Antimicrobial Resistance Patterns of ESBL-Producing Escherichia coli in Dogs from Thailand: Evaluation of Algal Extracts as Novel Antimicrobial Agents
Abstract
1. Introduction
2. Results
2.1. E. coli Detection and Antimicrobial Susceptibility Testing
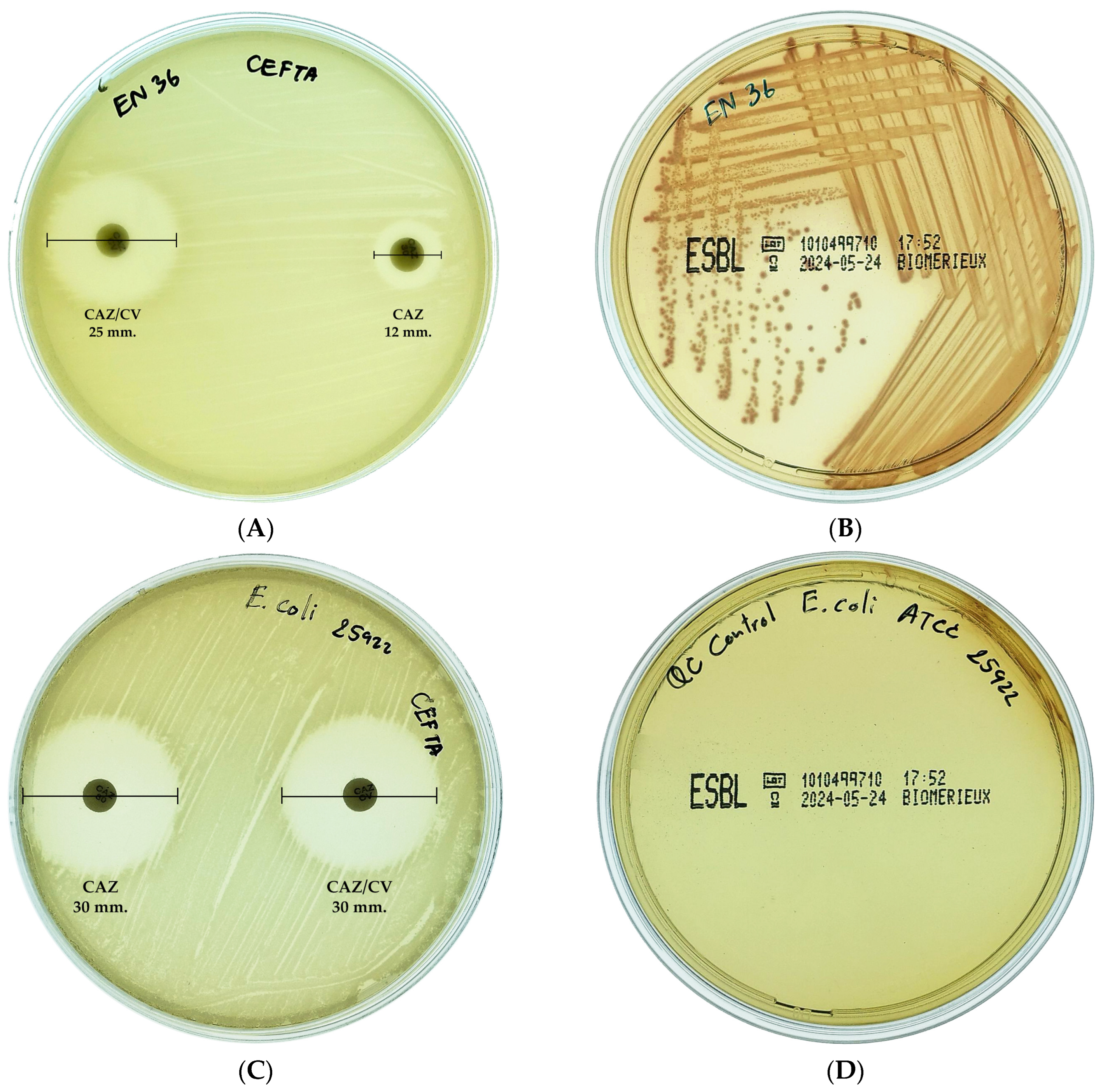
2.2. ESBL-Producing E. coli
2.3. Detection of Beta-Lactamase Genes
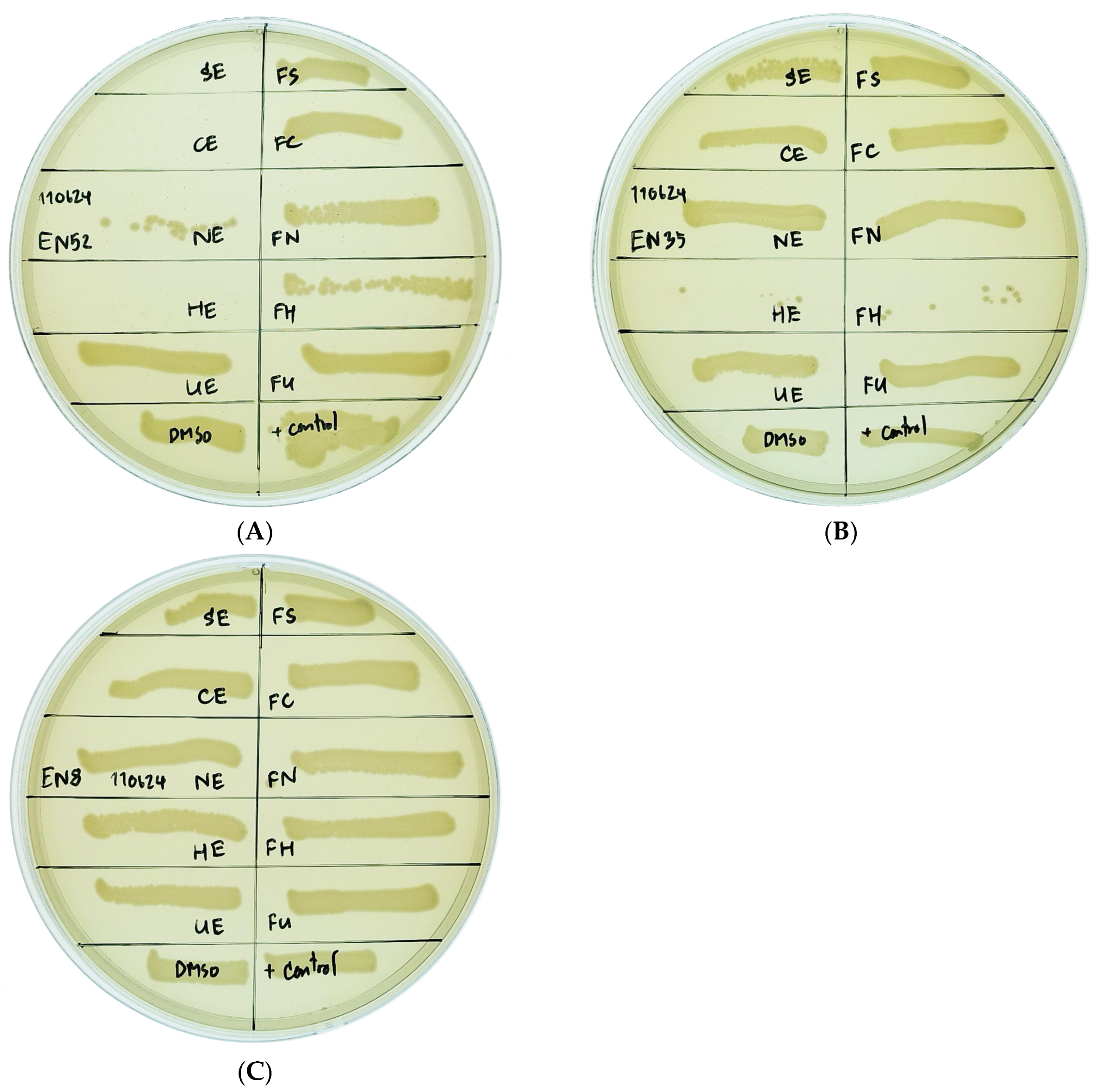
2.4. Effect of Algal Extract on Inhibition of Multidrug-Resistant E. coli Isolates
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Identification of E. coli
4.2. Antimicrobial Susceptibility Testing
4.3. Multiple Antibiotic Resistance (MAR) Index Calculation
4.4. Detection of ESBL-Producing E. coli
4.5. Detection of Beta-Lactamase Genes from MDR E. coli Isolates
| Genes | Primer Sequences (5′-3′) | Estimated Product Size (bp) | Reference |
|---|---|---|---|
| uidA | F-TGGTAATTACCGACGAAAACGGC | 147 | [38] |
| R-ACGCGTGGTTACAGTCTTGCG | |||
| blaTEM | F-ACGCTCACCGGCTCCAGATTAT | 445 | [41] |
| R-TCGCCGCATACACTATTCTCAGA | |||
| blaCTX-M | F-ATGTGAGYACCAGTAARGTGAT | 593 | [43] |
| R-TGGGTRAARTARGTSACCAGAAT | |||
| blaSHV | F-TGCTTTGTTATTCGGGCCAA | 747 | [44] |
| R-ATGCGTTATATTCGCTGTG |
4.6. Preparation of Crude Extracts
4.6.1. Biomass Preparation
4.6.2. Extraction Procedures
4.7. Antibacterial Activity of Algal Extracts Against Multidrug-Resistant E. coli Isolates
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Paul, D.; Gopal, J.; Kumar, M.; Manikandan, M. Nature to the natural rescue: Silencing microbial chats. Chem.-Biol. Interact. 2018, 280, 86–98. [Google Scholar] [CrossRef]
- Woolhouse, M.; Farrar, J. Policy: An intergovernmental panel on antimicrobial resistance. Nature 2014, 509, 555–557. [Google Scholar] [CrossRef]
- Wu, G.; Day, M.J.; Mafura, M.T.; Nunez-Garcia, J.; Fenner, J.J.; Sharma, M.; van Essen-Zandbergen, A.; Rodríguez, I.; Dierikx, C.; Kadlec, K.; et al. Comparative Analysis of ESBL-Positive Escherichia coli Isolates from Animals and Humans from the UK, The Netherlands and Germany. PLoS ONE 2013, 8, e75392. [Google Scholar] [CrossRef]
- Naghavi, M.; Mestrovic, T.; Gray, A.; Gershberg Hayoon, A.; Swetschinski, L.R.; Robles Aguilar, G.; Davis Weaver, N.; Ikuta, K.S.; Chung, E.; Wool, E.E.; et al. Global burden associated with 85 pathogens in 2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet Infect. Dis. 2024, 24, 868–895. [Google Scholar] [CrossRef]
- Hackmann, C.; Gastmeier, P.; Schwarz, S.; Lübke-Becker, A.; Bischoff, P.; Leistner, R. Pet husbandry as a risk factor for colonization or infection with MDR organisms: A systematic meta-analysis. J. Antimicrob. Chemother. 2021, 76, 1392–1405. [Google Scholar] [CrossRef]
- Mayer, A.M.S.; Guerrero, A.J.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Nakamura, F.; Fusetani, N. Marine Pharmacology in 2016–2017: Marine Compounds with Antibacterial, Antidiabetic, Antifungal, Anti-Inflammatory, Antiprotozoal, Antituberculosis and Antiviral Activities; Affecting the Immune and Nervous Systems, and Other Miscellaneous Mechanisms of Action. Mar. Drugs 2021, 19, 49. [Google Scholar] [CrossRef]
- Mir, R.; Salari, S.; Najimi, M.; Rashki, A. Determination of frequency, multiple antibiotic resistance index and resistotype of Salmonella spp. in chicken meat collected from southeast of Iran. Vet. Med. Sci. 2022, 8, 229–236. [Google Scholar] [CrossRef]
- Chen, C.; Li, Y.; Wu, Z.; Ruan, Y.; Long, T.; Wang, X.; Li, W.; Ren, H.; Liao, X.; Liu, Y.; et al. Cat and dog feces as reservoirs of diverse novel antibiotic resistance genes. Environ. Res. 2024, 261, 119690. [Google Scholar] [CrossRef]
- Hartantyo, S.H.P.; Chau, M.L.; Fillon, L.; Ariff, A.Z.B.M.; Kang, J.S.L.; Aung, K.T.; Gutiérrez, R.A. Sick pets as potential reservoirs of antibiotic-resistant bacteria in Singapore. Antimicrob. Resist. Infect. Control 2018, 7, 106. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, L.; Buldain, D.; Gortari Castillo, L.; Buchamer, A.; Chirino-Trejo, M.; Mestorino, N. Pet and Stray Dogs as Reservoirs of Antimicrobial-Resistant Escherichia coli. Int. J. Microbiol. 2021, 2021, 6664557. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Hu, Y.; Zhang, X.; Zhong, W.; Pan, S.; Wang, L.; Zhou, Z.; Liu, H.; Zhang, S.; Peng, G.; et al. Characteristics of MDR E. coli strains isolated from Pet Dogs with clinic diarrhea: A pool of antibiotic resistance genes and virulence-associated genes. PLoS ONE 2024, 19, e0298053. [Google Scholar] [CrossRef]
- Zhou, Y.; Ji, X.; Liang, B.; Jiang, B.; Li, Y.; Yuan, T.; Zhu, L.; Liu, J.; Guo, X.; Sun, Y. Antimicrobial Resistance and Prevalence of Extended Spectrum β-Lactamase-Producing Escherichia coli from Dogs and Cats in Northeastern China from 2012 to 2021. Antibiotics 2022, 11, 1506. [Google Scholar] [CrossRef]
- Moon, B.-Y.; Ali, M.S.; Kwon, D.-H.; Heo, Y.-E.; Hwang, Y.-J.; Kim, J.-I.; Lee, Y.J.; Yoon, S.-S.; Moon, D.-C.; Lim, S.-K. Antimicrobial Resistance in Escherichia coli Isolated from Healthy Dogs and Cats in South Korea, 2020–2022. Antibiotics 2024, 13, 27. [Google Scholar] [CrossRef]
- Schmidt, V.M.; Pinchbeck, G.L.; Nuttall, T.; McEwan, N.; Dawson, S.; Williams, N.J. Antimicrobial resistance risk factors and characterisation of faecal E. coli isolated from healthy Labrador retrievers in the United Kingdom. Prev. Vet. Med. 2015, 119, 31–40. [Google Scholar] [CrossRef]
- Buranasinsup, S.; Wiratsudakul, A.; Chantong, B.; Maklon, K.; Suwanpakdee, S.; Jiemtaweeboon, S.; Sakcamduang, W. Prevalence and characterization of antimicrobial-resistant Escherichia coli isolated from veterinary staff, pets, and pet owners in Thailand. J. Infect. Public Health 2023, 16, 194–202. [Google Scholar] [CrossRef]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Habib, I.; Mohteshamuddin, K.; Mohamed, M.-Y.I.; Lakshmi, G.B.; Abdalla, A.; Bakhit Ali Alkaabi, A. Domestic Pets in the United Arab Emirates as Reservoirs for Antibiotic-Resistant Bacteria: A Comprehensive Analysis of Extended-Spectrum Beta-Lactamase Producing Escherichia coli Prevalence and Risk Factors. Animals 2023, 13, 1587. [Google Scholar] [CrossRef]
- Liakopoulos, A.; Betts, J.; La Ragione, R.; van Essen-Zandbergen, A.; Ceccarelli, D.; Petinaki, E.; Koutinas, C.K.; Mevius, D.J. Occurrence and characterization of extended-spectrum cephalosporin-resistant Enterobacteriaceae in healthy household dogs in Greece. J. Med. Microbiol. 2018, 67, 931–935. [Google Scholar] [CrossRef]
- Carvalho, I.; Cunha, R.; Martins, C.; Martínez-Álvarez, S.; Safia Chenouf, N.; Pimenta, P.; Pereira, A.R.; Ramos, S.; Sadi, M.; Martins, Â.; et al. Antimicrobial Resistance Genes and Diversity of Clones among Faecal ESBL-Producing Escherichia coli Isolated from Healthy and Sick Dogs Living in Portugal. Antibiotics 2021, 10, 1013. [Google Scholar] [CrossRef]
- Yousfi, M.; Mairi, A.; Touati, A.; Hassissene, L.; Brasme, L.; Guillard, T.; De Champs, C. Extended spectrum β-lactamase and plasmid mediated quinolone resistance in Escherichia coli fecal isolates from healthy companion animals in Algeria. J. Infect. Chemother. 2016, 22, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Iseppi, R.; Di Cerbo, A.; Messi, P.; Sabia, C. Antibiotic Resistance and Virulence Traits in Vancomycin-Resistant Enterococci (VRE) and Extended-Spectrum β-Lactamase/AmpC-producing (ESBL/AmpC) Enterobacteriaceae from Humans and Pets. Antibiotics 2020, 9, 152. [Google Scholar] [CrossRef]
- Kawamura, K.; Sugawara, T.; Matsuo, N.; Hayashi, K.; Norizuki, C.; Tamai, K.; Kondo, T.; Arakawa, Y. Spread of CTX-Type Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolates of Epidemic Clone B2-O25-ST131 Among Dogs and Cats in Japan. Microb. Drug Resist. 2017, 23, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.S.; Song, W.; Park, H.-M.; Oh, J.-Y.; Chae, J.-C.; Jeong, S. Molecular Characterization of Fecal Extended-Spectrum β-Lactamase- and AmpC β-Lactamase-Producing Escherichia coli From Healthy Companion Animals and Cohabiting Humans in South Korea. Front. Microbiol. 2020, 11, 674. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Bethe, A.; Semmler, T.; Guenther, S.; Wieler, L.H. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 2012, 18, 646–655. [Google Scholar] [CrossRef]
- Carattoli, A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013, 303, 298–304. [Google Scholar] [CrossRef]
- Cui, L.; Zhao, X.; Li, R.; Han, Y.; Hao, G.; Wang, G.; Sun, S. Companion Animals as Potential Reservoirs of Antibiotic Resistant Diarrheagenic Escherichia coli in Shandong, China. Antibiotics 2022, 11, 828. [Google Scholar] [CrossRef]
- Sun, Y.; Zeng, Z.; Chen, S.; Ma, J.; He, L.; Liu, Y.; Deng, Y.; Lei, T.; Zhao, J.; Liu, J.H. High prevalence of blaCTX-M extended-spectrum β-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2010, 16, 1475–1481. [Google Scholar] [CrossRef]
- Doi, Y.; Adams-Haduch, J.; Peleg, A.; D’Agata, E. The role of horizontal gene transfer in the dissemination of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates in an endemic setting. Diagn. Microbiol. Infect. Dis. 2012, 74, 34–38. [Google Scholar] [CrossRef]
- Santoyo, S.; Rodríguez-Meizoso, I.; Cifuentes, A.; Jaime, L.; García-Blairsy Reina, G.; Señorans, F.J.; Ibáñez, E. Green processes based on the extraction with pressurized fluids to obtain potent antimicrobials from Haematococcus pluvialis microalgae. LWT-Food Sci. Technol. 2009, 42, 1213–1218. [Google Scholar] [CrossRef]
- Cherrington, C.A.; Hinton, M.; Mead, G.C.; Chopra, I. Organic Acids: Chemistry, Antibacterial Activity and Practical Applications. In Advances in Microbial Physiology; Rose, A.H., Tempest, D.W., Eds.; Academic Press: Cambridge, MA, USA, 1991; Volume 32, pp. 87–108. [Google Scholar]
- Pan-utai, W.; Boonpok, S.; Pornpukdeewattana, S. Combination of mechanical and chemical extraction of astaxanthin from Haematococcus pluvialis and its properties of microencapsulation. Biocatal. Agric. Biotechnol. 2021, 33, 101979. [Google Scholar] [CrossRef]
- Darwesh, O.M.; Mahmoud, R.H.; Abdo, S.M.; Marrez, D.A. Isolation of Haematococcus lacustris as source of novel anti-multi-antibiotic resistant microbes agents; fractionation and identification of bioactive compounds. Biotechnol. Rep. 2022, 35, e00753. [Google Scholar] [CrossRef]
- Rayamajhi, V.; Byeon, H.; An, Y.; Kim, T.; Lee, J.; Lee, J.; Lee, K.; Kim, C.; Shin, H.; Jung, S. Screening and Selection of Antibiotics for Enhanced Production of Astaxanthin by Haematococcus lacustris. Life 2024, 14, 977. [Google Scholar] [CrossRef]
- Yap, W.-F.; Tay, V.; Tan, S.-H.; Yow, Y.-Y.; Chew, J. Decoding Antioxidant and Antibacterial Potentials of Malaysian Green Seaweeds: Caulerpa racemosa and Caulerpa lentillifera. Antibiotics 2019, 8, 152. [Google Scholar] [CrossRef]
- Nagappan, T.; Vairappan, C.S. Nutritional and bioactive properties of three edible species of green algae, genus Caulerpa (Caulerpaceae). J. Appl. Phycol. 2014, 26, 1019–1027. [Google Scholar] [CrossRef]
- Nurkolis, F.; Taslim, N.A.; Qhabibi, F.R.; Kang, S.; Moon, M.; Choi, J.; Choi, M.; Park, M.N.; Mayulu, N.; Kim, B. Ulvophyte Green Algae Caulerpa lentillifera: Metabolites Profile and Antioxidant, Anticancer, Anti-Obesity, and In Vitro Cytotoxicity Properties. Molecules 2023, 28, 1365. [Google Scholar] [CrossRef]
- Lomartire, S.; Gonçalves, A.M.M. An Overview on Antimicrobial Potential of Edible Terrestrial Plants and Marine Macroalgae Rhodophyta and Chlorophyta Extracts. Mar. Drugs 2023, 21, 163. [Google Scholar] [CrossRef]
- Tantawiwat, S.; Tansuphasiri, U.; Wongwit, W.; Wongchotigul, V.; Kitayaporn, D. Development of multiplex PCR for the de-tection of total coliform bacteria for Escherichia coli and Clostridium perfringens in drinking water. Southeast Asian J. Trop. Med. Public Health 2005, 36, 162–169. [Google Scholar] [PubMed]
- CLSI Supplement M100; Performance Standards for Antimicrobial Susceptibility Testing. CLSI: Wayne, PA, USA, 2020.
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Monstein, H.J.; ÖStholm-Balkhed, Å.; Nilsson, M.V.; Nilsson, M.; Dornbusch, K.; Nilsson, L.E. Multiplex PCR amplification assay for the detection of SHV, TEM and CTX-M genes in Enterobacteriaceae. APMIS 2007, 115, 1400–1408. [Google Scholar] [CrossRef]
- Chaisaeng, S.; Chopjitt, P.; Kasemsiri, P.; Putthanachote, N.; Boueroy, P.; Takeuchi, D.; Akeda, Y.; Hamada, S.; Kerdsin, A. High prevalence of ESBL-producing E. coli phylogroup B2 clinical isolates in northeastern Thailand. BMC Microbiol. 2024, 24, 425. [Google Scholar] [CrossRef]
- Boyd David, A.; Tyler, S.; Christianson, S.; McGeer, A.; Muller Matthew, P.; Willey Barbara, M.; Bryce, E.; Gardam, M.; Nordmann, P.; Mulvey Michael, R. Complete Nucleotide Sequence of a 92-Kilobase Plasmid Harboring the CTX-M-15 Extended-Spectrum Beta-Lactamase Involved in an Outbreak in Long-Term-Care Facilities in Toronto, Canada. Antimicrob. Agents Chemother. 2004, 48, 3758–3764. [Google Scholar] [CrossRef] [PubMed]
- Paterson David, L.; Hujer Kristine, M.; Hujer Andrea, M.; Yeiser, B.; Bonomo Michael, D.; Rice Louis, B.; Bonomo Robert, A. Extended-Spectrum β-Lactamases in Klebsiella pneumoniae Bloodstream Isolates from Seven Countries: Dominance and Widespread Prevalence of SHV- and CTX-M-Type β-Lactamases. Antimicrob. Agents Chemother. 2003, 47, 3554–3560. [Google Scholar] [CrossRef] [PubMed]
- Pan-utai, W.; Iamtham, S.; Boonbumrung, S.; Mookdasanit, J. Improvement in the Sequential Extraction of Phycobiliproteins from Arthrospira platensis Using Green Technologies. Life 2022, 12, 1896. [Google Scholar] [CrossRef] [PubMed]
- Phomkaivon, N.; Pongponpai, P.; Kosawatpat, P.; Thongdang, B.; Pan-utai, W. Extraction, Characterisation and Evaluation of Antioxidant and Probiotic Growth Potential of Water-Soluble Polysaccharides from Ulva rigida Macroalgae. Foods 2024, 13, 1630. [Google Scholar] [CrossRef]
- Pan-utai, W.; Satmalee, P.; Saah, S.; Paopun, Y.; Tamtin, M. Brine-Processed Caulerpa lentillifera Macroalgal Stability: Physicochemical, Nutritional and Microbiological Properties. Life 2023, 13, 2112. [Google Scholar] [CrossRef]
- CLSI M07-A9; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. CLSI: Wayne, PA, USA, 2012.
| Antimicrobial Class | Antimicrobial Agent (µg) | No. of Resistant Strains (%) | No. of Intermediate Strains (%) | No. of Susceptible Strains (%) |
|---|---|---|---|---|
| Penicillin | Ampicillin (10) | 34 (79.07) | 0 (0) | 9 (20.93) |
| β-Lactam combination agents | Amoxicillin–clavulanate (20/10) | 16 (37.21) | 4 (9.3) | 23 (53.49) |
| Cephems | Cefepime (30) | 8 (18.60) | 4 (9.3) | 31 (72.09) |
| Cefotaxime (30) | 14 (32.56) | 2 (4.65) | 27 (62.79) | |
| Ceftriaxone (30) | 14 (32.56) | 1 (2.33) | 28 (65.12) | |
| Carbapenem | Meropenem (10) | 1 (2.33) | 7 (16.28) | 35 (81.40) |
| Aminoglycosides | Gentamicin (10) | 16 (37.21) | 1 (2.33) | 26 (60.47) |
| Amikacin (30) | 7 (16.28) | 5 (11.63) | 31 (72.09) | |
| Tetracycline | Tetracycline (30) | 28 (65.12) | 2 (4.65) | 13 (30.23) |
| Fluoroquinolones | Ciprofloxacin (5) | 27 (62.79) | 9 (20.93) | 7 (16.28) |
| Folate pathway antagonist | Trimethoprim–sulfamethoxazole (1.25/23.75) | 26 (60.47) | 2 (4.65) | 15 (34.88) |
| Isolate | Antibiotic Agents | MAR Index | Genes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CRO | CTX | FEP | AMC | AMP | TE | MEM | CIP | CN | AK | SXT | |||
| En1 | S | S | S | R | R | S | S | R | S | S | R | 0.36 | blaTEM |
| En2 | R | R | R | R | R | R | I | R | R | S | R | 0.82 | blaTEM |
| En3 ** | R | R | R | R | R | R | I | R | R | S | S | 0.73 | blaTEM |
| En6 * | S | S | S | S | S | S | S | S | R | S | S | 0.09 | ND |
| En7 | R | R | S | R | R | R | S | R | S | R | I | 0.64 | ND |
| En8 ** | R | R | R | R | R | R | S | R | R | S | R | 0.82 | blaCTX-M |
| En9 * | S | S | S | S | S | S | S | R | S | S | S | 0.09 | ND |
| En12 | S | S | S | S | R | R | S | R | S | R | R | 0.45 | blaTEM |
| En13 | S | S | S | R | R | R | S | R | S | S | S | 0.36 | blaTEM |
| En14 ** | R | R | I | R | R | R | S | R | R | S | R | 0.73 | blaTEM |
| En15 | I | R | I | R | R | R | S | R | R | S | R | 0.64 | blaTEM |
| En16 ** | R | R | S | R | R | R | I | R | R | R | R | 0.82 | blaTEM |
| En17 | R | R | R | R | R | R | I | R | R | R | R | 0.91 | ND |
| En19 * | S | S | S | S | S | S | S | I | S | S | S | 0.00 | ND |
| En20 * | S | S | S | I | R | S | S | I | S | S | R | 0.18 | blaTEM |
| En21 | S | S | S | I | R | R | S | I | S | S | R | 0.27 | blaTEM |
| En22 * | S | S | S | S | S | S | S | S | S | R | S | 0.09 | ND |
| En23 * | S | S | S | S | R | R | I | S | S | I | S | 0.18 | blaTEM |
| En24 ** | R | R | R | R | R | R | I | R | R | S | R | 0.82 | blaTEM |
| En25 | S | S | S | S | R | R | S | R | S | S | R | 0.36 | blaTEM |
| En26 | S | S | S | I | R | R | S | R | R | S | R | 0.45 | blaTEM |
| En27 * | S | S | S | S | S | S | S | I | S | S | I | 0.00 | ND |
| En28 * | S | S | S | S | R | I | S | I | S | S | R | 0.18 | blaTEM |
| En29 ** | R | I | S | R | R | R | R | R | I | I | S | 0.55 | blaCTX-M |
| En30 | S | S | S | S | R | S | S | S | R | S | R | 0.27 | blaTEM |
| En32 | S | I | I | R | R | R | S | R | R | S | R | 0.55 | blaCTX-M |
| En33 | R | R | R | R | R | R | I | R | R | I | R | 0.82 | ND |
| En34 ** | R | R | I | R | R | R | S | R | S | S | R | 0.64 | blaTEM |
| En35 | S | S | S | S | S | R | S | R | S | S | R | 0.27 | ND |
| En36 ** | R | R | R | S | R | R | S | R | R | S | S | 0.64 | blaCTX-M |
| En37 * | S | S | S | S | S | R | S | S | S | S | S | 0.09 | blaTEM |
| En38 | S | S | S | S | R | R | S | S | S | S | R | 0.27 | blaTEM |
| En41 * | S | S | S | S | S | S | S | S | S | S | S | 0.00 | blaTEM |
| En42 * | S | S | S | S | R | S | S | I | S | S | S | 0.09 | ND |
| En43 * | S | S | S | S | R | S | S | I | S | I | S | 0.09 | blaSHV |
| En45 | R | R | S | R | R | R | S | R | R | R | R | 0.73 | blaTEM |
| En46 ** | R | R | R | I | R | I | S | R | R | S | R | 0.64 | ND |
| En47 | S | S | S | S | R | R | S | R | S | S | R | 0.36 | blaTEM |
| En48 | S | S | S | S | R | R | S | R | S | S | R | 0.36 | blaTEM |
| En49 | S | S | S | S | R | R | S | R | S | S | R | 0.36 | ND |
| En50 * | S | S | S | S | R | S | S | I | S | S | S | 0.09 | ND |
| En51 * | S | S | S | S | S | S | S | I | S | I | S | 0.00 | ND |
| En52 | S | S | S | S | R | R | S | R | S | R | R | 0.45 | ND |
| Algal Extracts | Concentration in 12.5% DMSO (mg/mL) | Number of Multidrug-Resistant E. coli Isolates Affected by Algal Extracts | ||
|---|---|---|---|---|
| Inhibiting | Slightly Inhibiting | Non-Inhibiting | ||
| SE | 1.43 | 0 | 4 (13.79%) | 25 (86.21%) |
| HE | 1.55 | 3 (10.34%) | 9 (31.03%) | 17 (58.62%) |
| NE | 2.94 | 0 | 1 (3.45%) | 28 (96.55%) |
| UE | 3.21 | 0 | 0 | 29 (100%) |
| CE | 3.10 | 1 (3.45%) | 0 | 28 (96.55%) |
| FS | 3.28 | 0 | 0 | 29 (100%) |
| FH | 2.75 | 0 | 4 (13.79%) | 25 (86.21%) |
| FN | 1.65 | 0 | 0 | 29 (100%) |
| FU | 2.28 | 0 | 0 | 29 (100%) |
| FC | 1.40 | 0 | 1 (3.45%) | 28 (96.55%) |
| Algae | Abbreviation of | |
|---|---|---|
| Ethanol Extraction | Enzymatic Hydrolysis | |
| Arthrospira platensis | SE | FS |
| Haematococcus pluvialis | HE | FH |
| Nannochloropsis oculata | NE | FN |
| Ulva rigida | UE | FU |
| Caulerpa lentillifera | CE | FC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Satchasataporn, K.; Khunbutsri, D.; Chopjitt, P.; Sutjarit, S.; Pan-utai, W.; Meekhanon, N. Antimicrobial Resistance Patterns of ESBL-Producing Escherichia coli in Dogs from Thailand: Evaluation of Algal Extracts as Novel Antimicrobial Agents. Antibiotics 2025, 14, 377. https://doi.org/10.3390/antibiotics14040377
Satchasataporn K, Khunbutsri D, Chopjitt P, Sutjarit S, Pan-utai W, Meekhanon N. Antimicrobial Resistance Patterns of ESBL-Producing Escherichia coli in Dogs from Thailand: Evaluation of Algal Extracts as Novel Antimicrobial Agents. Antibiotics. 2025; 14(4):377. https://doi.org/10.3390/antibiotics14040377
Chicago/Turabian StyleSatchasataporn, Khomson, Duangdaow Khunbutsri, Peechanika Chopjitt, Samak Sutjarit, Wanida Pan-utai, and Nattakan Meekhanon. 2025. "Antimicrobial Resistance Patterns of ESBL-Producing Escherichia coli in Dogs from Thailand: Evaluation of Algal Extracts as Novel Antimicrobial Agents" Antibiotics 14, no. 4: 377. https://doi.org/10.3390/antibiotics14040377
APA StyleSatchasataporn, K., Khunbutsri, D., Chopjitt, P., Sutjarit, S., Pan-utai, W., & Meekhanon, N. (2025). Antimicrobial Resistance Patterns of ESBL-Producing Escherichia coli in Dogs from Thailand: Evaluation of Algal Extracts as Novel Antimicrobial Agents. Antibiotics, 14(4), 377. https://doi.org/10.3390/antibiotics14040377







