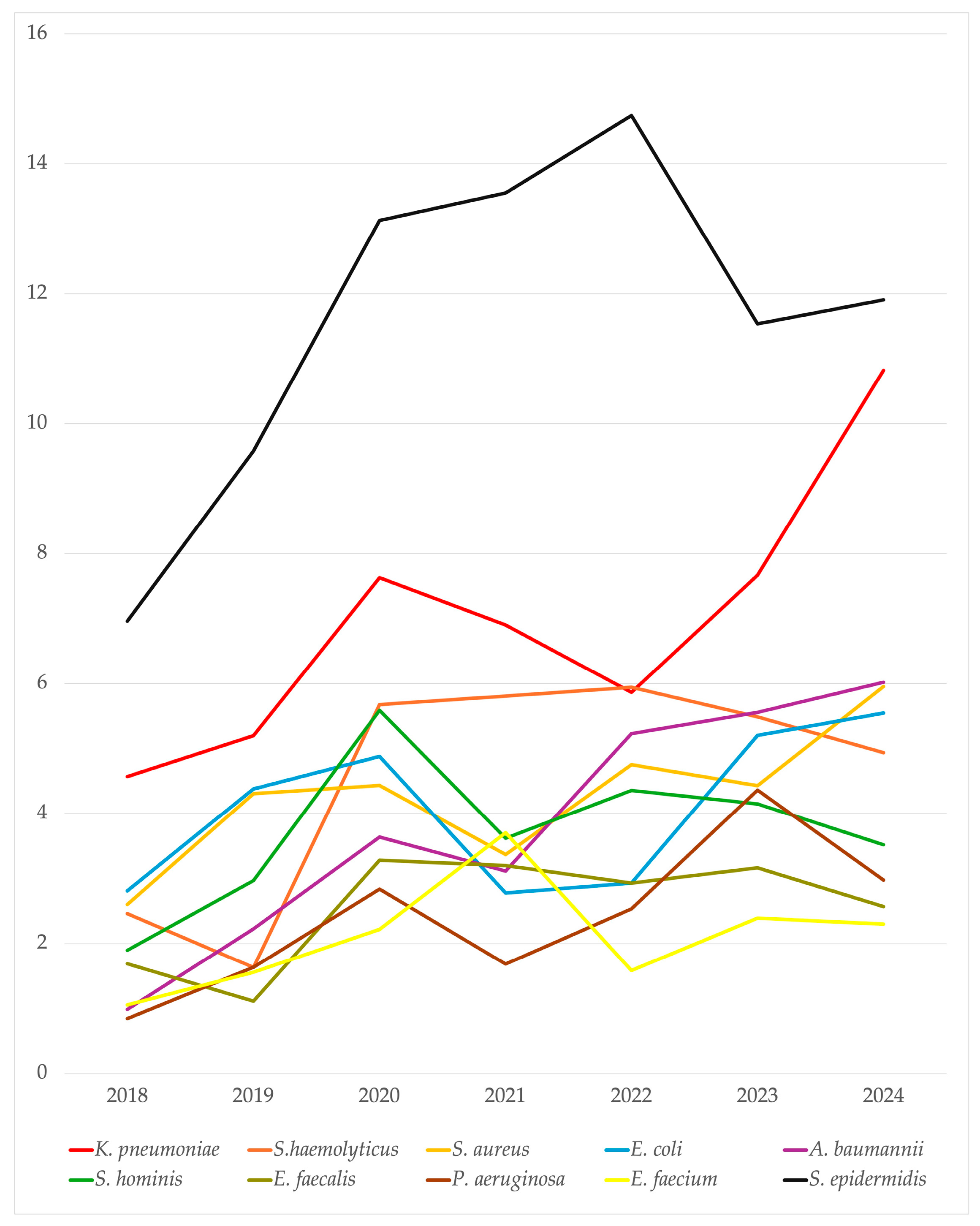
2.2. Microbiological Isolates
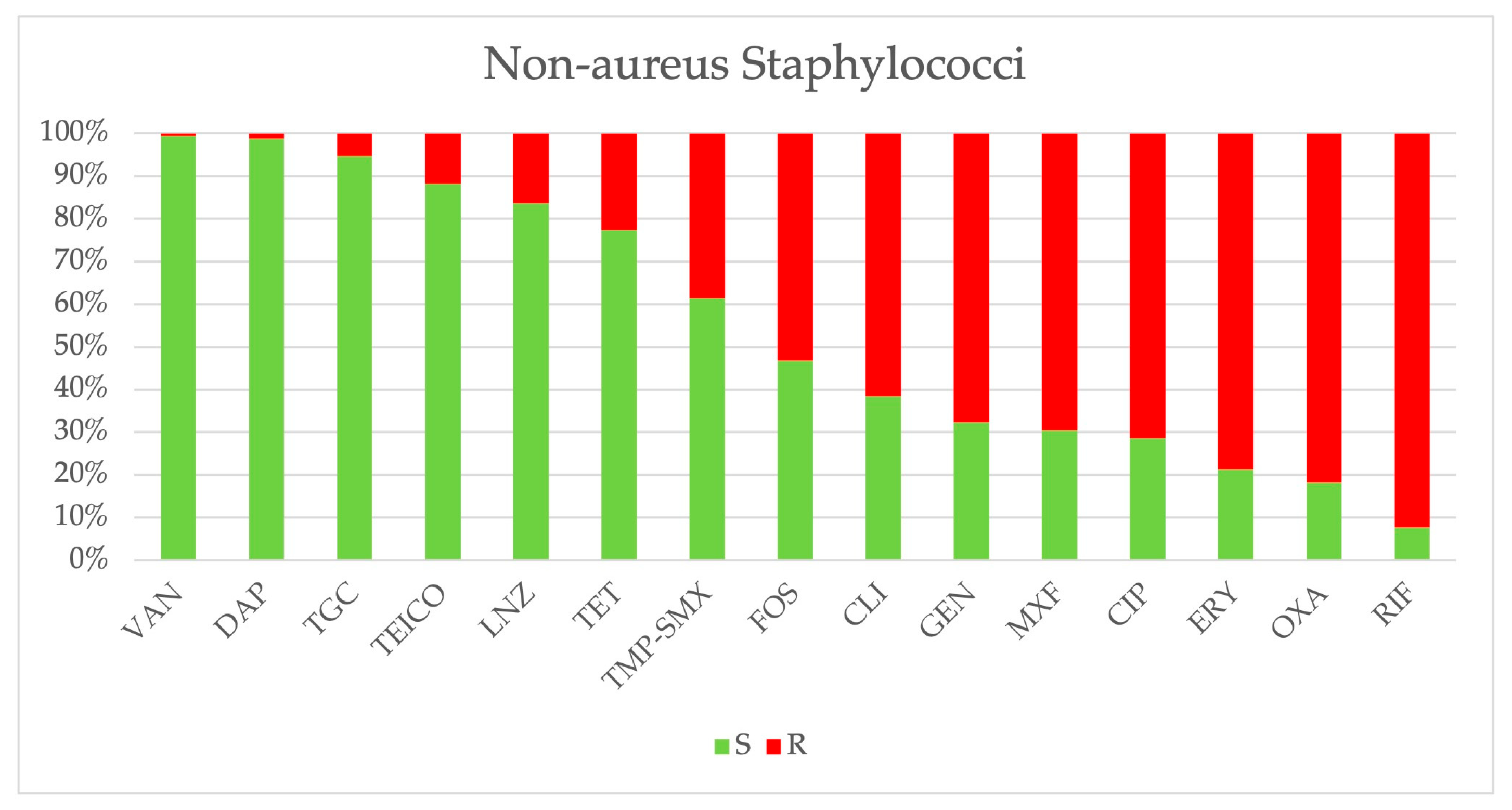
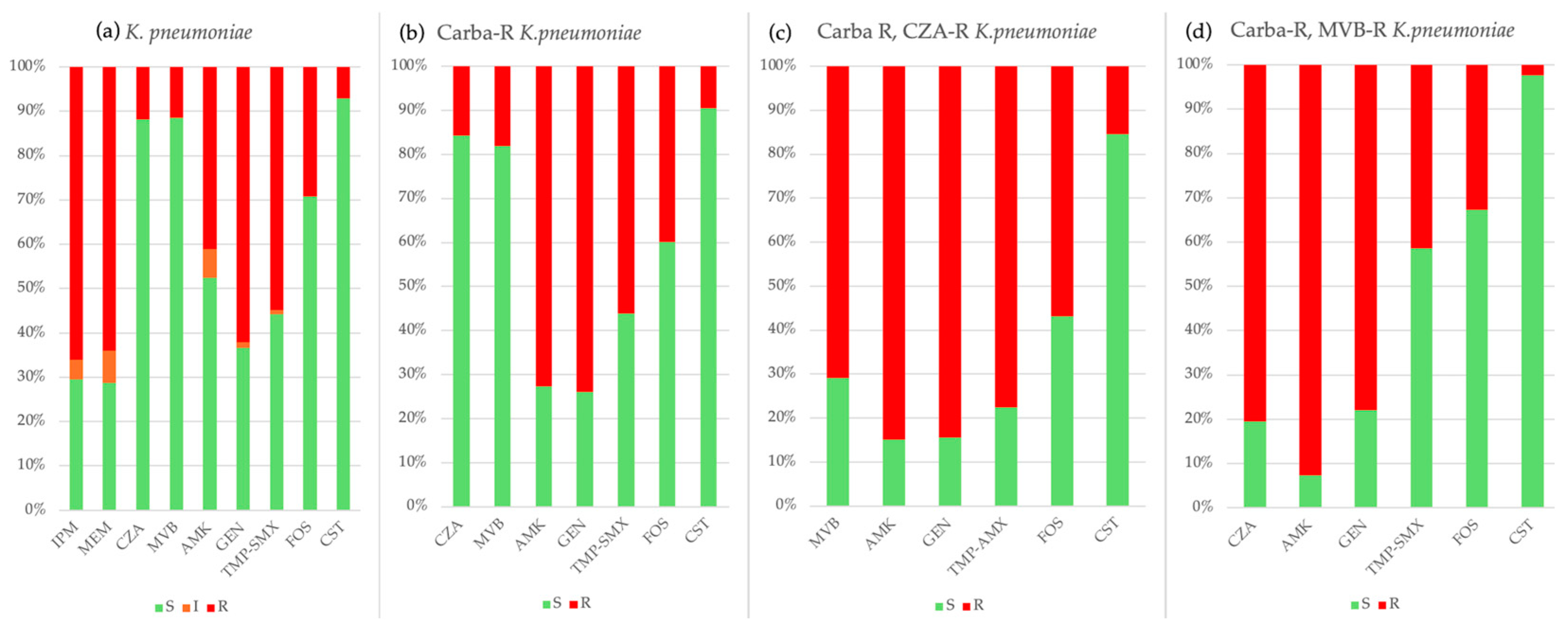
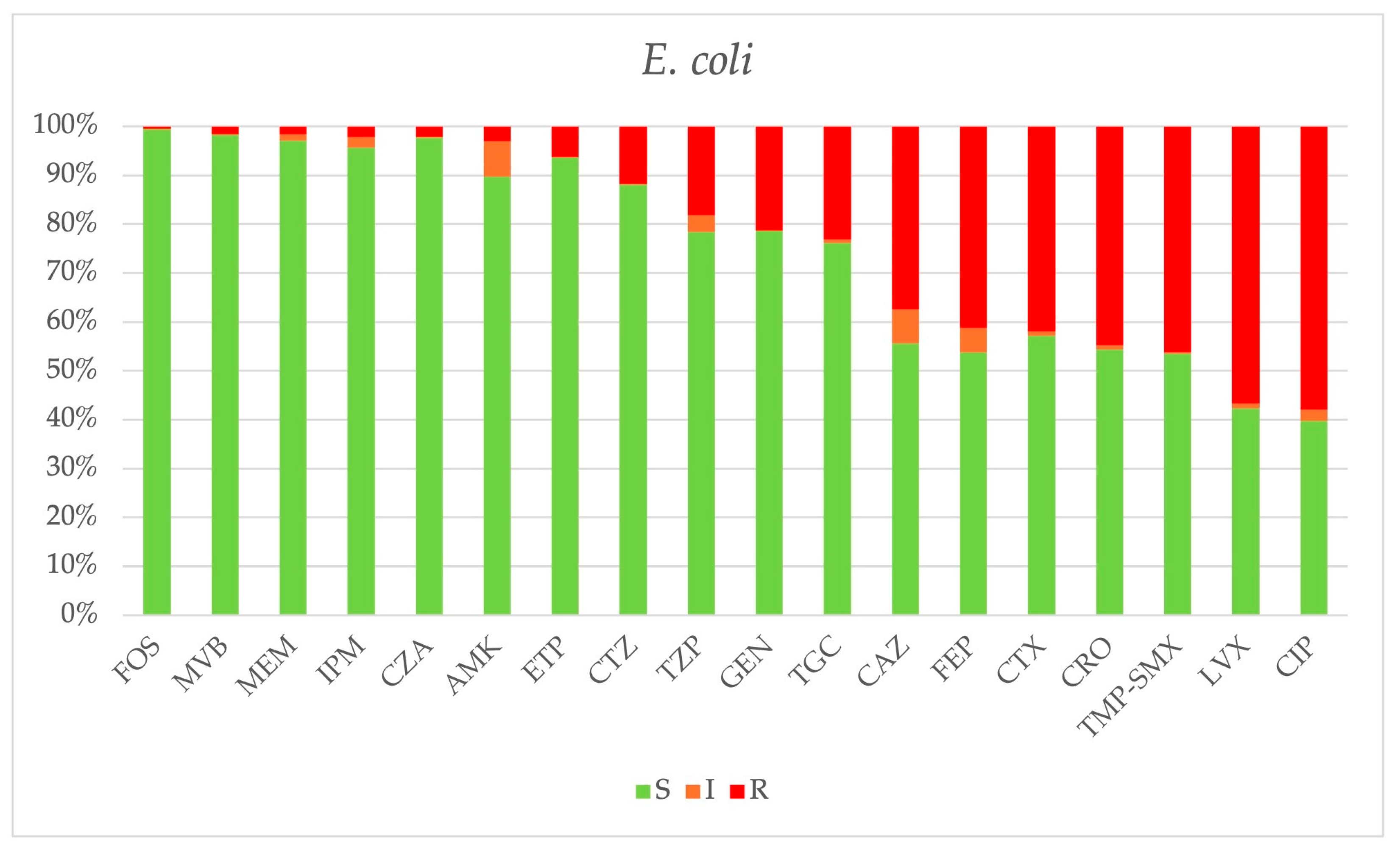
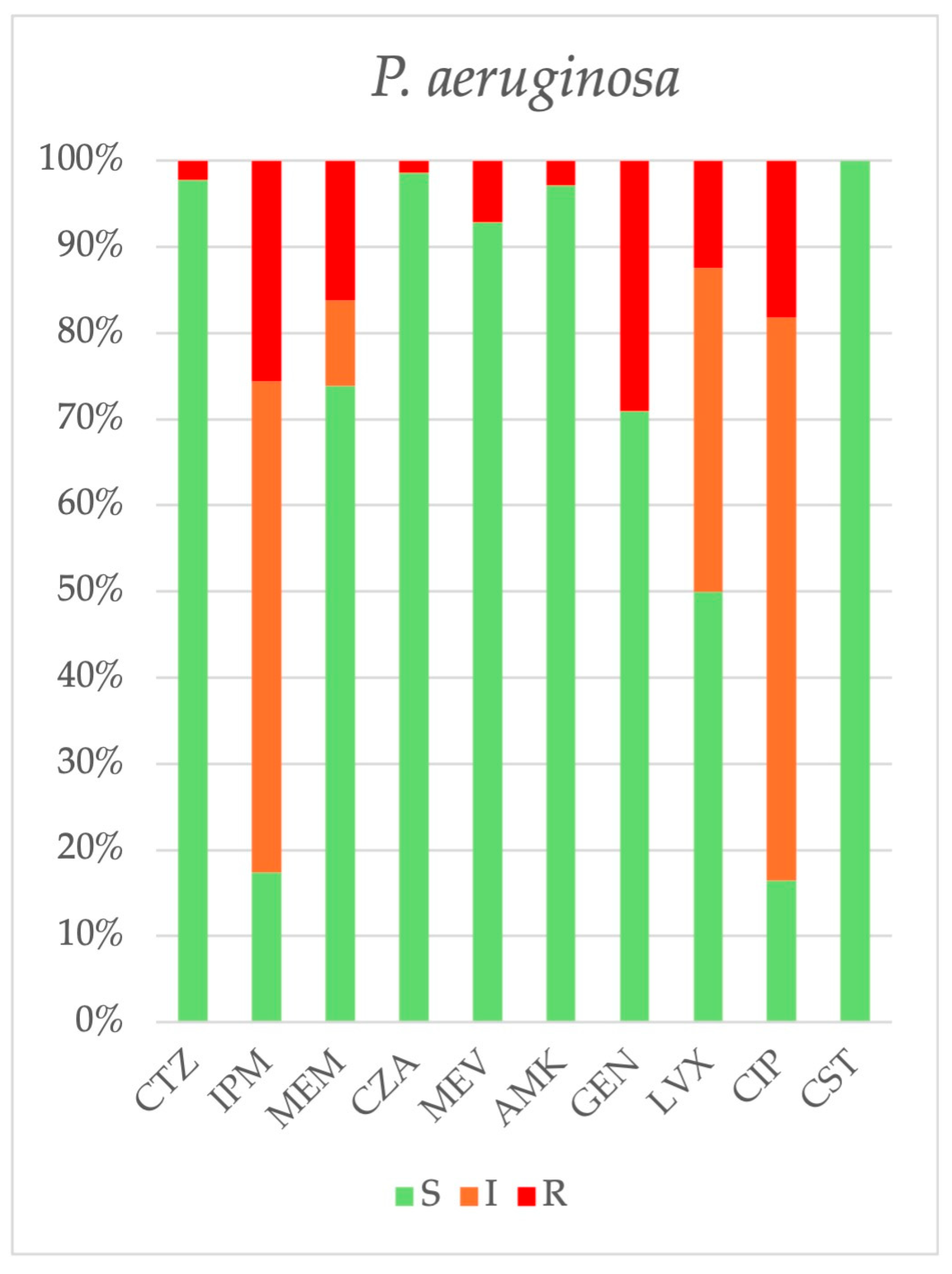
2.3. Antimicrobial Resistance
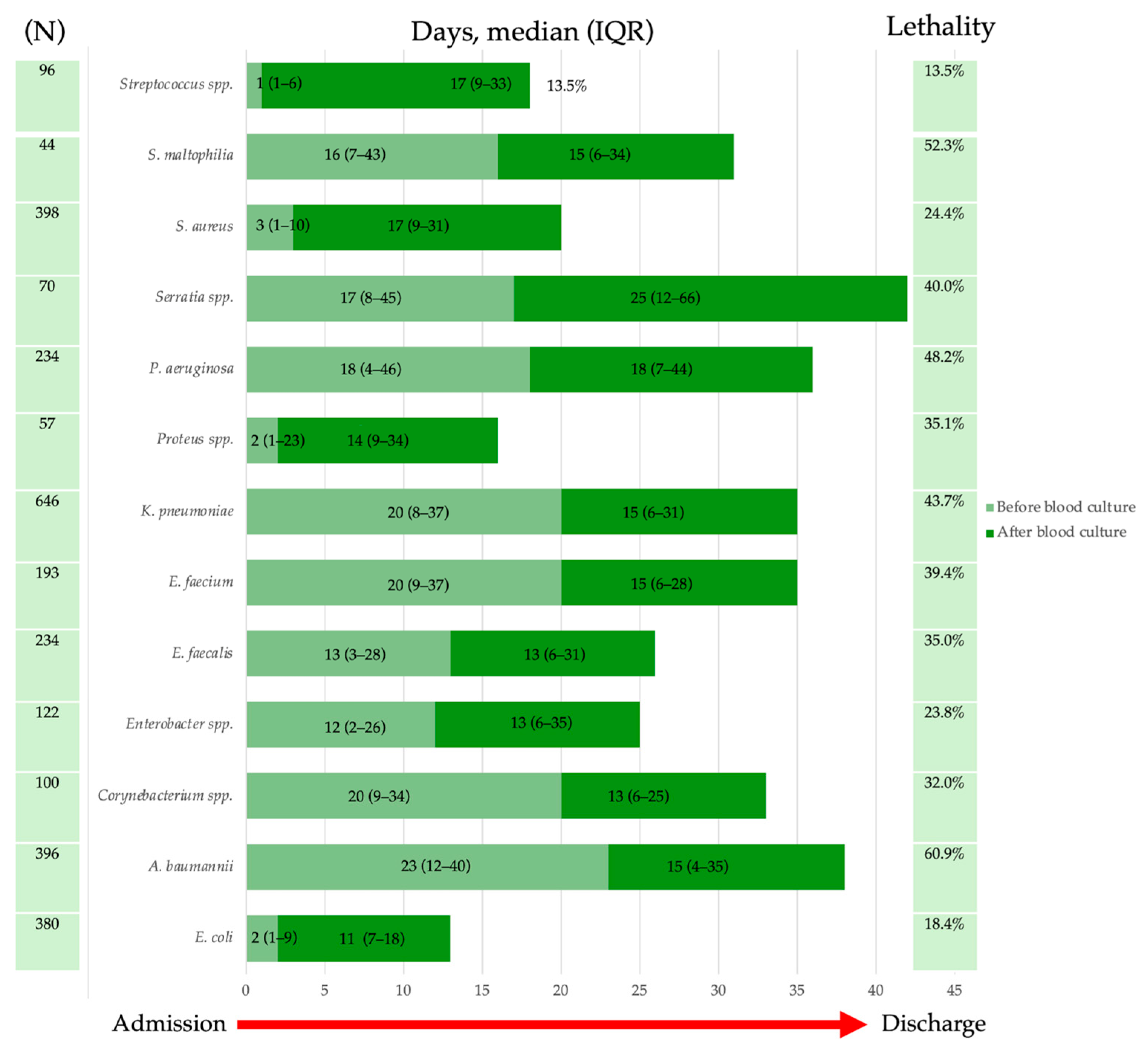
2.4. Lethality
3. Discussion
4. Limits
5. Materials and Methods
5.1. Data Collection
5.2. Blood Cultures
5.3. Statistical Analysis
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BSI | Bloodstream infections |
| AMR | Antimicrobial resistance |
| WHO | World Health Organization |
| ISS | Istituto Superiore di Sanità |
| MRSA | Methicillin-resistant S. aureus |
| VRE | Vancomycin-resistant Enterococcus spp. |
| IQR | Interquartile range |
| ICU | Intensive care unit |
| NICU | Neonatal intensive care unit |
| UTI | Urinary tract infection |
| IAI | Intrabdominal infection |
| CNSI | Central nervous system infection |
| OI | Osteoarticular infection |
| SSTI | Skin and soft tissue infection |
| CCI | Charlson comorbidity index |
| LOS | Length of stay |
| CVC | Central venous catheter |
| TMP-SMX | Trimethoprim-sulfamethoxazole |
| AMCLI | Associazione microbiologi clinici italiani |
| EUCAST | European committee on antimicrobial susceptibility testing |
| CLSI | Clinical and laboratory standard institute |
| AST | Antimicrobial susceptibility |
| CI | Confidence interval |
| cOR | Crude odd ratio |
| aOR | Adjusted odd ratio |
References
- WHO. Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 5 March 2025).
- WHO. Bacterial Priority Pathogens List. 2024. Available online: https://iris.who.int/bitstream/handle/10665/376776/9789240093461-eng.pdf?sequence=1 (accessed on 5 March 2025).
- Goto, M.; Al-Hasan, M.N. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect. 2013, 19, 501–509. [Google Scholar] [CrossRef] [PubMed]
- ECOC. Antimicrobial Resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report 2023. Available online: https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-eueea-ears-net-annual-epidemiological-report-2023?utm_source=chatgpt.com (accessed on 17 April 2025).
- Mun, S.J.; Kim, S.H.; Kim, H.T.; Moon, C.; Wi, Y.M. The epidemiology of bloodstream infection contributing to mortality: The difference between community-acquired, healthcare-associated, and hospital-acquired infections. BMC Infect Dis. 2022, 22, 336. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Antimicrobial Resistance ISS Report. Available online: https://www.epicentro.iss.it/antibiotico-resistenza/resistenza (accessed on 22 March 2025).
- Sangiorgio, G.; Calvo, M.; Stefani, S. Aztreonam and avibactam combination therapy for metallo-β-lactamase-producing gram-negative bacteria: A comprehensive review. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2024, in press. [Google Scholar] [CrossRef] [PubMed]
- Dashboard AMR—Rete MIC Sicilia. Available online: https://www.qualitasiciliassr.it/?q=rete-laboratori (accessed on 22 March 2025).
- Antimicrobial Resistance Surveillance in Europe 2023–2021 Data. Available online: https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2023-2021-data (accessed on 1 March 2025).
- Huemer, M.; Mairpady Shambat, S.; Brugger, S.D.; Zinkernagel, A.S. Antibiotic resistance and persistence-Implications for human health and treatment perspectives. EMBO Rep. 2020, 21, e51034. [Google Scholar] [CrossRef]
- Wang, Y.; Sholeh, M.; Yang, L.; Shakourzadeh, M.Z.; Beig, M.; Azizian, K. Global trends of ceftazidime-avibactam resistance in gram-negative bacteria: Systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 2025, 14, 10. [Google Scholar] [CrossRef]
- Liu, S.Y.; Chou, S.H.; Chuang, C.; Juan, C.H.; Ho, Y.C.; Ho, H.L.; Chen, L.; Lin, Y.T. Clinical and Microbiological characteristics of patients with ceftazidime/avibactam-resistant Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae strains. Ann. Clin. Microbiol. Antimicrob. 2025, 24, 26. [Google Scholar] [CrossRef]
- Sharma, R.; Dinesh, L. Acinetobacter baumannii: A Comprehensive Review of Global Epidemiology, Clinical Implications, Host Interactions, Mechanisms of Antimicrobial Resistance and Mitigation Strategies. Microb. Pathog 2025, 204, 107605. [Google Scholar] [CrossRef]
- Kanj, S.S.; Kantecki, M.; Arhin, F.F.; Gheorghe, M. Epidemiology and outcomes associated with MBL-producing Enterobacterales: A systematic literature review. Int. J. Antimicrob. Agents 2025, 65, 107449. [Google Scholar] [CrossRef]
- Nielsen, S.L.; Lassen, A.T.; Gradel, K.O.; Jensen, T.G.; Kolmos, H.J.; Hallas, J.; Pedersen, C. Bacteremia is associated with excess long-term mortality: A 12-year population-based cohort study. J. Infect. 2015, 70, 111–126. [Google Scholar] [CrossRef]
- Cauhapé, V.; Lamy, B.; Lotte, R.; Touitou, I.; Boyer, L.; Contenti, J.; Parisot, F.; Ruimy, R.; Carles, M.; Courjon, J. Lesson from the COVID-19 pandemic lockdown: A major change of hospital-diagnosed bacteremia epidemiology. Infect. Dis. Now. 2023, 53, 104709. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bauer, K.A.; Puzniak, L.A.; Yu, K.C.; Finelli, L.; Moise, P.; Ai, C.; Watts, J.A.; Gupta, V. Epidemiology and outcomes of culture-positive bloodstream pathogens prior to and during the SARS-CoV-2 pandemic: A multicenter evaluation. BMC Infect. Dis. 2022, 22, 841. [Google Scholar] [CrossRef] [PubMed]
- Gasch, O.; Badia-Cebada, L.; Carmezim, J.; Vaqué, M.; Pomar, V.; Moreno, E.; Marrón, A.; Jiménez-Martínez, E.; García-Quesada, M.J.; Garcia-Alarcón, X.; et al. Effects of the COVID-19 Pandemic on Incidence and Epidemiology of Catheter-Related Bacteremia, Spain. Emerg. Infect. Dis. 2022, 28, 2181–2189. [Google Scholar] [CrossRef] [PubMed]
- Pipitò, L.; Rubino, R.; D’Agati, G.; Bono, E.; Mazzola, C.V.; Urso, S.; Zinna, G.; Distefano, S.A.; Firenze, A.; Bonura, C.; et al. Antimicrobial Resistance in ESKAPE Pathogens: A Retrospective Epidemiological Study at the University Hospital of Palermo, Italy. Antibiotics 2025, 14, 186. [Google Scholar] [CrossRef]
- Lee, C.M.; Kim, C.J.; Kim, S.E.; Park, K.H.; Bae, J.Y.; Choi, H.J.; Jung, Y.; Lee, S.S.; Choe, P.G.; Park, W.B.; et al. Risk factors for early mortality in patients with carbapenem-resistant Acinetobacter baumannii bacteraemia. J. Glob. Antimicrob. Resist. 2022, 31, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Mortensen, V.H.; Søgaard, M.; Mygind, L.H.; Wolkewitz, M.; Kristensen, B.; Schønheyder, H.C. Incidence and mortality of hospital-acquired bacteraemia: A population-based cohort study applying a multi-state model approach. Clin. Microbiol. Infect. 2022, 28, 879.e9–879.e15. [Google Scholar] [CrossRef]
- Singh, G.; Loho, T.; Yulianti, M.; Aditianingsih, D.; Zakiyah, L.F.; Masse, S.F.; Triono, M.R. Factors associated with antibiotic resistance and survival analysis of severe pneumonia patients infected with Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa: A retrospective cohort study in Jakarta, Indonesia. SAGE Open Med. 2024, 12, 20503121241264097. [Google Scholar] [CrossRef]
- Varon, B.; Palacios-Baena, Z.R.; de Kraker, M.E.A.; Rodríguez-Baño, J.; Leibovici, L.; Paul, M. Universal Risk Factors for Mortality in Bloodstream Infections (UNIFORM): A systematic review and Delphi survey. Clin. Microbiol. Infect. 2024, 30, 453–461. [Google Scholar] [CrossRef]
- García-Rodríguez, J.F.; Mariño-Callejo, A. The factors associated with the trend in incidence of Bacteraemia and associated mortality over 30 years. BMC Infect. Dis. 2023, 23, 69. [Google Scholar] [CrossRef]
- Johnson, A.P.; Uttley, A.H.; Woodford, N.; George, R.C. Resistance to vancomycin and teicoplanin: An emerging clinical problem. Clin. Microbiol. Rev. 1990, 3, 280–291. [Google Scholar] [CrossRef]
- Serra, N.; Di Carlo, P.; Andriolo, M.; Mazzola, G.; Diprima, E.; Rea, T.; Anastasia, A.; Fasciana, T.M.A.; Pipitò, L.; Capra, G.; et al. Staphylococcus aureus and Coagulase-Negative Staphylococci from Bloodstream Infections: Frequency of Occurrence and Antimicrobial Resistance, 2018–2021. Life 2023, 13, 1356. [Google Scholar] [CrossRef]
- Mosquera, J.M.A.; Assis Reveiz, J.K.; Barrera, L.; Liscano, Y. Impact of Central Line-Associated Bloodstream Infections on Mortality and Hospital Stay in Adult Patients at a Tertiary Care Institution in Cali, Colombia, 2015–2018. J. Clin. Med. 2024, 13, 5376. [Google Scholar] [CrossRef] [PubMed]
- Alwazzeh, M.J.; Alnimr, A.; Al Nassri, S.A.; Alwarthan, S.M.; Alhajri, M.; AlShehail, B.M.; Almubarak, M.; Alghamdi, N.S.; Wali, H.A. Microbiological trends and mortality risk factors of central line-associated bloodstream infections in an academic medical center 2015–2020. Antimicrob. Resist. Infect. Control 2023, 12, 128. [Google Scholar] [CrossRef] [PubMed]
- ECDC. Point Prevalence Survey of Healthcare-Associated Infections and Antimicrobial Use in European Acute Care Hospitals—2022–2023. Available online: https://www.ecdc.europa.eu/en/publications-data/PPS-HAI-AMR-acute-care-europe-2022-2023 (accessed on 1 March 2025).
- Percorso Diagnostico, Infezioni del Torrente Circolatorio. Available online: https://www.amcli.it/wp-content/uploads/2023/05/13_PD_INFEZIONI-DEL-TORRENTE-CIRCOLATORIO_DEF23mag2023.pdf (accessed on 1 March 2025).
- Miller, J.M.; Binnicker, M.J.; Campbell, S.; Carroll, K.C.; Chapin, K.C.; Gonzalez, M.D.; Harrington, A.; Jerris, R.C.; Kehl, S.C.; Leal, S.M., Jr.; et al. Guide to Utilization of the Microbiology Laboratory for Diagnosis of Infectious Diseases: 2024 Update by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM). Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2024, ciae104. [Google Scholar] [CrossRef] [PubMed]
- Tan, R.; Liu, J.; Li, M.; Huang, J.; Sun, J.; Qu, H. Epidemiology and antimicrobial resistance among commonly encountered bacteria associated with infections and colonization in intensive care units in a university-affiliated hospital in Shanghai. J. Microbiol. Immunol. Infect. 2014, 47, 8–94. [Google Scholar] [CrossRef] [PubMed]
- Benkő, R.; Gajdács, M.; Matuz, M.; Bodó, G.; Lázár, A.; Hajdú, E.; Papfalvi, E.; Hannauer, P.; Erdélyi, P.; Pető, Z. Prevalence and Antibiotic Resistance of ESKAPE Pathogens Isolated in the Emergency Department of a Tertiary Care Teaching Hospital in Hungary: A 5-Year Retrospective Survey. Antibiotics 2020, 9, 624. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, Y.-C.; Kuo, S.-C.; Fang, C.-T.; Lauderdale, T.-L. Changing epidemiology and antimicrobial resistance of bacteria causing bacteremia in Taiwan: 2002–2020. Microbiol. Spectr. 2024, 12, e0060824. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rödenbeck, M.; Ayobami, O.; Eckmanns, T.; Pletz, M.W.; Bleidorn, J.; Markwart, R. Clinical epidemiology and case fatality due to antimicrobial resistance in Germany: A systematic review and meta-analysis, 1 January 2010 to 31 December 2021. Euro Surveill. 2023, 28, 2200672. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jernigan, J.A.; Hatfield, K.M.; Wolford, H.; Nelson, R.E.; Olubajo, B.; Reddy, S.C.; McCarthy, N.; Paul, P.; McDonald, L.C.; Kallen, A.; et al. Multidrug-Resistant Bacterial Infections in U.S. Hospitalized Patients, 2012–2017. N. Engl. J. Med. 2020, 382, 1309–1319. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Machuca, I.; Guzmán-Puche, J.; Pérez-Nadales, E.; Gracia-Ahufinger, I.; Mendez, A.; Cano, A.; Castón, J.J.; Domínguez, A.; Torre-Cisneros, J.; Martínez-Martínez, L. Community-acquired bacteraemia by Klebsiella pneumoniae producing KPC-3 and resistant to ceftazidime/avibactam. J. Glob. Antimicrob. Resist. 2022, 30, 399–402. [Google Scholar] [CrossRef]
- Hobson, C.A.; Pierrat, G.; Tenaillon, O.; Bonacorsi, S.; Bercot, B.; Jaouen, E.; Jacquier, H.; Birgy, A. Klebsiella pneumoniae Carbapenemase Variants Resistant to Ceftazidime-Avibactam: An Evolutionary Overview. Antimicrob. Agents Chemother. 2022, 66, e0044722. [Google Scholar] [CrossRef]
- Cui, Q.; Wang, C.; Wang, Q.; Qin, J.; Li, M.; Ding, B.; Shen, Z. Ceftazidime/Avibactam Resistance in Carbapenemase-Producing Klebsiella pneumoniae. Emerg. Infect. Dis. 2023, 29, 2398–2400. [Google Scholar] [CrossRef] [PubMed]
- Boattini, M.; Bianco, G.; Bastos, P.; Comini, S.; Corcione, S.; Almeida, A.; Costa, C.; De Rosa, F.G.; Cavallo, R. Prevalence and mortality of ceftazidime/avibactam-resistant KPC-producing Klebsiella pneumoniae bloodstream infections (2018–2022). Eur. J. Clin. Microbiol. Infect. Dis. 2024, 43, 155–166. [Google Scholar] [CrossRef]
- Oliva, A.; Campogiani, L.; Savelloni, G.; Vitale, P.; Lodi, A.; Sacco, F.; Imeneo, A.; Volpicelli, L.; Polani, R.; Raponi, G.; et al. Clinical Characteristics and Outcome of Ceftazidime/Avibactam-Resistant Klebsiella pneumoniae Carbapenemase-Producing Klebsiella pneumoniae Infections: A Retrospective, Observational, 2-Center Clinical Study. Open Forum Infect. Dis. 2023, 10, ofad327. [Google Scholar] [CrossRef]
- Gaibani, P.; Lombardo, D.; Bussini, L.; Bovo, F.; Munari, B.; Giannella, M.; Bartoletti, M.; Viale, P.; Lazzarotto, T.; Ambretti, S. Epidemiology of Meropenem/Vaborbactam Resistance in KPC-Producing Klebsiella pneumoniae Causing Bloodstream Infections in Northern Italy, 2018. Antibiotics 2021, 10, 536. [Google Scholar] [CrossRef] [PubMed]
- Gaibani, P.; Re, M.C.; Campoli, C.; Viale, P.L.; Ambretti, S. Bloodstream infection caused by KPC-producing Klebsiella pneumoniae resistant to ceftazidime/avibactam: Epidemiology and genomic characterization. Clin. Microbiol. Infect. 2020, 26, 516.e1–516.e4. [Google Scholar] [CrossRef] [PubMed]
- Galani, I.; Papoutsaki, V.; Karaiskos, I.; Moustakas, N.; Galani, L.; Maraki, S.; Mavromanolaki, V.E.; Legga, O.; Fountoulis, K.; Platsouka, E.D.; et al. In vitro activities of omadacycline, eravacycline, cefiderocol, apramycin, and comparator antibiotics against Acinetobacter baumannii causing bloodstream infections in Greece, 2020–2021: A multicenter study. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 843–852. [Google Scholar] [CrossRef]
- Liu, P.Y.; Ko, W.C.; Lee, W.S.; Lu, P.L.; Chen, Y.H.; Cheng, S.H.; Lu, M.C.; Lin, C.Y.; Wu, T.S.; Yen, M.Y.; et al. In vitro activity of cefiderocol, cefepime/enmetazobactam, cefepime/zidebactam, eravacycline, omadacycline, and other comparative agents against carbapenem-non-susceptible Pseudomonas aeruginosa and Acinetobacter baumannii isolates associated from bloodstream infection in Taiwan between 2018–2020. J. Microbiol. Immunol. Infect. Wei Mian Yu Gan ran za zhi 2022, 55, 888–895. [Google Scholar] [CrossRef]
- Rodríguez-Aguirregabiria, M.; Lázaro-Perona, F.; Cacho-Calvo, J.B.; Arellano-Serrano, M.S.; Ramos-Ramos, J.C.; Rubio-Mora, E.; Díaz-Almirón, M.; Asensio-Martín, M.J. Challenges Facing Two Outbreaks of Carbapenem-Resistant Acinetobacter baumannii: From Cefiderocol Susceptibility Testing to the Emergence of Cefiderocol-Resistant Mutants. Antibiotics 2024, 13, 784. [Google Scholar] [CrossRef]
- Kritsotakis, E.I.; Lagoutari, D.; Michailellis, E.; Georgakakis, I.; Gikas, A. Burden of multidrug and extensively drug-resistant ESKAPEE pathogens in a secondary hospital care setting in Greece. Epidemiol. Infect. 2022, 150, e170. [Google Scholar] [CrossRef]
- Blondeau, J.; Charles, M.K.; Loo, V.; Adam, H.; Gonzalez Del Vecchio, M.; Ghakis, C.; O’Callaghan, E.; El Ali, R. A nested cohort 5-year Canadian surveillance of Gram-negative antimicrobial resistance for optimized antimicrobial therapy. Sci. Rep. 2023, 13, 14142. [Google Scholar] [CrossRef]
- Lan, M.; Dongmei, K.; Guodong, S.; Haifeng, Y.; Guofeng, C.; Mengting, C.; Xiaoyun, F. Risk factors for bacteremic pneumonia and mortality (28-day mortality) in patients with Acinetobacter baumannii bacteremia. BMC Infect. Dis. 2024, 24, 448. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ge, L.; Chen, L.; Komarow, L.; Hanson, B.; Reyes, J.; Cober, E.; Alenazi, T.; Zong, Z.; Xie, Q.; et al. Clinical Outcomes and Bacterial Characteristics of Carbapenem-resistant Acinetobacter baumannii Among Patients from Different Global Regions. Clin. Infect. Dis. 2024, 78, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Hassoun-Kheir, N.; Guedes, M.; Ngo Nsoga, M.T.; Argante, L.; Arieti, F.; Gladstone, B.P.; Kingston, R.; Naylor, N.R.; Pezzani, M.D.; Pouwels, K.B.; et al. A systematic review on the excess health risk of antibiotic-resistant bloodstream infections for six key pathogens in Europe. Clin. Microbiol. Infect. 2024, 30 (Suppl. S1), S14–S25. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Liu, M.; Wu, J.; Chen, S.; Yang, H.; Long, J.; Duan, G. Mortality and genetic diversity of antibiotic-resistant bacteria associated with bloodstream infections: A systemic review and genomic analysis. BMC Infect. Dis. 2024, 24, 1385. [Google Scholar] [CrossRef]
- Pipitò, L.; Puccio, R.; Marrali, D.; Mancuso, A.; Gagliano, M.C.; Gaudiano, R.; Piccione, M.; Iaria, C.; Cascio, A. Sepsis in Patients Hospitalized in Sicily, Italy, over the Period of 2016–2020: A Retrospective Study. J. Clin. Med. 2024, 13, 2239. [Google Scholar] [CrossRef]
- Clinical Breakpoints—Breakpoints and Guidance. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 27 March 2024).
- Werfen Biwer. Available online: https://www.werfen.com/it/it/healthcare-it-solutions/biwer (accessed on 27 March 2025).
- Hindler, J.F.; Stelling, J. Analysis and Presentation of Cumulative Antibiograms: A New Consensus Guideline from the Clinical and Laboratory Standards Institute. Clin. Infect. Dis. 2007, 44, 867–873. [Google Scholar] [CrossRef]
- CLSI. Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data. Available online: https://clsi.org/standards/products/microbiology/documents/m39/ (accessed on 19 April 2025).

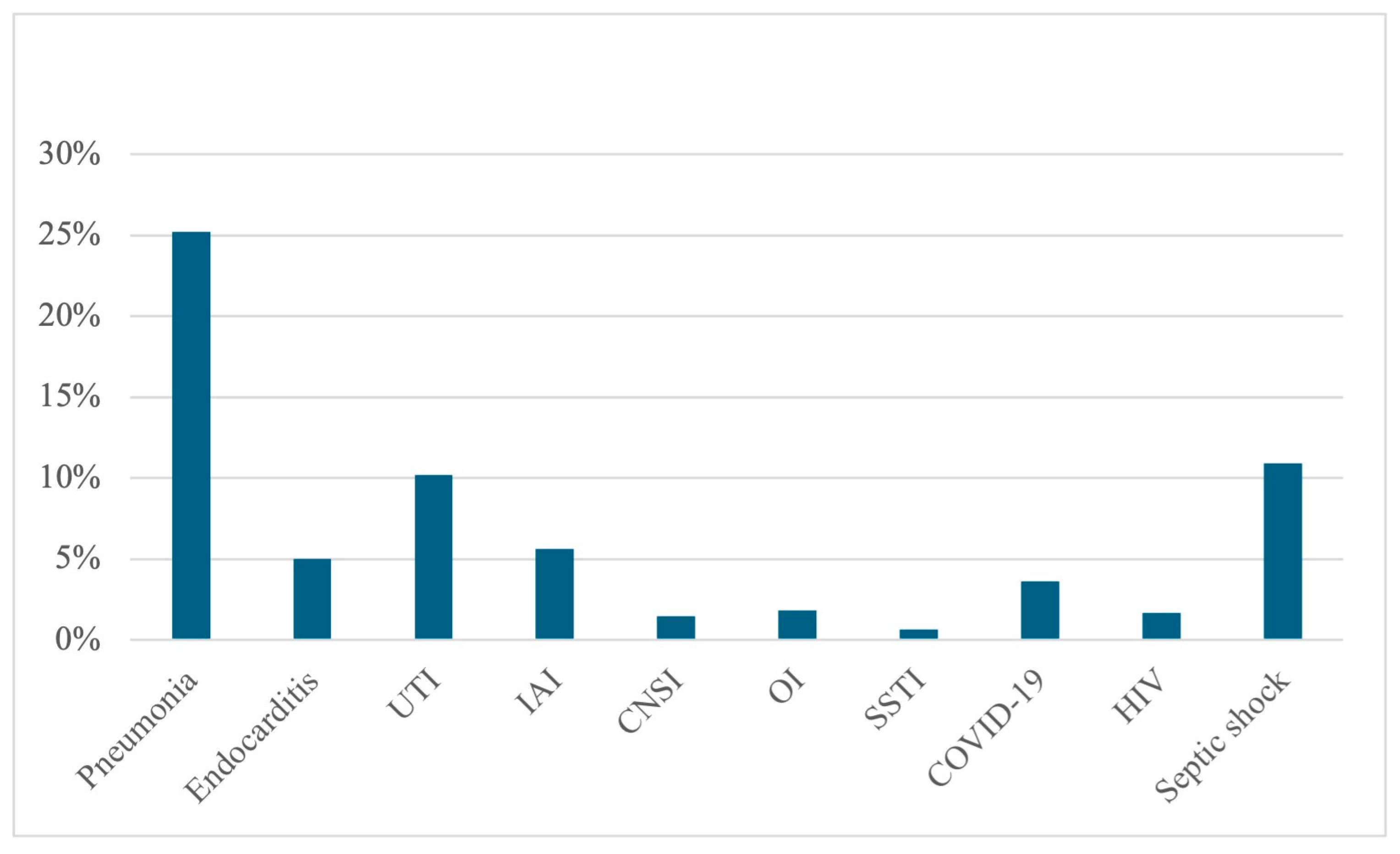
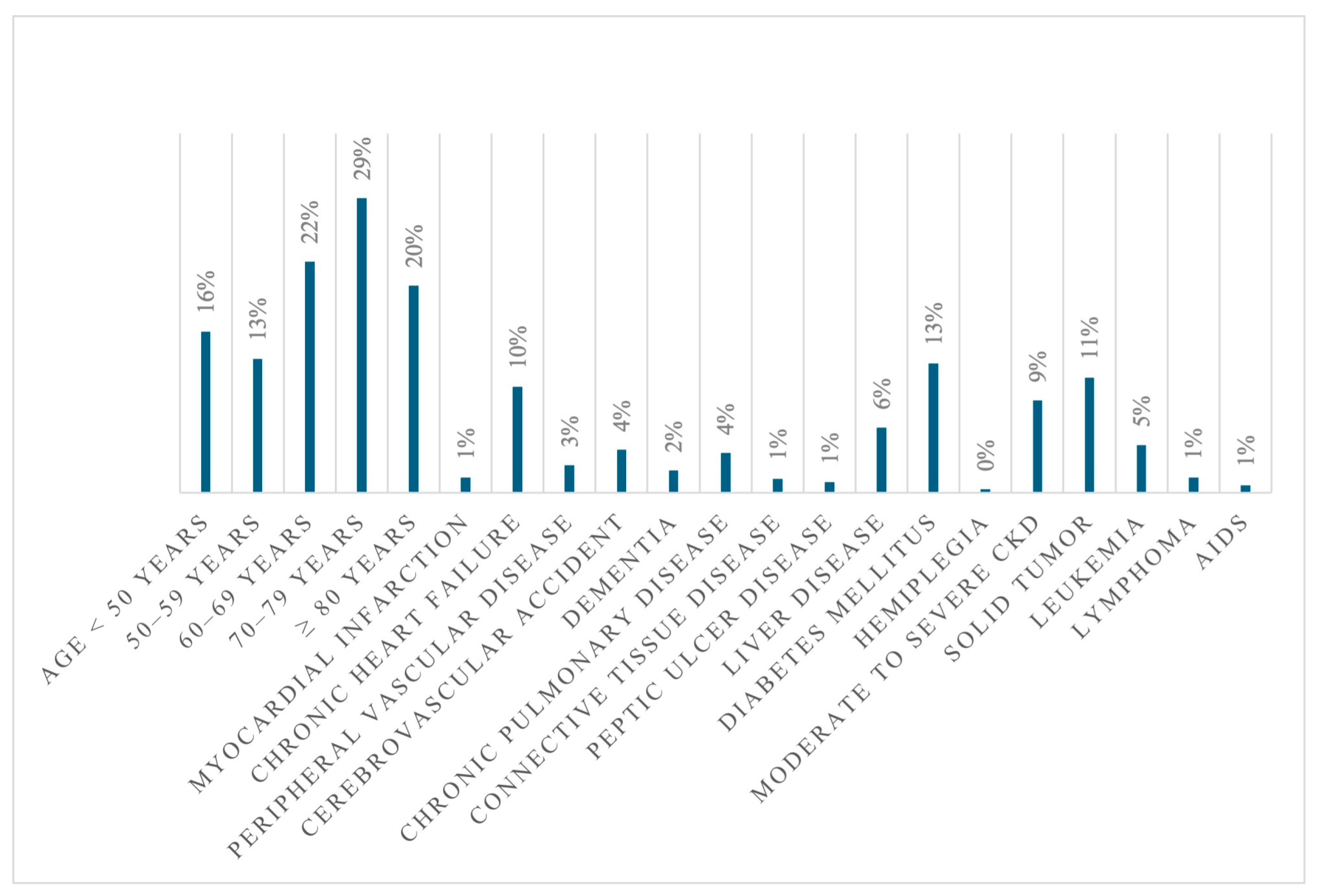

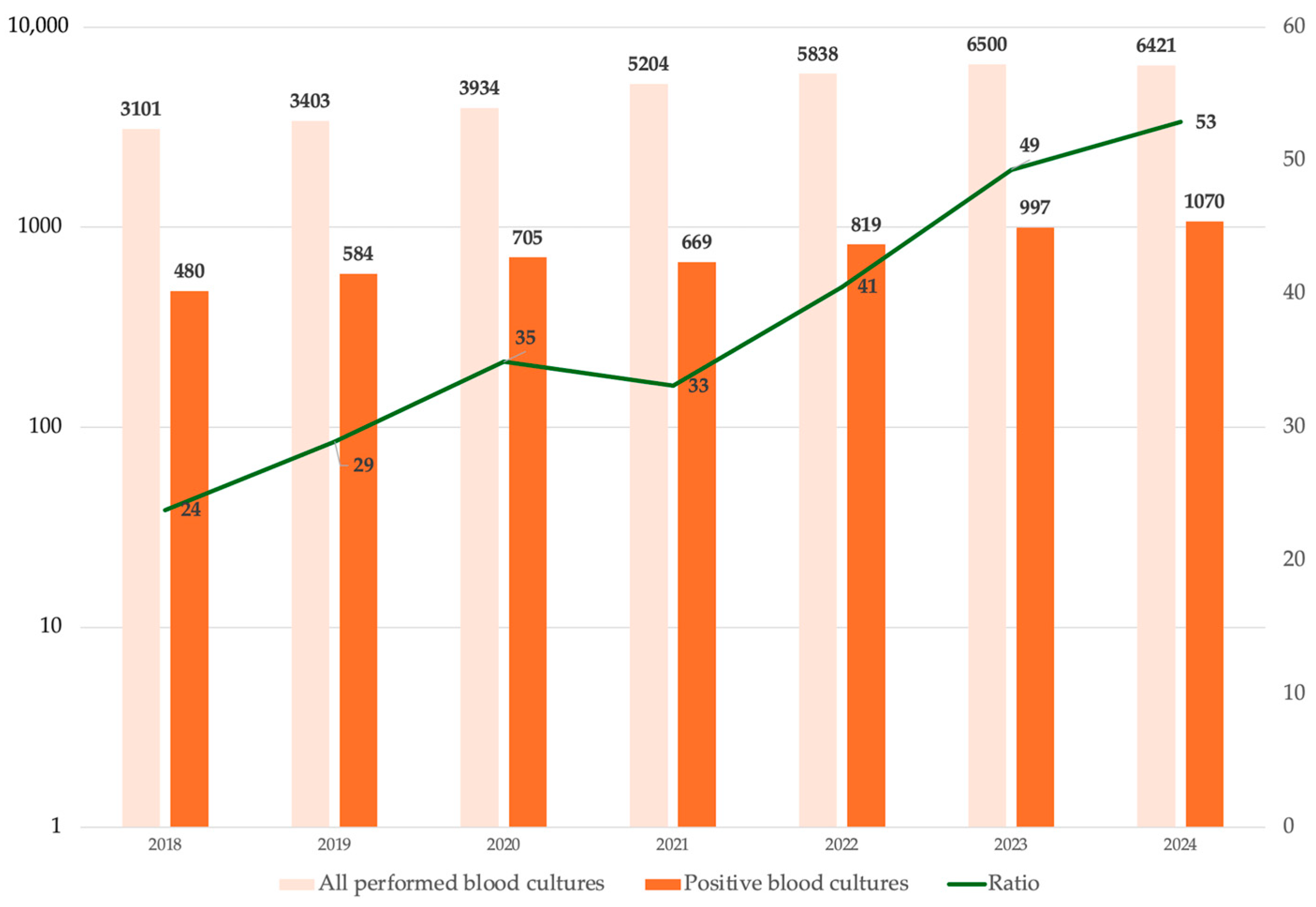
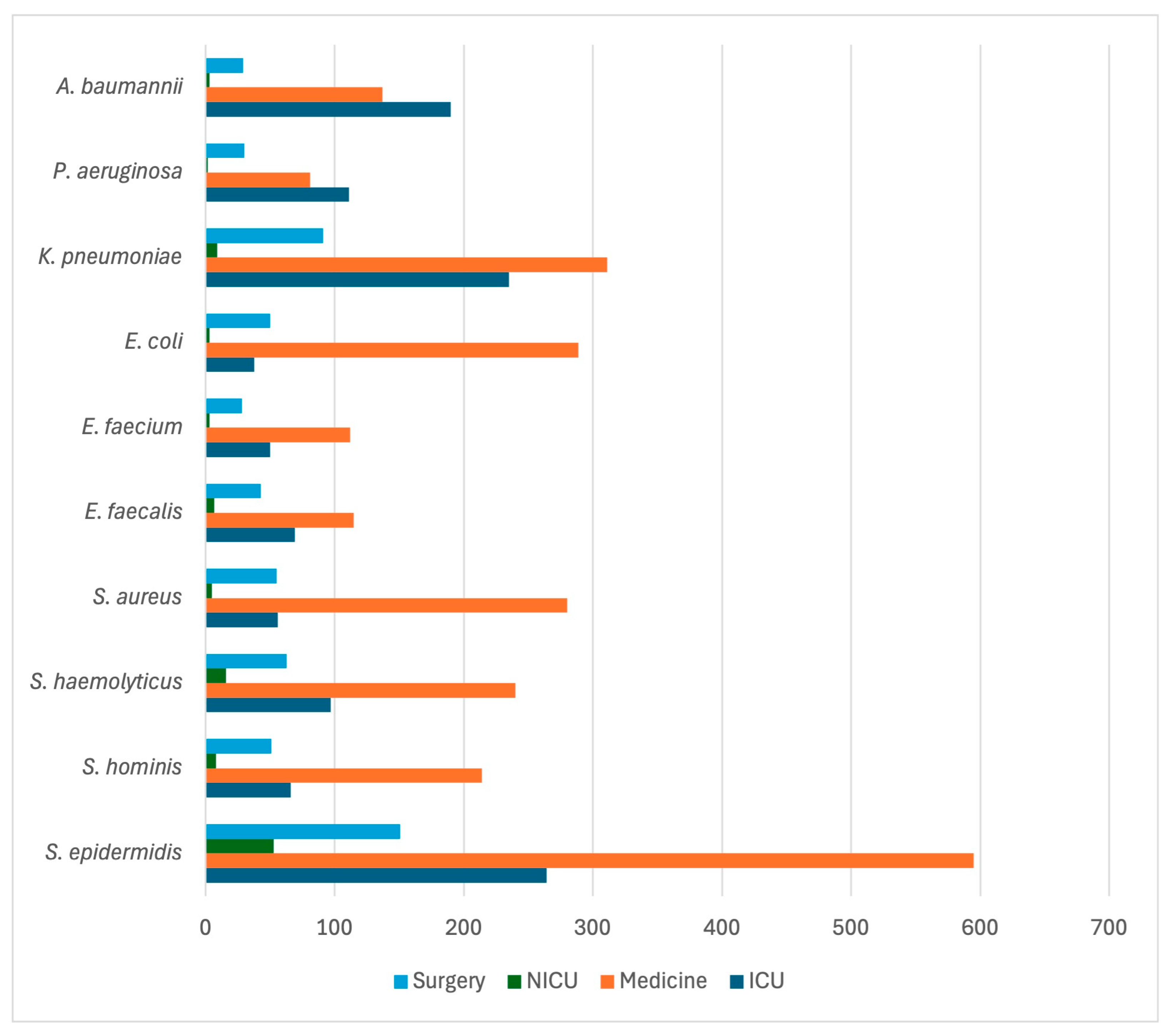

| Variable | Alive at Discharge N = 2168 | Dead at Discharge N = 799 | cOR | p |
|---|---|---|---|---|
| Male | 60.56% (1313) | 54.82% (438) | 0.79 | 0.005 |
| ICU | 3.69% (80) | 55.19% (441) | 32.15 | <0.001 |
| LOS (mean ± SD) | 30.81 ± 26.13 | 31.98 ± 31.13 | 0.308 | |
| CCI | 3.38 ± 2.12 | 3.54 ± 1.72 | 0.048 | |
| Septic shock | 3.74% (81) | 30.41% (243) | 11.26 | <0.001 |
| Pneumonia | 22.69% (492) | 33.04% (256) | 1.61 | <0.001 |
| Endocarditis | 5.67% (123) | 3.25% (26) | 0.56 | 0.007 |
| UTI | 12.64% (274) | 3.63% (29) | 0.26 | <0.001 |
| IAI | 6.78% (147) | 2.50% (20) | 0.35 | <0.001 |
| CNSI | 1.15% (25) | 2.38% (19) | 2.09 | 0.014 |
| OI | 2.35% (51) | 0.50% (4) | 0.21 | <0.001 |
| SSTI | 0.64% (14) | 0.62% (5) | 0.952 | |
| COVID-19 | 3.04% (66) | 5.26% (42) | 1.77 | 0.004 |
| HIV | 1.89% (41) | 1.13% (9) | 0.151 | |
| Number of isolates | 1.90 ± 1.91 | 2.79 ± 3.21 | <0.001 |
| Bacteria | Overall N = 5324 | Medicine 2989 | Surgery 729 | ICU 1471 | NICU 135 |
|---|---|---|---|---|---|
| Staphylococcus spp. S. epidermidis S. aureus S. haemolyticus S. hominis S. capitis S. pettenkoferi S. caprae S. lugdunensis S. warneri S. cohnii S. pasteuri S. simulans S. saprophyticus S. equorum S. condimenti S. intermedius S. lentus S. xylosus S. sciuri S. kloosi Other CONS Other COPS Others | 2510 (47.1%) 1063 (20.0%) 398 (7.5%) 416 (7.8%) 339 (6.4%) 125 (2.3%) 52 (1.0%) 24 (0.4%) 12 (0.2%) 13 (0.2%) 14 (0.2%) 4 (0.1%) 4 (0.1%) 3 (<0.1%) 2 (<0.1%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) 13 (0.2%) 2 (<0.1%) 20 (0.4%) | 1499 (50.1%) 595 (19.9%) 281 (9.4%) 240 (8.0%) 214 (7.1%) 75 (2.5%) 28 (0.9%) 15 (0.5%) 9 (0.3%) 6 (0.2%) 8 (0.3%) 1 (<0.1%) 4 (0.1%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) 0 (0.0%) 0 (0.0%) 6 (0.2%) 1 (<0.1%) 10 (0.3%) | 347 (47.6%) 151 (20.7%) 55 (7.5%) 63 (8.6%) 51 (7.0%) 9 (1.2%) 6 (0.8%) 0 (0.0%) 1 (0.1%) 1 (0.1%) 2 (0.3%) 1 (0.1%) 0 (0.0%) 0 (0.0%) 1 (0.1%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 1 (0.1%) 2 (0.3%) 0 (0.0%) 3 (0.4%) | 572 (38.9%) 264 (17.9%) 57 (3.9%) 97 (6.6%) 66 (4.5%) 39 (2.6%) 18 (1.2%) 9 (0.6%) 2 (0.1%) 2 (0.1%) 4 (0.3%) 1 (0.1%) 0 (0.0%) 1 (0.1%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 1 (0.1%) 0 (0.0%) 5 (0.3%) 0 (0.0%) 6 (0.4%) | 92 (68.1%) 53 (39.2%) 5 (3.7%) 16 (11.8%) 8 (5.9%) 2 (1.5%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 4 (3.0%) 0 (0.0%) 1 (0.7%) 0 (0.0%) 1 (0.7%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 1 (0.7%) 1 (0.7%) |
| Enterococcus spp. E. faecalis E. faecium E. avium E. casseliflavus E. durans E. gallinarum E. hirae E. raffinosus | 440 (8.3%) 234 (4.4%) 193 (3.6%) 3 (<0.1%) 3 (<0.1%) 1 (<0.1%) 3 (<0.1%) 1 (<0.1%) 2 (<0.1%) | 237 (7.9%) 115 (3.8%) 112 (3.7%) 3 (0.1%) 2 (0.1%) 1 (<0.1%) 3 (0.1%) 0 (0.0%) 1 (<0.1%) | 72 (9.9%) 43 (5.9%) 28 (3.8%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 1 (0.1%) 0 (0.0%) | 121 (8.2%) 69 (4.7%) 50 (3.4%) 0 (0.0%) 1 (0.1%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 1 (0.1%) | 10 (7.4%) 7 (5.2%) 3 (2.2%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| E. coli | 380 (7.1%) | 289 (9.7%) | 50 (6.8%) | 38 (2.6%) | 3 (2.2%) |
| Pseudomonas spp. P. aeruginosa P. oryzihabitans P. putida P. stutzeri Others | 234 (4.4%) 224 (4.2%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) 7 (0.1%) | 84 (2.8%) 81 (2.7%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) 0 (0.0%) | 31 (4.2%) 30 (4.1%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 1 (0.1%) | 117 (7.9%) 111 (7.5%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 6 (0.4%) | 2 (1.5%) 2 (1.5%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| Klebsiella spp. K. pneumoniae K. oxytoca K. ozaenae Others | 693 (13.0%) 646 (12.1%) 29 (0.5%) 4 (0.1%) 13 (0.2%) | 344 (11.5%) 311 (10.4%) 23 (0.8%) 2 (0.1%) 8 (0.3%) | 98 (13.4%) 91 (12.5%) 4 (0.5%) 2 (0.3%) 1 (0.1%) | 241 (16.4%) 235 (16.0%) 2 (0.1%) 0 (0.0%) 4 (0.3%) | 10 (7.4%) 9 (6.7%) 0 (0.0%) 0 (0.0%) 1 (0.7%) |
| Enterobacter spp. E. cloacae E. aerogenes E. hormaechei E. kobei E. asburiae E. gergoviae E. sakazakii Others | 122 (2.3%) 75 (1.4%) 22 (0.4%) 15 (0.3%) 2 (<0.1%) 2 (<0.1%) 1 (<0.1%) 1 (<0.1%) 4 (0.1%) | 64 (2.1%) 39 (1.3%) 13 (0.4%) 6 (0.2%) 0 (0.0%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) 3 (0.1%) | 22 (3.0%) 17 (2.3%) 3 (0.4%) 2 (0.3%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) | 30 (2.0%) 14 (0.9%) 6 (0.4%) 6 (0.4%) 1 (0.1%) 1 (0.1%) 0 (0.0%) 1 (0.1%) 1 (0.1%) | 6 (4.4%) 5 (3.7%) 0 (0.0%) 1 (0.7%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| Citrobacter spp. C. koseri C. freundii C. farmeri C. amalonaticus | 10 (0.2%) 5 (0.1%) 3 (<0.1%) 1 (<0.1%) 1 (<0.1%) | 6 (0.2%) 4 (0.1%) 2 (0.1%) 0 (0.0%) 0 (0.0%) | 2 (0.3%) 0 (0.0%) 0 (0.0%) 1 (0.1%) 1 (0.1%) | 2 (0.1%) 1 (0.1%) 1 (0.1%) 0 (0.0%) 0 (0.0%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| Morganella morgannii | 6 (0.1%) | 3 (0.1%) | 0 (0.0%) | 3 (0.2%) | 0 (0.0%) |
| Serratia spp. S. marcensces S. liquefaciens S. plymuthica Others | 70 (1.3%) 52 (1.1%) 4 (0.1%) 3 (<0.1%) 11 (0.2%) | 17 (0.6%) 12 (0.4%) 1 (< 0.1%) 3 (0.1%) 2 (0.1%) | 13 (1.8%) 12 (1.6%) 0 (0.0%) 0 (0.0%) 1 (0.1%) | 39 (2.6%) 28 (1.9%) 3 (0.2%) 0 (0.0%) 8 (0.5%) | 1 (0.7%) 1 (0.7%) 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| Acinetobacter spp. A. baumannii A. lwoffii Others | 415 (7.8%) 396 (7.4%) 7 (0.1%) 12 (0.2%) | 165 (5.5%) 154 (5.1%) 3 (0.1%) 8 (0.3%) | 35 (4.8%) 34 (4.7%) 1 (0.1%) 0 (0.0%) | 214 (14.5%) 208 (14.1%) 2 (0.1%) 4 (0.3%) | 1 (0.7%) 0 (0.0%) 1 (0.7%) 0 (0.0%) |
| Proteus spp. P. mirabilis P. vulgaris P. hauseri | 57 (1.0%) 54 (1.0%) 2 (<0.1%) 1 (<0.1%) | 35 (1.2%) 34 (1.1%) 0 (0.0%) 1 (<0.1%) | 9 (1.2%) 8 (1.1%) 1 (0.1%) 0 (0.0%) | 13 (0.9%) 12 (0.8%) 1 (0.1%) 0 (0.0%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| Providencia spp. P. stuartii P. rustigianii | 19 (0.3%) 18 (0.3%) 1 (<0.1%) | 7 (0.2%) 6 (0.2%) 1 (<0.1%) | 3 (0.4%) 3 (0.4%) 0 (0.0%) | 9 (0.6%) 9 (0.6%) 0 (0.0%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| Stenotrophomonas maltophilia | 44 (0.8%) | 16 (0.5%) | 5 (0.7%) | 22 (1.5%) | 1 (0.7%) |
| Streptococcus spp. S. bovis S. gallolyticus S. anginosus S. pneumoniae S. agalactiae S. constellatus S. dysgalactiae S. pyogens S. sanguinis S. parasanguinis S. acidominimus S. cristatus S. equi S. gordonii S. salivarius S. intermedius S. mitis S. oralis Others | 96 (1.8%) 10 (0.2%) 8 (0.1%) 8 (0.1%) 6 (0.1%) 5 (0.1%) 4 (0.1%) 2 (<0.1%) 5 (0.1%) 3 (<0.1%) 3 (<0.1%) 2 (<0.1%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) 4 (0.1%) 15 (0.3%) 9 (0.2%) 8 (0.1%) | 73 (2.4%) 10 (0.3%) 6 (0.2%) 5 (0.2%) 6 (0.2%) 3 (0.1%) 3 (0.1%) 1 (<0.1%) 5 (0.2%) 0 (0.0%) 3 (0.1%) 2 (0.1%) 0 (0.0%) 1 (<0.1%) 1 (<0.1%) 0 (0.0%) 3 (0.1%) 10 (0.3%) 8 (0.3%) 6 (0.2%) | 14 (1.9%) 0 (0.0%) 2 (0.3%) 2 (0.3%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 1 (0.1%) 0 (0.0%) 3 (0.4%) 0 (0.0%) 0 (0.0%) 1 (0.1%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 1 (0.1%) 2 (0.3%) 1 (0.1%) 1 (0.1%) | 7 (0.5%) 0 (0.0%) 0 (0.0%) 1 (0.1%) 0 (0.0%) 1 (0.1%) 1 (0.1%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 1 (0.1%) 0 (0.0%) 2 (0.1%) 0 (0.0%) 1 (0.1%) | 2 (1.5%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 1 (0.7%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 1 (0.7%) 0 (0.0%) 0 (0.0%) |
| Corynebacterium spp. C. striatum C. afermentas C. jeikeium C. amycolatum C. coyleae C. falseni C. imitans C. matruchotii C. mucifaciens C. propinquum C. urealyticum Others | 100 (1.9%) 71 (1.3%) 6 (0.1%) 6 (0.1%) 5 (0.1%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) 2 (<0.1%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) 4 (0.1%) | 68 (2.3%) 50 (1.7%) 3 (0.1%) 3 (0.1%) 2 (0.1%) 1 (<0.1%) 0 (0.0%) 1 (<0.1%) 2 (0.1%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) 3 (0.1%) | 11 (1.5%) 8 (1.1%) 2 (0.3%) 0 (0.0%) 1 (0.1%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) | 20 (1.3%) 13 (0.9%) 1 (0.1%) 3 (0.2%) 2 (0.1%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 1 (0.1%) | 1 (0.7%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 1 (0.7%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| Propionibacterium spp. P. acnes Others | 5 (0.1%) 4 (0.1%) 1 (<0.1%) | 2 (0.1%) 2 (0.1%) 0 (0.0%) | 1 (0.1%) 0 (0.0%) 1 (0.1%) | 2 (0.1%) 2 (0.1%) 0 (0.0%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| Haemophilus spp H. parainfluenzae H. influenzae | 4 (0.1%) 2 (<0.1%) 2 (<0.1%) | 3 (0.1%) 2 (0.1%) 1 (<0.1%) | 1 (0.1%) 0 (0.0%) 1 (0.1%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| Listeria monocytogenes | 14 (0.3%) | 8 (0.3%) | 1 (0.1%) | 3 (0.2%) | 2 (1.5%) |
| Brucella spp. | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Salmonella typhi | 3 (<0.1%) | 3 (0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Salmonella spp. | 4 (0.1%) | 3 (0.1%) | 1 (0.1%) | 0 (0.0%) | 0 (0.0%) |
| Vibrio spp. | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Nocardia spp. | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Achromobacter spp. A. xilosoxidans Others | 20 (0.4%) 14 (0.3%) 6 (0.1%) | 7 (0.2%) 5 (0.2%) 2 (0.1%) | 2 (0.3%) 0 (0.0%) 2 (0.3%) | 11 (0.7%) 9 (0.6%) 2 (0.1%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| Aeromonas spp. A. caviae A. veronii A. sobria | 5 (0.1%) 2 (<0.1%) 2 (<0.1%) 1 (<0.1%) | 5 (0.2%) 2 (0.1%) 2 (0.1%) 1 (<0.1%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| Alcaligenes faecalis | 2 (<0.1%) | 1 (<0.1%) | 1 (0.1%) | 0 (0.0%) | 0 (0.0%) |
| Bacillus cereus | 2 (<0.1%) | 2 (0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Bacillus licheniformis | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Bacteroides spp. B. fragilis B. caccae B. theiyaiotamicron | 15 (0.3%) 11 (0.2%) 2 (<0.1%) 2 (<0.1%) | 9 (0.3%) 6 (0.2%) 1 (<0.1%) 2 (0.1%) | 4 (0.5%) 4 (0.5%) 0 (0.0%) 0 (0.0%) | 2 (0.1%) 1 (0.1%) 1 (0.1%) 0 (0.0%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| Brevibacterium spp. B. casei Others | 3 (<0.1%) 2 (<0.1%) 1 (<0.1%) | 3 (0.1%) 2 (0.1%) 1 (<0.1%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| Fusobacterium spp. F. necrophorum F. nucleatum | 3 (<0.1%) 2 (<0.1%) 1 (<0.1%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) | 2 (0.3%) 2 (0.3%) 0 (0.0%) | 1 (0.1%) 0 (0.0%) 1 (0.1%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| Clostridioides spp. C. paraputrificum C. perfringens C. septicum C. sporogenes | 6 (0.1%) 2 (<0.1%) 2 (<0.1%) 1 (<0.1%) 1 (<0.1%) | 5 (0.2%) 2 (0.1%) 1 (<0.1%) 1 (<0.1%) 1 (<0.1%) | 1 (0.1%) 0 (0.0%) 1 (0.1%) 0 (0.0%) 0 (0.0%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| Brevundimonas vesicularis | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Burkholderia spp. | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Cedecea davisae | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 1 (0.1%) | 0 (0.0%) |
| Delftia acidovorans | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Dermabacter hominis | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Escherichia hermannii | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Gemella haemolysans | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Hafnia alvei | 4 (0.1%) | 2 (0.1%) | 0 (0.0%) | 2 (0.1%) | 0 (0.0%) |
| Lactobacillus rhamnosus | 2 (<0.1%) | 0 (0.0%) | 1 (0.1%) | 1 (0.1%) | 0 (0.0%) |
| Lactococcus garvieae | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (0.7%) |
| Leclercia adecarboxylata | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Micrococcus luteus | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Moraxella spp. M. osloensis M. atlantae | 3 (<0.1%) 2 (<0.1%) 1 (<0.1%) | 3 (0.1%) 2 (0.1%) 1 (<0.1%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) |
| Neisseria flavescens | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Ochrobactrum anthropii | 2 (<0.1%) | 1 (<0.1%) | 1 (0.1%) | 0 (0.0%) | 0 (0.0%) |
| Oligella ureolytica | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Pantoea agglomerans | 2 (<0.1%) | 1 (<0.1%) | 1 (0.1%) | 0 (0.0%) | 0 (0.0%) |
| Raoultella ornithinolytica | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Rodococcus equi | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Roseomonas spp. | 3 (<0.1%) | 3(0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Rothia spp. R. mucilaginosa Others | 2 (<0.1%) 1 (<0.1%) 1 (<0.1%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) | 0 (0.0%) 0 (0.0%) 0 (0.0%) | 2 (1.5%) 1 (0.7%) 1 (0.7%) |
| Stomatococcus spp. | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (0.7%) |
| Tissierella praeacuta | 1 (<0.1%) | 1 (<0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Non-identified | 3 (<0.1%) | 3 (0.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Bacteria | N = 5324 | 2018 N = 501 | 2019 N = 580 | 2020 N = 733 | 2021 N = 624 | 2022 N = 819 | 2023 N = 1078 | 2024 N = 989 |
|---|---|---|---|---|---|---|---|---|
| Staphylococci excluding S. aureus | 2113 | 198 | 221 | 322 | 283 | 352 | 404 | 333 |
| Oxacillin | 1716/2100 (81.7%) | 158/196 (80.6%) | 179/221 (81.0%) | 260/319 (81.5%) | 232/283 (82.0%) | 289/352 (82.1%) | 331/398 (83.2%) | 267/331 (80.7%) |
| Ciprofloxacin | 1498/2097 (71.4%) | 148/198 (74.7%) | 157/219 (71.7%) | 230/320 (71.9%) | 202/283 (71.4%) | 246/352 (69.9%) | 295/399 (73.9%) | 220/326 (67.5%) |
| Moxifloxacin | 1428/2054 (69.5%) | 126/184 (68.5%) | 147/210 (70.0%) | 225/318 (70.7%) | 198/282 (70.2%) | 244/350 (69.7%) | 282/392 (71.9%) | 206/318 (64.8%) |
| TMP-SMX | 805/2088 (38.5%) | 60/196 (30.6%) | 81/219 (37.0%) | 117/318 (36.8%) | 133/282 (47.2%) | 139/350 (39.7%) | 158/398 (39.7%) | 117/325 (36.0%) |
| Gentamicin | 1421/2100 (67.7%) | 118/197 (59.9%) | 141/219 (64.4%) | 216/320 (67.5%) | 206/283 (72.8%) | 249/352 (70.7%) | 269/398 (67.6%) | 222/331 (67.0%) |
| Tetracycline | 472/2090 (22.6%) | 48/196 (24.5%) | 52/219 (23.7%) | 90/318 (28.3%) | 78/281 (27.7%) | 57/348 (16.4%) | 66/398 (16.6%) | 81/330 (24.5%) |
| Tigecycline | 108/2062 (5.2%) | 18/184 (9.8%) | 7/208 (3.4%) | 17/316 (5.4%) | 16/280 (5.7%) | 20/350 (5.7%) | 6/398 (1.5%) | 24/326 (7.4%) |
| Clindamycin | 1280/2082 (61.5%) | 105/193 (54.4%) | 119/211 (56.4%) | 181/317 (57.1%) | 181/281 (64.4%) | 217/351 (61.8%) | 262/398 (65.8%) | 215/331 (64.9%) |
| Erythromycin | 1652/2101 (78.6%) | 141/196 (71.9%) | 168/221 (76.0%) | 245/320 (76.6%) | 235/283 (83.0%) | 288/352 (81.8%) | 326/398 (81.9%) | 249/331 (75.2%) |
| Teicoplanin | 242/2046 (11.8%) | 4/191 (2.1%) | 19/215 (8.8%) | 33/313 (10.5%) | 47/280 (16.8%) | 71/328 (21.6%) | 33/394 (8.4%) | 35/325 (10.8%) |
| Vancomycin | 12/2110 (0.6%) | 1/198 (0.5%) | 1/221 (0.4%) | 3/321 (0.9%) | 2/283 (0.7%) | 1/352 (0.3%) | 4/402 (1.0%) | 0/333 (0.0%) |
| Linezolid | 343/2100 (16.3%) | 26/195 (13.3%) | 34/214 (15.9%) | 39/319 (12.2%) | 48/283 (17.0%) | 66/352 (18.7%) | 70/404 (17.3%) | 60/333 (18.0%) |
| Daptomycin | 26/2084 (1.2%) | 2/186 (1.1%) | 3/210 (1.4%) | 2/319 (0.6%) | 5/283 (1.8%) | 2/352 (0.6%) | 7/401 (1.7%) | 5/333 (1.5%) |
| Rifampicin | 849/920 (92.3%) | 52/115 (45.2%) | 65/68 (95.6%) | 116/118 (98.3%) | 128/128 (100%) | 162/163 (99.4%) | 179/180 (99.4%) | 147/148 (99.3%) |
| Fosfomycin | 1105/2075 (53.2%) | 52/186 (27.9%) | 88/209 (42.1%) | 144/318 (45.3%) | 176/283 (62.2%) | 195/351 (55.5%) | 251/398 (63.1%) | 199/330 (60.3%) |
| K. pneumoniae | 646 | 66 | 73 | 89 | 75 | 74 | 123 | 146 |
| TZP | 516/645 (80.0%) | 55/66 (83.3%) | 57/73 (78.1%) | 70/89 (78.6%) | 66/75 (88.0%) | 60/74 (81.1%) | 89/123 (72.3%) | 119/145 (82.1%) |
| Cefepime | 542/644 (84.2%) | 55/66 (83.3%) | 60/73 (82.2%) | 77/89 (86.5%) | 67/75 (89.3%) | 62/72 (86.1%) | 98/123 (79.7%) | 123/146 (84.2%) |
| Ceftazidime | 557/646 (86.2%) | 56/66 (83.3%) | 61/73 (83.6%) | 77/89 (86.5%) | 68/75 (90.7%) | 65/74 (87.8%) | 105/123 (85.4%) | 125/146 (85.6%) |
| CZA | 58/491 (11.8%) | NT | 0/26 (0.0%) | 3/52 (5.8%) | 12/74 (16.2%) | 3/72 (4.2%) | 18/123 (14.6%) | 22/144 (15.3%) |
| CZT | 313/432 (72.4%) | NT | NT | 16/22 (72.7%) | 65/74 (87.8%) | 51/68 (75.0%) | 78/122 (63.9%) | 103/146 (70.5%) |
| Ertapenem | 470/644 (73.0%) | 49/66 (74.2%) | 52/73 (71.2%) | 68/88 (77.3%) | 65/75 (86.7%) | 55/74 (74.3%) | 79/123 (64.2%) | 102/145 (70.3%) |
| Imipenem | 415/641 (64.7%) | 36/65 (55.4%) | 39/73 (53.4%) | 60/87 (69.0%) | 61/75 (81.3%) | 53/74 (71.6%) | 73/123 (59.3%) | 93/144 (64.6%) |
| Meropenem | 404/642 (62.9%) | 36/66 (54.5%) | 41/73 (56.2%) | 57/88 (64.8%) | 60/75 (80.0%) | 52/72 (72.2%) | 74/123 (60.2%) | 84/145 (57.9%) |
| MEV | 41/362 (11.3%) | NT | NT | NT | 0/31 (0.0%) | 0/66 (0.0%) | 21/122 (17.2%) | 20/143 (14.0%) |
| Gentamicin | 401/646 (62.1%) | 39/66 (59.1%) | 36/73 (49.3%) | 64/89 (71.9%) | 57/75 (76.0%) | 47/74 (63.5%) | 59/123 (48.0%) | 99/146 (67.8%) |
| Ciprofloxacin | 536/646 (83.0%) | 54/66 (81.8%) | 59/73 (80.8%) | 77/89 (86.5%) | 66/75 (88.0%) | 65/74 (87.8%) | 93/123 (75.6%) | 122/146 (83.6%) |
| TMP-SMX | 354/645 (54.9%) | 35/66 (53.0%) | 33/73 (45.2%) | 50/88 (56.8%) | 40/75 (53.3%) | 33/74 (44.6%) | 64/123 (52.0%) | 99/146 (67.8%) |
| Fosfomycin | 188/644 (29.2%) | 19/66 (28.8%) | 23/73 (31.5%) | 12/88 (13.6%) | 17/75 (22.7%) | 26/74 (35.1%) | 49/123 (39.8%) | 42/145 (29.0%) |
| Colistin | 45/626 (7.2%) | 3/62 (4.8%) | 4/72 (5.5%) | 14/85 (16.5%) | 11/73 (15.1%) | 4/71 (5.6%) | 8/121 (6.6%) | 1/142 (0.7%) |
| S. aureus | 398 | 38 | 62 | 49 | 38 | 60 | 71 | 80 |
| Oxacillin | 117/398 (29.4%) | 16/38 (42.1%) | 20/62 (32.2%) | 24/49 (49.0%) | 9/38 (23.7%) | 11/60 (18.3%) | 14/71 (19.7%) | 23/80 (28.7%) |
| Ciprofloxacin | 108/396 (27.3%) | 18/38 (47.4%) | 21/62 (33.9%) | 16/48 (33.3%) | 10/38 (26.3%) | 10/60 (16.7%) | 15/70 (21.4%) | 18/80 (22.5%) |
| Moxifloxacin | 103/386 (26.7%) | 16/35 (45.7%) | 21/62 (33.9%) | 16/48 (33.3%) | 10/36 (27.8%) | 10/60 (16.7%) | 13/67 (19.4%) | 17/78 (21.8%) |
| TMP-SMX | 19/395 (4.8%) | 5/37 (13.5%) | 5/62 (8.0%) | 1/48 (2.1%) | 4/38 (10.5%) | 0/59 (0.0%) | 1/71 (1.4%) | 3/80 (3.7%) |
| Gentamicin | 61/396 (15.4%) | 8/38 (21.0%) | 14/62 (22.6%) | 7/48 (14.6%) | 7/38 (18.4%) | 6/60 (10.0%) | 9/70 (12.9%) | 10/80 (12.5%) |
| Tetracycline | 46/396 (11.6%) | 4/38 (10.5%) | 7/62 (11.3%) | 11/48 (22.9%) | 6/38 (15.8%) | 5/60 (8.3%) | 8/70 (11.4%) | 5/80 (6.2%) |
| Tigecycline | 5/392 (1.3%) | 2/35 (5.7%) | 1/61 (1.6%) | 1/48 (2.0%) | 0 (0.0%) | 0 (0.0%) | 1/70 (1.4%) | NT |
| Clindamycin | 137/395 (34.7%) | 11/37 (29.7%) | 26/62 (41.9%) | 15/48 (31.2%) | 14/38 (36.8%) | 24/60 (40.0%) | 18/70 (25.7%) | 29/80 (36.2%) |
| Erythromycin | 146/396 (36.9%) | 14/38 (36.8%) | 26/62 (41.9%) | 19/48 (39.6%) | 15/38 (39.5%) | 23/60 (38.3%) | 19/70 (27.1%) | 30/80 (37.5%) |
| Teicoplanin | 0/396 (0.0%) | 0/38 (0.0%) | 0/62 (0.0%) | 0/48 (0.0%) | 0/38 (0.0%) | 0/60 (0.0%) | 0/70 (0.0%) | 0/80 (0.0%) |
| Vancomycin | 3/398 (0.7%) | 0/38 (0.0%) | 2/62 (3.2%) | 0/49 (0.0%) | 0/38 (0.0%) | 0/60 (0.0%) | 1/71 (1.4%) | 0/80 (0.0%) |
| Linezolid | 1/397 (0.2%) | 0/37 (0.0%) | 0/62 (0.0%) | 0/49 (0.0%) | 0/38 (0.0%) | 0/60 (0.0%) | 0/71 (0.0%) | 1/80 (1.2%) |
| Daptomycin | 6/396 (1.5%) | 0/98 (0.0%) | 0/62 (0.0%) | 1/49 (2.0%) | 2/38 (5.3%) | 1/60 (1.7%) | 2/71 (2.8%) | 0/80 (0.0%) |
| Rifampicin | 29/43 (67.4%) | 5/18 (27.8%) | 6/7 (85.7%) | 2/2 (100%) | 4/4 (100%) | 1/1 (100%) | 4/4 (100%) | 7/7 (100%) |
| Fosfomycin | 27/394 (6.8%) | 5/36 (13.9%) | 4/62 (6.4%) | 3/48 (6.2%) | 3/38 (7.9%) | 4/60 (6.7%) | 5/70 (7.1%) | 3/80 (3.7%) |
| A. baumannii | 396 | 24 | 42 | 49 | 38 | 67 | 90 | 86 |
| Meropenem | 383/395 (97.0%) | 23/24 (95.8%) | 38/42 (90.5%) | 44/48 (91.7%) | 37/38 (97.4%) | 67/67 (100%) | 90/90 (100%) | 84/86 (97.7%) |
| Gentamicin | 349/393 (88.8%) | 23/24 (95.8%) | 38/42 (90.5%) | 43/46 (93.5%) | 37/38 (97.4%) | 53/67 (79.1%) | 73/89 (82.0%) | 82/86 (95.3%) |
| Ciprofloxacin | 379/394 (96.2%) | 24/24 (100%) | 39/40 (97.5%) | 45/46 (97.8%) | 37/38 (97.4%) | 62/62 (100%) | 88/88 (100%) | 84/84 (100%) |
| TMP-SMX | 347/395 (87.8%) | 22/24 (91.7%) | 37/42 (88.1%) | 35/48 (72.9%) | 30/38 (78.9%) | 60/67 (89.5%) | 84/90 (93.3%) | 79/86 (91.9%) |
| Colistin | 8/391 (2.0%) | 0/24 (0.0%) | 3/42 (7.1%) | 0/46 (0.0%) | 3/38 (7.9%) | 0/66 (0.0%) | 1/89 (1.1%) | 1/86 (1.2%) |
| E. coli | 380 | 42 | 59 | 55 | 31 | 37 | 74 | 82 |
| TZP | 69/380 (18.1%) | 5/42 (11.9%) | 10/59 (16.9%) | 4/55 (7.3%) | 4/31 (12.9%) | 8/37 (21.6%) | 21/74 (28.4%) | 17/82 (20.7%) |
| Cefepime | 145/379 (38.2%) | 11/42 (26.2%) | 23/59 (39.0%) | 24/55 (43.6%) | 11/31 (35.5%) | 12/37 (32.4%) | 29/74 (39.2%) | 35/81 (43.2%) |
| Ceftazidime | 142/379 (37.5%) | 10/42 (23.8%) | 20/58 (34.5%) | 22/55 (40.0%) | 9/31 (29.0%) | 14/37 (37.8%) | 31/74 (41.9%) | 36/82 (43.9%) |
| CZA | 5/225 (2.2%) | NT | NT | 0/10 (0.0%) | 0/30 (0.0%) | 1/31 (3.2%) | 2/73 (2.7%) | 2/81 (2.5%) |
| CZT | 26/221 (11.8%) | NT | NT | 0/6 (0.0%) | 2/30 (6.7%) | 3/31 (9.7%) | 9/73 (12.3%) | 12/81 (14.8%) |
| Ertapenem | 24/378 (6.3%) | 2/42 (4.8%) | 2/59 (3.4%) | 0/55 (0.0%) | 1/31 (3.2%) | 4/37 (10.8%) | 8/74 (10.8%) | 7/80 (8.7%) |
| Imipenem | 8/376 (2.1%) | 2/42 (4.8%) | 0/59 (0.0%) | 0/55 (0.0%) | 0/30 (0.0%) | 1/37 (2.7%) | 4/74 (5.4%) | 1/82 (1.2%) |
| Meropenem | 6/380 (1.6%) | 2/42 (4.8%) | 0/59 (0.0%) | 0/55 (0.0%) | 1/31 (3.2%) | 1/36 (2.8%) | 1/74 (1.3%) | 1/81 (1.2%) |
| MVB | 2/127 (1.6%) | NT | NT | NT | 0/1 (0%) | 1/17 (5.9%) | 1/55 (1.8%) | 0/54 (0.0%) |
| Gentamicin | 81/380 (21.3%) | 4/42 (9.5%) | 11/59 (18.6%) | 19/55 (34.5%) | 8/31 (25.8%) | 8/37 (21.6%) | 13/74 (17.6%) | 18/82 (21.9%) |
| Ciprofloxacin | 216/380 (56.8%) | 26/42 (61.9%) | 37/59 (62.7%) | 32/55 (58.2%) | 15/31 (48.4%) | 17/37 (45.9%) | 39/74 (52.7%) | 50/82 (61.0%) |
| TMP-SMX | 174/376 (46.3%) | 19/42 (45.2%) | 30/59 (50.8%) | 25/54 (46.3%) | 14/31 (45.2%) | 16/37 (43.2%) | 37/73 (50.7%) | 33/80 (41.2%) |
| Fosfomycin | 2/376 (0.5%) | 1/42 (2.4%) | 0/59 (0.0%) | 0/52 (0.0%) | 0/30 (0.0%) | 0/37 (0.0%) | 1/74 (1.3%) | 0/82 (0.0%) |
| Colistin | 5/348 (1.4%) | 4/38 (10.5%) | 0/54 (0.0%) | 0/51 (0.0%) | 1/28 (3.6%) | 0/34 (0.0%) | 0/65 (0.0%) | 0/78 (0.0%) |
| Tigecycline | 65/281 (23.1%) | 0/42 (0.0%) | 0/59 (0.0%) | 10/48 (20.8%) | 0/1 (0.0%) | 4/24 (16.7%) | 25/52 (48.1%) | 26/55 (47.3%) |
| E. faecalis | 234 | 24 | 16 | 37 | 37 | 37 | 46 | 37 |
| Ampicillin | 0/86 (0.0%) | 0/1 (0.0%) | NT | 0/1 (0.0%) | NT | 0/3 (0.0%) | 0/46 (0.0%) | 0/37 (0.0%) |
| Imipenem | 23/234 (9.8%) | 0/24 (0.0%) | 0/16 (0.0%) | 3/34 (8.8%) | 11/26 (42.3%) | 6/31 (19.3%) | 3/43 (7.0%) | 0/37 (0.0%) |
| Gentamicin | 112/234 (47.9%) | 15/24 (62.5%) | 9/16 (56.2%) | 19/37 (51.3%) | 18/37 (48.6%) | 18/37 (48.6%) | 21/46 (45.6%) | 12/37 (32.4%) |
| Teicoplanin | 0/234 (0.0%) | (0.0%) | (0.0%) | (0.0%) | (0.0%) | (0.0%) | (0.0%) | (0.0%) |
| Vancomycin | 0/233 (0%) | (0.0%) | (0.0%) | (0.0%) | (0.0%) | (0.0%) | (0.0%) | (0.0%) |
| Linezolid | 0/234 (0%) | (0.0%) | (0.0%) | (0.0%) | (0.0%) | (0.0%) | (0.0%) | (0.0%) |
| Tigecycline | 1/233 (0.4%) | 0/24 (0.0%) | 0/16 (0.0%) | 1/37 (2.7%) | 0/37 (0.0%) | 0/37 (0.0%) | 0/46 (0.0%) | 0/36 (0.0%) |
| P. aeruginosa | 224 | 12 | 22 | 33 | 19 | 32 | 67 | 39 |
| TZP | 33/224 (14.7%) | 5/12 (41.7%) | 1/22 (4.5%) | 6/33 (18.2%) | 3/19 (15.8%) | 2/32 (6.2%) | 13/67 (19.4%) | 3/39 (7.7%) |
| Cefepime | 41/219 (18.7%) | 6/12 (50.0%) | 2/21 (9.5%) | 8/33 (24.2%) | 4/19 (21.0%) | 2/32 (6.2%) | 14/63 (22.2%) | 5/39 (12.8%) |
| Ceftazidime | 43/122 (35.2%) | 8/12 (66.7%) | 1/22 (4.5%) | 6/33 (18.2%) | 4/19 (21.0%) | 5/32 (15.6%) | 14/65 (21.5%) | 5/39 (12.8%) |
| CZA | 2/145 (1.4%) | NT | NT | 0/3 (0.0%) | 0/16 (0.0%) | 1/31 (3.2%) | 1/60 (1.7%) | 0/35 (0.0%) |
| CTZ | 3/136 (2.2%) | NT | NT | 0/3 (0.0%) | 0/16 (0.0%) | 0/25 (0.0%) | 2/58 (3.4%) | 1/34 (2.9%) |
| Imipenem | 56/217 (25.8%) | 7/12 (58.3%) | 6/22 (27.3%) | 10/33 (30.3%) | 4/19 (21.0%) | 5/32 (15.6%) | 16/61 (26.2%) | 8/38 (21.0%) |
| Meropenem | 36/222 (16.2%) | 7/12 (58.3%) | 6/21 (28.6%) | 6/33 (18.2%) | 3/19 (15.8%) | 5/32 (15.6%) | 8/66 (12.1%) | 1/39 (2.6%) |
| Tobramycin | 26/221 (11.8%) | 8/12 (66.7%) | 3/22 (13.6%) | 7/33 (21.2%) | 0/19 (0.0%) | 3/31 (9.7%) | 4/67 (6.0%) | 1/37 (2.7%) |
| Amikacin | 3/106 (2.8%) | NT | 0/1 | NT | NT | NT | 3/67 | 0/39 |
| Gentamicin | 16/58 (27.6%) | 8/12 (66.7%) | 3/22 (13.6%) | 5/21 (23.8%) | NT | NT | 0/2 (0%) | 0/1 (0%) |
| Ciprofloxacin | 41/223 (18.4%) | 8/12 (66.7%) | 4/22 (18.2%) | 11/33 (33.3%) | 4/19 (21.0%) | 3/32 (9.4%) | 8/67 (11.9%) | 3/38 (7.9%) |
| Colistin | 2/218 (0.9%) | 0/12 (0.0%) | 0/22 (0.0%) | 0/30 (0.0%) | 0/19 (0.0%) | 0/32 (0.0%) | 2/65 (3.1%) | 0/38 (0.0%) |
| E. faecium | 193 | 16 | 20 | 27 | 42 | 20 | 36 | 32 |
| Ampicillin | 56/67 (83.6%) | NT | NT | NT | NT | NT | 32/36 (88.9%) | 24/31 (77.4%) |
| Imipenem | 171/192 (89.0%) | 13/16 (81.2%) | 17/20 (85.0%) | 26/27 (96.3%) | 37/42 (88.1%) | 19/20 (95.0%) | 33/36 (91.7%) | 26/31 (83.9%) |
| Gentamicin | 112/192 (58.3%) | 6/16 (37.5%) | 12/20 (60.0%) | 15/27 (55.5%) | 23/42 (54.8%) | 17/20 (85.0%) | 21/36 (58.3%) | 18/31 (58.1%) |
| Teicoplanin | 52/192 (27.1%) | 0/16 (0.0%) | 0/20 (0.0%) | 4/27 (14.8%) | 9/42 (21.4%) | 8/20 (40.0%) | 14/36 (38.9%) | 17/31 (54.8%) |
| Vancomycin | 52/193 (26.9%) | 0/16 (0.0%) | 1/20 (5.0%) | 4/27 (14.8%) | 9/42 (21.4%) | 7/20 (35.0%) | 14/36 (38.9%) | 17/32 (53.1%) |
| Linezolid | 6/193 (3.1%) | 0/16 (0.0%) | 1/20 (5.0%) | 0/27 (0.0%) | 0/42 (0.0%) | 0/20 (0.0%) | 3/36 (8.3%) | 2/32 (6.2%) |
| Q-D | 46/104 (44.2%) | NT | NT | 0/2 (0.0%) | 1/28 (3.6%) | 0/14 (0.0%) | 19/34 (55.9%) | 26/26 (100%) |
| Enterobacter spp. | 122 | 20 | 15 | 11 | 8 | 25 | 30 | 13 |
| Cefepime | 33/122 (27.0%) | 5/20 (25.0%) | 3/15 (20.0%) | 5/11 (45.4%) | 3/8 (37.5%) | 5/25 (20.0%) | 10/30 (33.3%) | 2/13 (15.4%) |
| Meropenem | 8/122 (6.5%) | 0/20 (0.0%) | 1/15 (6.7%) | 2/11 (18.2%) | 0/8 (0.0%) | 0/25 (0.0%) | 4/30 (13.3%) | 1/13 (7.7%) |
| Gentamicin | 20/122 (16.4%) | 3/20 (15.0%) | 2/15 (13.3%) | 4/11 (36.4%) | 3/8 (37.5%) | 2/25 (8.0%) | 5/30 (16.7%) | 1/13 (7.7%) |
| Ciprofloxacin | 27/122 (22.1%) | 8/20 (40.0%) | 4/15 (26.7%) | 4/11 (36.4%) | 2/8 (25.0%) | 2/25 (8.0%) | 5/30 (16.7%) | 2/13 (15.4%) |
| TMP-SMX | 27/122 (22.1%) | 7/20 (35.0%) | 3/15 (20.0%) | 4/11 (36.4%) | 3/8 (37.5%) | 3/25 (12.0%) | 5/30 (16.7%) | 2/13 (15.4%) |
| Fosfomycin | 15/122 (12.3%) | 0/20 (0.0%) | 2/15 (13.3%) | 3/11 (27.3%) | 0/8 (0.0%) | 3/25 (12.0%) | 4/30 (13.3%) | 3/13 (23.0%) |
| Colistin | 4/118 (3.4%) | 0/19 (0.0%) | 0/15 (0.0%) | 0/11 (0.0%) | 0/6 (0.0%) | 0/25 (0.0%) | 4/29 (13.8%) | 0/13 (0.0%) |
| Pathogens | All Wards | ICU | Medical Wards | Surgery | NICU |
|---|---|---|---|---|---|
| A. baumannii | 241/396 (60.9%) | 183/208 (88.0%) | 55/154 (35.7%) | 3/34 (8.8%) | 0 |
| S. maltophilia | 23/44 (52.3%) | 20/22 (90.9%) | 3/16 (18.8%) | 0/5 (0.0%) | 0/1 (0.0%) |
| P. aeruginosa | 108/224 (48.2%) | 87/111 (78.4%) | 14/81 (17.3%) | 7/30 (23.3%) | 0/2 (0.0%) |
| K. pneumoniae | 282/646 (43.7%) | 185/235 (78.7%) | 81/311 (26.0%) | 10/91 (11.0%) | 6/9 (66.7%) |
| Serratia spp. | 28/70 (40.0%) | 28/38 (73.7%) | 0/18 (0.0%) | 0/13 (0.0%) | 0/1 (0.0%) |
| E. faecium | 76/193 (39.4%) | 42/50 (84.0%) | 32/112 (28.6%) | 2/28 (7.1%) | 0/3 (0.0%) |
| Proteus spp. | 20/57 (35.1%) | 11/13 (84.6%) | 8/27 (22.9%) | 1/9 (11.1%) | 0 |
| E. faecalis | 82/234 (35.0%) | 55/69 (79.7%) | 22/115 (19.1%) | 4/43 (9.3%) | 1/7 (14.3%) |
| Corynebacterium spp. | 32/100 (32.0%) | 15/20 (75.0%) | 17/68 (25.0%) | 0/11 (0.0%) | 0/1 (0.0%) |
| S. aureus | 97/398 (24.4%) | 48/57 (84.2%) | 46/281 (16.4%) | 3/55 (5.5%) | 0/5 (0.0%) |
| Enterobacter spp. | 29/122 (23.8%) | 23/30 (76.7%) | 4/64 (6.3%) | 1/22 (4.5%) | 1/6 (16.7%) |
| E. coli | 70/380 (18.4%) | 38/38 (100%) | 29/289 (10.0%) | 3/50 (6.0%) | 0/3 (0.0%) |
| Streptococcus spp. | 13/96 (13.5%) | 6/7 (85.7%) | 7/73 (9.6%) | 0/14 (0.0%) | 0/2 (0.0%) |
| Variables | aOR | CI | p |
|---|---|---|---|
| Age | 1.030 | 1.019–1.040 | <0.001 |
| Male | 0.750 | 0.602–0.934 | 0.010 |
| ICU | 33.450 | 24.585–45.512 | <0.001 |
| CCI | 1.011 | 0.934–1.094 | 0.791 |
| Septic shock | 5.819 | 4.203–8.058 | 0.791 |
| Pneumonia | 1.964 | 1.542–2.502 | <0.001 |
| Endocarditis | 1.062 | 0.638–1.769 | 0.817 |
| CNSI | 2.840 | 1.309–6.162 | 0.008 |
| COVID-19 | 1.404 | 0.799–0.455 | 0.436 |
| Number of isolates | 0.955 | 0.908–1.006 | 0.082 |
| A. baumannii | 1.655 | 1.143–2.397 | 0.008 |
| K. pneumoniae | 1.393 | 1.032–1.881 | 0.030 |
| P. aeruginosa | 1.241 | 0.760–2.025 | 0.388 |
| S. maltophilia | 1.497 | 0.547–4.092 | 0.432 |
| Constant | 0.017 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pipitò, L.; Bono, E.; Mazzola, C.V.; Rubino, R.; Anastasia, A.; Distefano, S.A.; Firenze, A.; Giammanco, G.M.; Bonura, C.; Cascio, A. Outcome of Bloodstream Infections Caused by Antibiotic-Resistant Bacteria: A 7-Year Retrospective Study at the University Hospital of Palermo, Italy. Antibiotics 2025, 14, 464. https://doi.org/10.3390/antibiotics14050464
Pipitò L, Bono E, Mazzola CV, Rubino R, Anastasia A, Distefano SA, Firenze A, Giammanco GM, Bonura C, Cascio A. Outcome of Bloodstream Infections Caused by Antibiotic-Resistant Bacteria: A 7-Year Retrospective Study at the University Hospital of Palermo, Italy. Antibiotics. 2025; 14(5):464. https://doi.org/10.3390/antibiotics14050464
Chicago/Turabian StylePipitò, Luca, Eleonora Bono, Chiara Vincenza Mazzola, Raffaella Rubino, Antonio Anastasia, Salvatore Antonino Distefano, Alberto Firenze, Giovanni M. Giammanco, Celestino Bonura, and Antonio Cascio. 2025. "Outcome of Bloodstream Infections Caused by Antibiotic-Resistant Bacteria: A 7-Year Retrospective Study at the University Hospital of Palermo, Italy" Antibiotics 14, no. 5: 464. https://doi.org/10.3390/antibiotics14050464
APA StylePipitò, L., Bono, E., Mazzola, C. V., Rubino, R., Anastasia, A., Distefano, S. A., Firenze, A., Giammanco, G. M., Bonura, C., & Cascio, A. (2025). Outcome of Bloodstream Infections Caused by Antibiotic-Resistant Bacteria: A 7-Year Retrospective Study at the University Hospital of Palermo, Italy. Antibiotics, 14(5), 464. https://doi.org/10.3390/antibiotics14050464








