Antimicrobial and Metal Resistance Genes in Bacteria Isolated from Mine Water in Austria
Abstract
1. Introduction
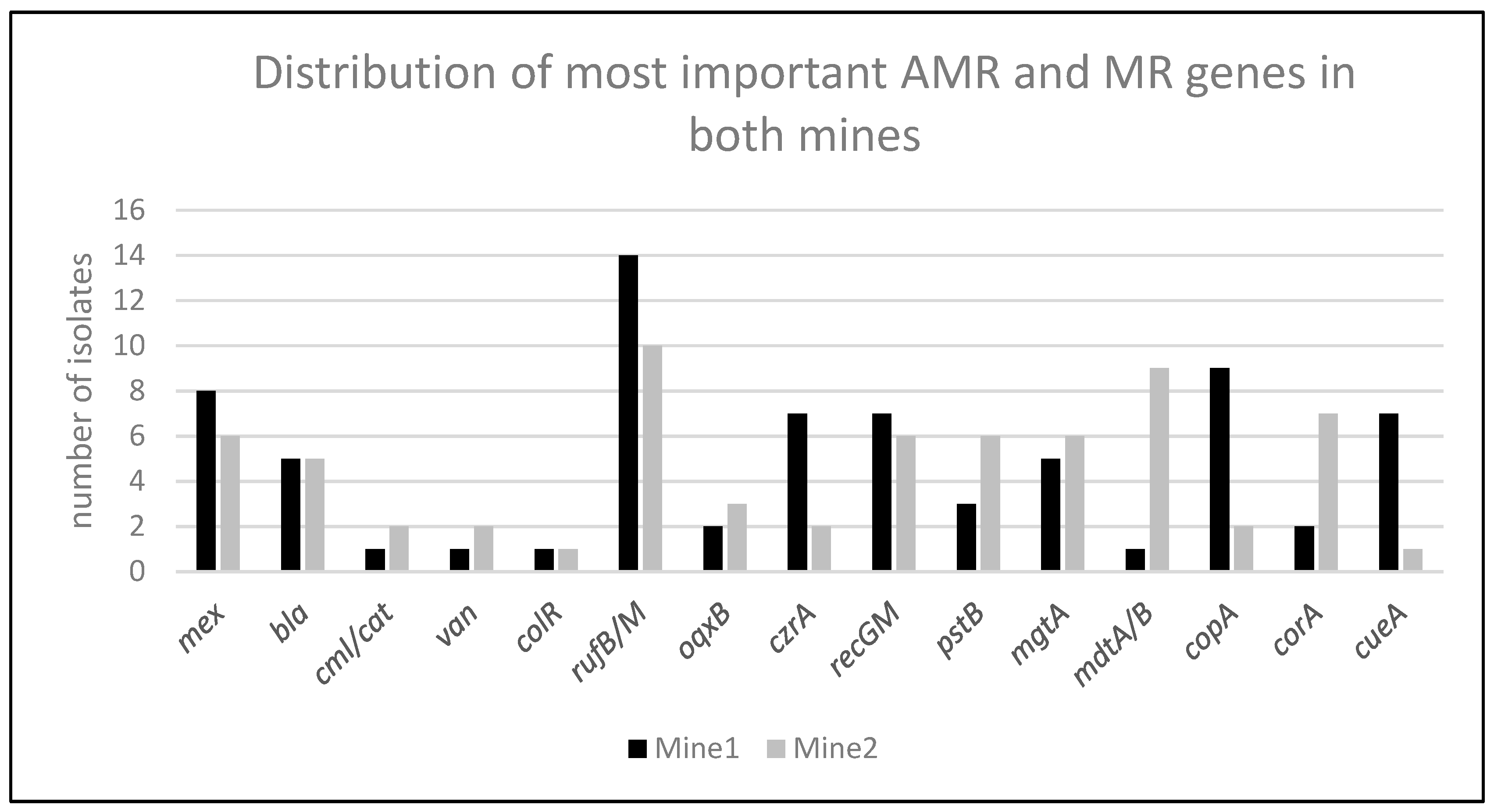
2. Results
3. Discussion
4. Materials and Methods
- Mine 1: This site is a former lead and silver mining location. Zinc-containing ore was mined extensively from the 18th century until 1927, when the mine was closed. The old mine is now used as a tourist attraction and for cheese ripening. Four different sampling sites were selected: three water-containing sites in the old deep mining sites and one in the cheese ripening tunnel.
- Mine 2: This site is known for iron mining. Five sampling locations were chosen, including water from former deep mining areas, metal-containing rock, and two different sludge water sites.
- Bacterial identification. The identity of these bacterial species was determined by performing a BLASTn (assessed 19 April 2024) [29] search on the 16S rRNA gene sequence. Whenever this gene was not intact, the largest partial sequence was taken as in input for BLASTn.
- In silico identification of antimicrobial resistance genes. After WGS, the genomes were screened for known antimicrobial resistance genes using ABRicate (version1.0.1) [30], against NCBI (5286 sequences) [31], CARD (2631 sequences) [32], Resfinder (3077 sequences) [33], ARG-ANNOT (2223 sequences) [34], and MEGARES (6635 sequences) [35,36], databases downloaded on 17 April 2024) according to defaults.
- Metal resistance genes. Annotated genomes were analyzed for the presence of several heavy metal resistance genes. Metal resistance genes were identified performing BLASTn (accessed 23 May 2024) between the nucleotide sequences of the coding sequences and the MEGARES 3.0 database (downloaded on 23 May 2024), in the categories “Metals” and “Multi-compound” [35,36].
- Sequence availability. Sequences are publicly available under PRJNA1224714.
- Statistical analysis. The Fisher’s exact test was used to study differences between antimicrobial resistance and metal resistance genes within the isolates.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Escamilla-Rodríguez, A.; Carlos-Hernández, S.; Díaz-Jiménez, L. Evidence of Resistance of Heavy Metals from Bacteria Isolated from Natural Waters of a Mining Area in Mexico. Water 2021, 13, 2766. [Google Scholar] [CrossRef]
- Fashola, M.O.; Ngole-Jeme, V.M.; Babalola, O.O. Heavy Metal Pollution from Gold Mines: Environmental Effects and Bacterial Strategies for Resistance. Int. J. Environ. Res. Public Health 2016, 13, 1047. [Google Scholar] [CrossRef] [PubMed]
- Kusi, J.; Ojewole, C.O.; Ojewole, A.E.; Nwi-Mozu, I. Antimicrobial Resistance Development Pathways in Surface Waters and Public Health Implications. Antibiotics 2022, 11, 821. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Richmond, M.H.; John, M. Co-transduction by a staphylococcal phage of the genes responsible for penicillinase synthesis and resistance to mercury salts. Nature 1964, 202, 1360–1361. [Google Scholar] [CrossRef]
- Stehling, E.G.; Furlan, J.P.R.; Lopes, R.; Chodkowski, J.; Stopnisek, N.; Savazzi, E.A.; Shade, A. The relationship between water quality and the microbial virulome and resistome in urban streams in Brazil. Environ. Pollut. 2024, 348, 123849. [Google Scholar] [CrossRef] [PubMed]
- Bosch, J.; Bezuidenhout, C.; Coertze, R.; Molale-Tom, L. Metal- and antibiotic-resistant heterotrophic plate count bacteria from a gold mine impacted river: The Mooi River system, South Africa. Environ. Sci. Pollut. Res. 2023, 30, 31605–31619. [Google Scholar] [CrossRef]
- Österreichisches Montan-Handbuch. 2024. Available online: https://www.bmf.gv.at/themen/bergbau/bergbau-in-oesterreich/bergbau-in-oesterreich.html (accessed on 26 February 2025).
- Hermann, R.; Baumgartner, R.J.; Sarc, R.; Ragossnig, A.; Wolfsberger, T.; Eisenberger, M.; Budiscshowsky, A.; Pomberger, R. Landfill mining in Austria: Foundations for an integrated ecological and economic assessment. Waste Manag. Res. 2014, 32, 48–58. [Google Scholar] [CrossRef]
- Bustamante, M.; Mei, S.; Daras, I.M.; van Doorn, G.S.; Falcao Salles, J.; de Vos, M.G.J. An eco-evolutionary perspective on antimicrobial resistance in the context of One Health. iScience 2024, 28, 111534. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Meléndez-Sánchez, E.R.; Martínez-Prado, M.A.; Núñez-Ramírez, D.M.; Rojas-Contreras, J.A.; Lópes-Miranda, J.; Medina-Torres, L. Review: Biotechnological Potential of As- and Zn-Resistant Autochthonous Microorganisms from Mining Process. Water Air Soil Pollut. 2021, 232, 332. [Google Scholar] [CrossRef]
- Kormos, D.; Lin, K.; Pruden, A.; Marr, L.C. Critical review of antibiotic resistance genes in the atmosphere. Environ. Sci. Process. Impacts 2022, 24, 870–883. [Google Scholar] [CrossRef]
- Thomas, J.C., IV; Oladeinde, A.; Kieran, T.J.; Finger, J.W., Jr.; Bayona-Vasquez, N.J.; Cartee, J.C.; Beasley, J.C.; Seaman, J.C.; McArthur, J.V.; Rhodes, O.E., Jr.; et al. Co-occurrence of antibiotic, biocide, and heavy metal resistance genes in bacteria from metal and radionuclide contaminated soils at the Savannah River Site. Microbiol. Biotechnol. 2020, 13, 1179–1200. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I.; Roderick, I.M. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 2007, 271, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Dash, H.R.; Chakraborty, J. Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollution. Appl. Microbiol. Biotechnol. 2016, 100, 2967–2984. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.F.; Liu, G.P.; Zhang, F.; Li, Z.M.; Yang, X.L.; Yang, C.D.; Shen, J.L.; He, J.Z.; Li, B.L.; Zeng, J.G. Natural selenium stress influences the changes of antibiotic resistome in seleniferous forest soils. Environ. Microbiome 2022, 17, 26. [Google Scholar] [CrossRef]
- Strejcek, M.; Smrhova, T.; Junkova, P.; Uhlik, O. Whole-cell MALDI-TOF MS versus 16S rRNA gene analysis for identification and dereplication of recurrent bacterial isolates. Front. Microbiol. 2018, 9, 1294. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic Note: A place for DNA-DNA Reassociation and 16S rRNA Sequence Analysis in the Present Species Definition in Bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Farias, P.; Espírito Santo, C.; Branco, R.; Francisco, R.; Santos, S.; Hansen, L.; Sorensen, S.; Morais, P.V. Natural hot spots for gain of multiple resistances: Arsenic and antibiotic resistances in heterotrophic, aerobic bacteria from marine hydrothermal vent fields. Appl. Environ. Microbiol. 2015, 81, 2534–2543. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lo Giudice, A.; Casella, P.; Bruni, V.; Michaud, L. Response of bacterial isolates from Antarctic shallow sediments towards heavy metals, antibiotics and polychlorinated biphenyls. Ecotoxicology 2013, 22, 240–250. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J.V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Gillieatt, B.F.; Coleman, N.V. Unravelling the mechanisms of antibiotic and heavy metal resistance co-selection in environmental bacteria. FEMS Microbiol. Rev. 2024, 48, fuae017. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Coșeriu, R.L.; Mare, A.D.; Toma, F.; Vintilă, C.; Ciurea, C.N.; Togănel, R.O.; Cighir, A.; Simion, A.; Man, A. Uncovering the Resistance Mechanisms in Extended-Drug-Resistant Pseudomonas aeruginosa Clinical Isolates: Insights from Gene Expression and Phenotypic Tests. Microorganisms 2023, 11, 2211. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Akbar, A.; Sinegani, S.; Younessi, N. Antibiotic resistance of bacteria isolated from heavy metal-polluted soils with different land uses. J. Glob. Antimicrob. Resist. 2017, 10, 247–255. [Google Scholar] [CrossRef]
- Murray, L.M.; Hayes, A.; Snape, J.; Kasprzyk-Hordern, B.; Gaze, W.H.; Murray, A.K. Co-selection for antibiotic resistance by environmental contaminants. npj Antimicrob. Resist. 2024, 2, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Bolotin, E.; Hershberg, R. Horizontally acquired genes are often shared between closely related bacterial species. Front. Microbiol. 2017, 8, 1536. [Google Scholar] [CrossRef]
- Maguvu, T.E.; Frias, R.J.; Hernandez-Rosasm, A.I.; Holtz, B.A.; Niederholzer, F.J.A.; Duncan, R.A.; Yaghmour, M.A.; Culumber, C.M.; Gordon, P.E.; Vieira, F.C.F.; et al. Phylogenomic analyses and comparative genomics of Pseudomonas syringae associated with almond (Prunus dulcis) in California. PLoS ONE 2024, 19, e0297867. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, R.; Lu, P.; Chen, F.; Huang, Y.; Ding, H.; Cheng, T. Groundwater resistant gene accumulation in mining-agriculture complex zones: Insights from metagenomic analysis of subterranean mineral and terrestrial agricultural interactions. Environ. Res. 2024, 263, 120138. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.Y.; Gao, F.Z.; He, L.Y.; Zhang, M.; Liu, Y.S.; Qi, J.; Ying, G.G. Prevalence of antibiotic resistance genes in mining-impacted farmland environments. Ecotoxicol. Environ. Saf. 2025, 289, 117651. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Seemann, T. ABRicate: Mass Screening of Contigs for Antiobiotic Resistance Genes. 2016. Available online: https://github.com/tseemann/abricate (accessed on 26 February 2025).
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, e00361-20. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Doster, E.; Lakin, S.M.; Dean, C.J.; Wolfe, C.; Young, J.G.; Boucher, C.; Belk, K.E.; Noyes, N.R.; Morley, P.S. MEGARes 2.0: A database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 2020, 48, D561–D569. [Google Scholar] [CrossRef]
- Bonin, N.; Doster, E.; Worley, H.; Pinnell, L.J.; Bravo, J.E.; Ferm, P.; Marini, S.; Prosperi, M.; Noyes, N.; Morley, P.S.; et al. MEGARes and AMR++, v3.0: An updated comprehensive database of antimicrobial resistance determinants and an improved software pipeline for classification using high-throughput sequencing. Nucleic Acids Res. 2023, 51, D744–D752. [Google Scholar] [CrossRef] [PubMed]
| Isolate | NCBI | CARD | Resfinder | ARGANNOT | MEGARES | Metal Resistance Genes |
|---|---|---|---|---|---|---|
| 027916_Erz42 | - | mexF, mexK, mexQ | - | - | emhC, mexE, mexF, mexK, mexQ | actP, recGM, acn, fpvA, czrA, ruvB, modB, ruvBM, ctpC, oscA |
| 027917_Erz81an | - | mexF, mexK, mexQ | - | - | emhC, mexE, mexF, mexK, mexQ | acn, modB, recGM, oscA, copS, actP, ruvB, czrA, ctpC, ruvBM |
| 027918_Erz4110C | - | mexF, mexK, mexQ | - | - | emhC, mexE, mexF, mexK, mexQ | mntH, nhaB, acrD, cueO, mgtA (2), terC, pstC, pstB (2), pitA, mdtB, corA, glpF |
| 027919_Erz51 | - | mexF | - | - | emhC, mexE, mexF, ttgB | cinA, ruvBM, mdtC, ruvB, irlR |
| 027920_Erz4210C | - | mexF | - | - | emhC, mexE, mexF, ttgB | cinA, ruvB, irlR, ruvBM, mdtC |
| 027921_Erz910C | - | mexF | - | - | emhC, mexE, mexF, ttgB | cadR, recGM, ruvB, ctpC, fpvA, cueA, copR, cinA |
| 027922_Erz62 | cmlV, cpt, vanR-O | parY, cmlV, novA, vanRO | cmlV_1 | (Phe)cmlV, (Phe)cpt_strepv | cmlV, cpt, novA, parY, vanRO | dnaK, acn, czcP, ideR |
| 027923_Erz66 | cmlV, cpt, vanR-O | parY, cmlV, novA, vanRO | cmlV_1 | Phe)cmlV, (Phe)cpt_strep | cmlV, cpt, novA, parY, vanRO | dnaK, acn, czcP, ideR |
| 027924_Erz102 | - | - | - | - | - | dnaK |
| 027925_Arz111 | - | - | - | - | - | - |
| 027926_ARZ121 | - | mexF, CpxR, mexW | - | - | cpxAR, emhC, mexE, mexF, ttgB | czrA, arsB, recGM, czrA, copR (3), ruvB, acn, copA, copD, cueA |
| 027927_ARZ123 | blaSGM-1 | - | blaSGM-1 | (Bla)blaSGM-1 | blaSGM | copA, cnrA, nccA (3), ruvB, cutO, actPC, cueA |
| 027928_ARZ131 | blaSGM-1 | - | blaSGM-1 | (Bla)blaSGM-1 | blaSGM | nccA (3), ruvB, cnrA, copA, cutO, actPC, cueA |
| 027929_ARZ152 | - | - | - | - | - | - |
| 027930_ARZ153 | mphM, rphC | mphM, rphB | mphM. rphC | - | mphB, rph | alu1P |
| 027931_ARZ154 | - | - | - | - | - | - |
| 027932_ARZ161 | - | - | - | - | - | - |
| 027933_ARZ162 | - | mexF, mexW | - | - | emhC, mexE, mexF, ttgB | ruvB, recGM, cadR, czrA, cueA, irlR, arsB, copA, acn, copR (2), mertT, merA, merB, copS, arsBM, pstB, ctpC |
| 027934_ARZ163 | - | - | - | - | - | actP, acn |
| 027935_ARZ171 | - | - | - | - | - | - |
| 027936_Arz172 | catA10, rph | rphB | cat_4 | (Rif)rphD, (Phe)catA_variant1 | catA, rph | alu1P, merR1 |
| 027937_ARZ201 | - | mexF, mexQ | - | - | emhC, mexE, mexF, mexQ, ttgB | ruvB, mgtA, arsB, recGM, acn, copR, ruvBM, actP, copA, copB, czrA |
| 027938_ARZ204 | - | mexF, mexK, mexW | - | - | emhC, mexE, mexF, mexK, ttgB | copA, copB, copR (2), ruvB, copD, copS, recGM, mgtA, cueA |
| 027939_ARZ205 | - | mexF, mexQ | - | - | emhC, mexE, mexF, mexQ, ttgB | ruvBM, ruvB, recGM, copR, ACN, czrA, copB, copA, actP, arsB, mgtA |
| 027940_ARZ232 | - | - | - | - | - | mco |
| 027941_Erz41an | blaRAHN-1, oqxB9 | crp, hns, oqxB | blaRAHN-1_1, oqxB_1 | (Bla)blaRAHN-1, (Flq)OqxBgb | cpxAR, crp, hns, rahN, sdeB | cueO, copA, acrD (2), mgtA (2), pitA, pstB, pstA, pstS, modC, mdtB, corA |
| 027942_Erz51an | oqxB17 | crp, hns, oqxB | oqxB_1 | (Flq)OqxBgb | cpxAR, crp, hns, sdeB | cueO, acrD, mntH, mgtA (2), terC, pitA, pstB (2), pstC, nhaB, mdtB, corA, zntA, glpF |
| 027943_Erz61an | blaFOX-2, blaOXA-427, cphA1 | blaFOX-2, blaOXA-427, cphA5 | blaFOX-2_1, blaOXA-427_1, cphA1_1 | (Bla)blaFOX-2, (Bla)blaOXA-12, (Bla)cphA5 | cphA, blaFOX, blaOXA | ruvB, ruvBM, recGM |
| 027944_Erz71an | blaFOX-2, blaOXA-427, cphA5 | blaFOX-2, blaOXA-427, cphA5 | blaFOX-2_1, blaOXA-427_1, cphA5_1 | (Bla)blaFOX-2, (Bla)blaOXA-12, (Bla)cphA5 | cphA, blaFOX, blaOXA | zipB, cusA, ruvBM, recGM |
| 027945_Erz91an | - | crp, hns, emrR | - | - | crp, emrR, hns | acn, sitABCD, mdtB, mdtC, acrD, cueO, pstB, corC, fetB, corA |
| 027946_Arz131an | iri | rpoB2, iri | - | (Rif)iri | iri | dnaK, acn |
| 027947_Arz143an | bla1, fosB_gen, satA_Ba | bla1, fosB | fosB1_1 | (Bla)bla-1, (Fcyn)fosBx1 | bla1, fosB, sat | - |
| 027948_Arz231an | bciI, bla1, fosB_gen, satA_Ba, vanZ-F | bla1, bciI, fosB, vanZF | blaZ_12, fosB1_1 | (Bla)bla-1, (Bla)bla2, (Fcyn)fosBx1, (Gly)vanZF-Pp | bciI, bla1, blaZ, fosB, sat, vanYF | - |
| 027949_Arz21110Can | - | mexF, cpxR. mexW | - | - | cpxAR, emhC, mexE, mexF, ttgB | pstB, rcGM, merR, merP, merC, merA, arsBM, copR, copB, ruvB, ctpC, czrA, cueA, zraR |
| 027950_Arz2310C | - | mexF, mexK, mexW | - | - | emhC, mexE, mexF, mexK, ttgB | ruvB, actP1, cueA (2), copR, recGM, merR2, merT, merP, merF, merA, merD, merE, czrA, copAM, can |
| 027951_Arz13210C | - | mexF | - | - | emhC, mexE, mexF | copR (2), copS, copD, copAM, pbrA, merT (2), merA (3), ctpC (2), zraR, ruvB, czrA, mrdH, ruvBM, merE, merD, merF (2), merP (2), merR2, ctpV, merR, oscA |
| 027952_Arz21110C | - | - | - | - | - | ctpG, merT |
| 027953_Arz21210C | - | - | - | - | - | czcP |
| 027954_Erz4110Can | bla-C, oqxB19, qnrB39 | crp, hns, oqxB | oqxB_1, qnrB39_1 | (Flq)oqxBgb | blaC, cpxAR, crp, hns, qnrB, sdeB, sdeX, sdeY | znuC, sitABCD, mgtA, baeR, mdtC, mdtB, pstB, acrD, zntA, corA, pstA, pstC, copR, FETB, copA, corC |
| 027955_Arz18110Can | bla-C, oqxB11 | crp, hns, oqxB | oqxB_1 | (Flq)oqxBgb | blaC, cpxAR, crp, hns, sdeB, sdeX, sdeY | baeR, mdtC, mdtB, pstB (2), acrD, ruvB, modC, corC, mgtA, corA, pstA, pstC |
| 027956_Erz9210Can | oqxB9 | crp, hns, oqxB | oqxB_1 | (Flq)oqxBgb | crp, hns, sdeB | mntH, nhaB, acrD, cueO, mgtA, terC, pstC, pstB (2), pitA, mgtA, mdtB, corA, glpF |
| 027957_Erz9110Can | mcr-9.1 | crp, hns, mcr-9, acrB, baeR, emrR, marA | mcr-9_1 | (bla) E. coli, (Col)mcr-9.1 | acrB, cpxAR, crp, emrR, hns, marA, mcr, pbp2 | mdtA, mdtB, mdtC, baeS, baeR, ACRD, arsBM, modC, corC, mgtA, pstS, pstC, corB, dsbA, corA |
| 027958_Arz14110Can | mcr-9.1, qnrB96 | crp, hns, mcr-9, acrB, baeR, cpxA, emrR, marA, msbA | mcr-9.1, qnrB96 | (Col)mcr-9.1 | acrB, baeR, cpxAR, crp, emrR, hns, marA, mcr, msbA | acrD (2), baeR, baeS, mdtC, mdtB, mdtA, cutE, cusA, arsBM, mgtA, pstC, pstS, cueO, corA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prochaska, J.; Reitner, H.; Benold, C.; Stadtschnitzer, A.; Choijilsuren, B.; Sofka, D.; Hilbert, F.; Pacífico, C. Antimicrobial and Metal Resistance Genes in Bacteria Isolated from Mine Water in Austria. Antibiotics 2025, 14, 262. https://doi.org/10.3390/antibiotics14030262
Prochaska J, Reitner H, Benold C, Stadtschnitzer A, Choijilsuren B, Sofka D, Hilbert F, Pacífico C. Antimicrobial and Metal Resistance Genes in Bacteria Isolated from Mine Water in Austria. Antibiotics. 2025; 14(3):262. https://doi.org/10.3390/antibiotics14030262
Chicago/Turabian StyleProchaska, Jakob, Heinz Reitner, Christian Benold, Alfred Stadtschnitzer, Buyantogtokh Choijilsuren, Dmitrij Sofka, Friederike Hilbert, and Cátia Pacífico. 2025. "Antimicrobial and Metal Resistance Genes in Bacteria Isolated from Mine Water in Austria" Antibiotics 14, no. 3: 262. https://doi.org/10.3390/antibiotics14030262
APA StyleProchaska, J., Reitner, H., Benold, C., Stadtschnitzer, A., Choijilsuren, B., Sofka, D., Hilbert, F., & Pacífico, C. (2025). Antimicrobial and Metal Resistance Genes in Bacteria Isolated from Mine Water in Austria. Antibiotics, 14(3), 262. https://doi.org/10.3390/antibiotics14030262






