Artificial Intelligence in Bacterial Infections Control: A Scoping Review
Abstract
1. Introduction
2. Results
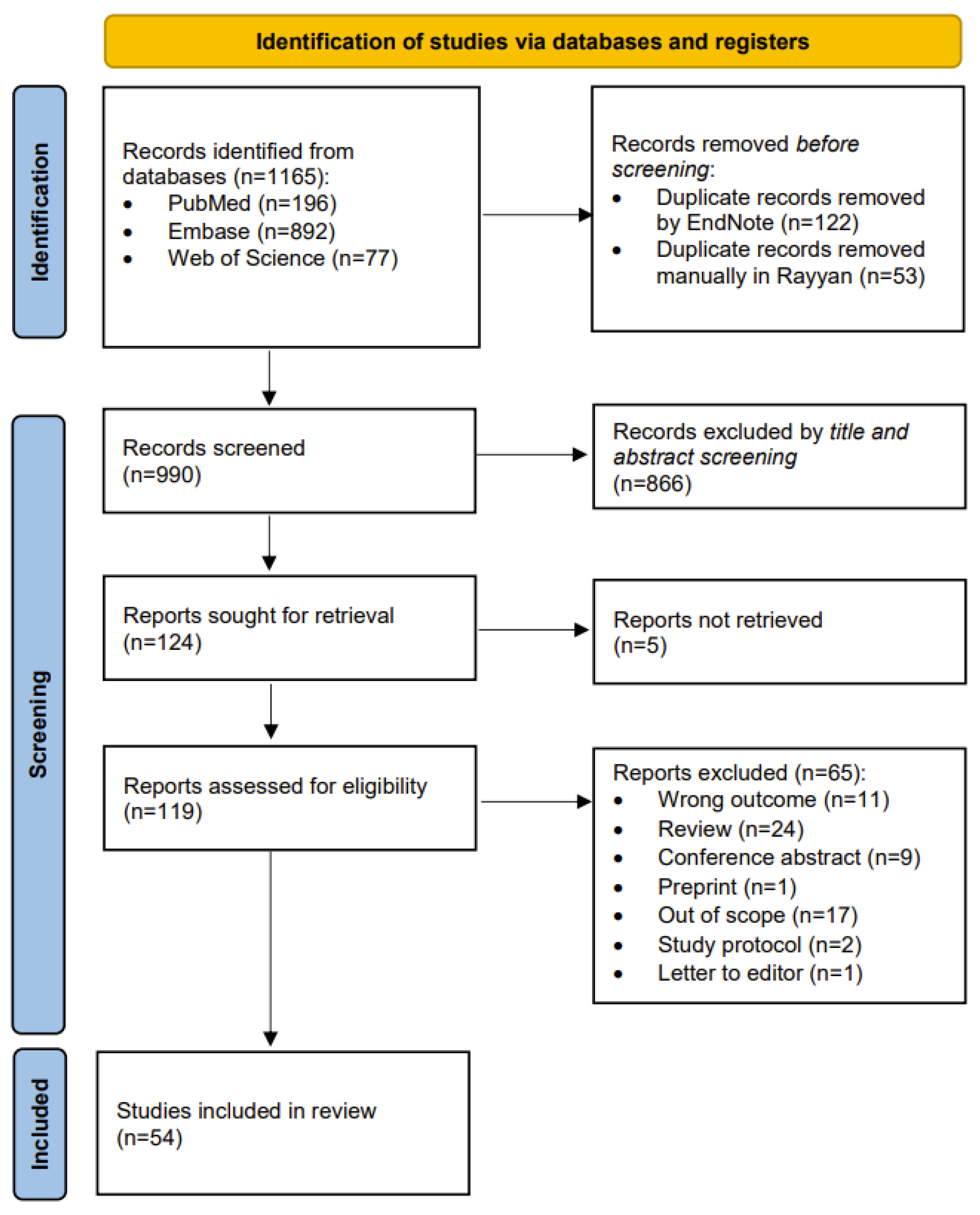
2.1. Search Results
2.2. Qualitative Synthesis
2.2.1. Scope of Countries
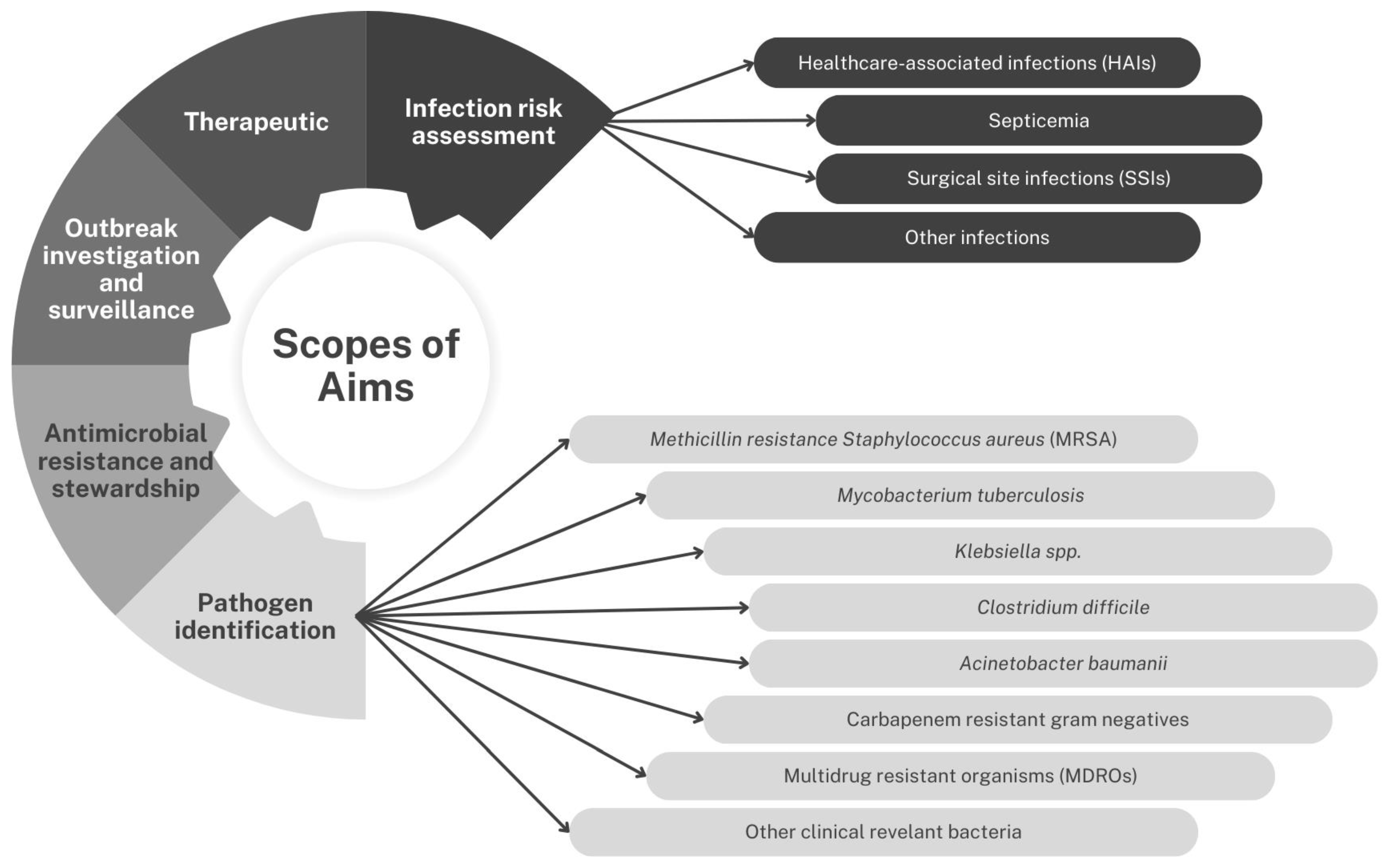
2.2.2. Scope of Aim
Pathogen Identification
- Methicillin Resistance Staphylococcus aureus (MRSA)
- Mycobacteria tuberculosis
- Klebsiella spp.
- Clostridium difficile
- Acinetobacter baumannii
- Multidrug-Resistant Organisms (MDROs)
- Carbapenem-resistant Gram-negative organisms
- Other clinically relevant bacteria
Infection Risk Assessment
- Healthcare-associated infections (HAIs)
- Septicemia
- Surgical site infection (SSI)
- Other infections
- Therapeutic Options
- Outbreak investigation and surveillance
- Antimicrobial resistance and stewardship
2.2.3. Scope of AI Type
Machine Learning (ML)
Hybrid Models: Machine Learning (ML) and Deep Learning (DL)
Deep Learning (DL)
Computational Biology and Machine Learning
Knowledge Discovery and Semantic Analysis (KD and SA)
Machine Learning (ML), Deep Learning (DL), and Natural Language Processing (NLP)
2.2.4. Advantages
2.2.5. Limitations
3. Discussion
4. Materials and Methods
4.1. Data Sources and Search Strategy
4.2. Study Selection and Eligibility Criteria
4.3. Data Extraction and Synthesis
4.4. Standardization
4.5. Data Cleaning and Handling Ambiguous and Missing Data
4.6. Data Synthesis and Generation of Thematic Scopes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| IPC | infection prevention and control |
| AMR | antimicrobial resistance |
| HAIs | healthcare-associated infections |
| ECDC | European Centre for Disease Prevention and Control |
| WHO | World Health Organization |
| CAIs | community-associated infections |
| AI | artificial intelligence |
| ML | Machine Learning |
| DL | Deep Learning |
| LR | Logistic Regression |
| RF | Random Forest |
| XGBoost | Extreme Gradient Boosting |
| ANN | Artificial Neural Network |
| SVM | Support Vector Machine |
| CB | Computational Biology |
| KBBN | Knowledge-Based Bayesian Network |
| HMM | hidden Markov model |
| KD&SA | Knowledge Discovery and Semantic Analysis |
| DTs | Decision Trees |
| CART | Classification and Regression Tree |
| GNNs | Graph Neural Networks |
| MOCA-I | Multi-Objective Classification Algorithm for Imbalanced Data |
| BN | Bayesian Network |
| HA-UTIs | hospital-acquired urinary tract infections |
| ICU | Intensive Care Unit |
| CNN | Convolutional Neural Network |
| D-LSTM | Deep Long Short-Term Memory Neural Network |
| 1D-CNN | One-Dimensional Convolutional Neural Network |
| CDI | Clostridioides Difficile Infection |
| NLP | Natural Language Processing |
| HAVM | healthcare-associated ventriculitis and meningitis |
| EDS-HAT | Enhanced Detection System for Healthcare-Associated Transmission |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| KNN | K-nearest neighbor |
| MRSA | Methicillin-Resistant Staphylococcus Aureus |
| BPNN | Backpropagation Neural Network |
| MDRO | multidrug-resistant organism |
| SSIs | surgical site infections |
| NN | Neural Network |
| IPMD | Integrated Promoter Markov Discriminant |
| LSTM | Long Short-Term Memory |
| CAP | community-acquired pneumonia |
| VRE | Vancomycin-Resistant Enterococci |
| CRE | Carbapenem-Resistant Enterobacterales |
| EHR | Electronic Health Record |
| MTBC | Mycobacterium Tuberculosis Complex |
| MTC | Mycobacterium Tuberculosis Complex |
| LTBI | Latent TB |
| IKPLAS | Invasive Klebsiella Pneumoniae Liver Abscess Syndrome |
| DM | diabetes mellitus |
| CRKP | Carbapenem-Resistant Klebsiella Pneumoniae |
| MDR | multidrug-resistant |
| HO-CDI | hospital-onset Clostridioides difficile infection |
| CR-GNB | Carbapenem-Resistant Gram-Negative Bacterial Bloodstream |
| CROs | Carbapenem-Resistant Organisms |
| CPOs | Carbapenemase-Producing Organisms |
| HPCF | Helicobacter Pylori Coccoid Form |
| WGS | whole genome sequencing |
| AMP | antimicrobial peptide |
| GBMs | Gradient Boosting Machines |
| WARNING | Worldwide Antimicrobial Resistance National/International Network Group |
| MGEs | Mobile Genetic Elements |
| GLASS | Global Antimicrobial Resistance and Use Surveillance System |
| IoT | Internet of Things |
| RoB | Risk of Bias Assessment |
| OSF | Open Science Framework |
| PRISMA-ScR | Preferred Reporting Items for Systematic Reviews and Meta-Analyses Extension for Scoping Reviews |
References
- Gilbert, G.L.; Kerridge, I. Hospital Infection Prevention and Control (IPC) and Antimicrobial Stewardship (AMS): Dual Strategies to Reduce Antibiotic Resistance (ABR) in Hospitals. In Ethics and Drug Resistance: Collective Responsibility for Global Public Health; Springer Nature: Berlin/Heidelberg, Germany, 2020; pp. 89–108. [Google Scholar]
- Storr, J.; Twyman, A.; Zingg, W.; Damani, N.; Kilpatrick, C.; Reilly, J.; Price, L.; Egger, M.; Grayson, M.L.; Kelley, E. Core components for effective infection prevention and control programmes: New WHO evidence-based recommendations. Antimicrob. Resist. Infect. Control 2017, 6, 6. [Google Scholar] [CrossRef]
- Zingg, W.; Storr, J.; Park, B.J.; Ahmad, R.; Tarrant, C.; Castro-Sanchez, E.; Tomczyk, S.; Kilpatrick, C.; Allegranzi, B.; Cardo, D.; et al. Implementation research for the prevention of antimicrobial resistance and healthcare-associated infections; 2017 Geneva infection prevention and control (IPC)-think tank (part 1). Antimicrob. Resist. Infect. Control 2019, 8, 87. [Google Scholar] [CrossRef]
- WHO. IPC and Antimicrobial Resistance (AMR). Available online: https://www.who.int/teams/integrated-health-services/infection-prevention-control/ipc-and-antimicrobial-resistance#:~:text=Strong%20IPC%20is%20a%20the,hygiene%20and%20better%20hospital%20hygiene (accessed on 9 June 2024).
- Scardoni, A.; Balzarini, F.; Signorelli, C.; Cabitza, F.; Odone, A. Artificial intelligence-based tools to control healthcare associated infections: A systematic review of the literature. J. Infect. Public. Health 2020, 13, 1061–1077. [Google Scholar] [CrossRef]
- Nuckchady, D.C. Incidence, Risk Factors, and Mortality From Hospital-Acquired Infections at a Hospital in Mauritius. Cureus 2021, 13, e19962. [Google Scholar] [CrossRef]
- Sikora, A.; Zahra, F. Nosocomial Infections. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Klumpp, D.J.; Rycyk, M.T.; Chen, M.C.; Thumbikat, P.; Sengupta, S.; Schaeffer, A.J. Uropathogenic Escherichia coli induces extrinsic and intrinsic cascades to initiate urothelial apoptosis. Infect. Immun. 2006, 74, 5106–5113. [Google Scholar] [CrossRef]
- Swanson, J.; Jeanes, A. Infection control in the community: A pragmatic approach. Br. J. Community Nurs. 2011, 16, 282–288. [Google Scholar] [CrossRef]
- Guan, X.-d.; He, L.-x.; Hu, B.-j.; Hu, J.; Huang, X.; Lai, G.; Li, Y.; Liu, Y.; Ni, Y.; Qiu, H. Laboratory diagnosis, clinical management and infection control of the infections caused by extensively drug-resistant Gram-negative bacilli: A Chinese consensus statement. Clin. Microbiol. Infect. 2016, 22, S15–S25. [Google Scholar] [CrossRef]
- Van Cutsem, G.; Isaakidis, P.; Farley, J.; Nardell, E.; Volchenkov, G.; Cox, H. Infection control for drug-resistant tuberculosis: Early diagnosis and treatment is the key. Clin. Infect. Dis. 2016, 62, S238–S243. [Google Scholar] [CrossRef]
- Wilson, J. Infection Control in Clinical Practice Updated Edition; Elsevier Health Sciences: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Arzilli, G.; De Vita, E.; Pasquale, M.; Carloni, L.M.; Pellegrini, M.; Di Giacomo, M.; Esposito, E.; Porretta, A.D.; Rizzo, C. Innovative Techniques for Infection Control and Surveillance in Hospital Settings and Long-Term Care Facilities: A Scoping Review. Antibiotics 2024, 13, 77. [Google Scholar] [CrossRef]
- Yang, L.; Lu, S.; Zhou, L. The Implications of Artificial Intelligence on Infection Prevention and Control: Current Progress and Future Perspectives. China CDC Wkly. 2024, 6, 901–904. [Google Scholar] [CrossRef]
- Maleki Varnosfaderani, S.; Forouzanfar, M. The Role of AI in Hospitals and Clinics: Transforming Healthcare in the 21st Century. Bioengineering 2024, 11, 337. [Google Scholar] [CrossRef]
- Tsitou, V.-M.; Rallis, D.; Tsekova, M.; Yanev, N. Microbiology in the era of artificial intelligence: Transforming medical and pharmaceutical microbiology. Biotechnol. Biotechnol. Equip. 2024, 38, 2349587. [Google Scholar] [CrossRef]
- Chindelevitch, L.; Jauneikaite, E.; Wheeler, N.E.; Allel, K.; Ansiri-Asafoakaa, B.Y.; Awuah, W.A.; Bauer, D.C.; Beisken, S.; Fan, K.; Grant, G. Applying data technologies to combat AMR: Current status, challenges, and opportunities on the way forward. arXiv 2022, arXiv:2208.04683. [Google Scholar]
- Sharma, M.; Savage, C.; Nair, M.; Larsson, I.; Svedberg, P.; Nygren, J.M. Artificial Intelligence Applications in Health Care Practice: Scoping Review. J. Med. Internet Res. 2022, 24, e40238. [Google Scholar] [CrossRef]
- Theodosiou, A.A.; Read, R.C. Artificial intelligence, machine learning and deep learning: Potential resources for the infection clinician. J. Infect. 2023, 87, 287–294. [Google Scholar] [CrossRef]
- Rabhi, S.; Jakubowicz, J.; Metzger, M.-H. Deep learning versus conventional machine learning for detection of healthcare-associated infections in French clinical narratives. Methods Inf. Med. 2019, 58, 31–41. [Google Scholar] [CrossRef]
- Jeon, K.; Kim, J.M.; Rho, K.; Jung, S.H.; Park, H.S.; Kim, J.S. Performance of a Machine Learning-Based Methicillin Resistance of Staphylococcus aureus Identification System Using MALDI-TOF MS and Comparison of the Accuracy according to SCCmec Types. Microorganisms 2022, 10, 1903. [Google Scholar] [CrossRef]
- Çaǧlayan, Ç.; Barnes, S.L.; Pineles, L.L.; Harris, A.D.; Klein, E.Y. A Data-Driven Framework for Identifying Intensive Care Unit Admissions Colonized With Multidrug-Resistant Organisms. Front. Public Health 2022, 10, 853757. [Google Scholar] [CrossRef]
- Ötleş, E.; Balczewski, E.A.; Keidan, M.; Oh, J.; Patel, A.; Young, V.B.; Rao, K.; Wiens, J. Clostridioides difficile infection surveillance in intensive care units and oncology wards using machine learning. Infect. Control Hosp. Epidemiol. 2023, 44, 1776–1781. [Google Scholar] [CrossRef]
- Aggarwal, S.; Dhall, A.; Patiyal, S.; Choudhury, S.; Arora, A.; Raghava, G.P.S. An ensemble method for prediction of phage-based therapy against bacterial infections. Front. Microbiol. 2023, 14, 1148579. [Google Scholar] [CrossRef]
- Althomsons, S.P.; Winglee, K.; Heilig, C.M.; Talarico, S.; Silk, B.; Wortham, J.; Hill, A.N.; Navin, T.R. Using Machine Learning Techniques and National Tuberculosis Surveillance Data to Predict Excess Growth in Genotyped Tuberculosis Clusters. Am. J. Epidemiol. 2022, 191, 1936–1943. [Google Scholar] [CrossRef]
- Aminian, M.; Couvin, D.; Shabbeer, A.; Hadley, K.; Enberg, S.; Rastogi, N.; Bennett, K.P. Predicting mycobacterium tuberculosis complex clades using knowledge-based bayesian networks. BioMed Res. Int. 2014, 2014, 398484. [Google Scholar] [CrossRef]
- Atkinson, A.; Ellenberger, B.; Piezzi, V.; Kaspar, T.; Salazar-Vizcaya, L.; Endrich, O.; Leichtle, A.B.; Marschall, J. Extending outbreak investigation with machine learning and graph theory: Benefits of new tools with application to a nosocomial outbreak of a multidrug-resistant organism. Infect. Control Hosp. Epidemiol. 2023, 44, 246–252. [Google Scholar] [CrossRef]
- Azé, J.; Sola, C.; Zhang, J.; Lafosse-Marin, F.; Yasmin, M.; Siddiqui, R.; Kremer, K.; Van Soolingen, D.; Refrégier, G. Genomics and machine learning for taxonomy consensus: The mycobacterium tuberculosis complex paradigm. PLoS ONE 2015, 10, e0130912. [Google Scholar] [CrossRef]
- Bournez, C.; Riool, M.; de Boer, L.; Cordfunke, R.A.; de Best, L.; van Leeuwen, R.; Drijfhout, J.W.; Zaat, S.A.J.; van Westen, G.J.P. CalcAMP: A New Machine Learning Model for the Accurate Prediction of Antimicrobial Activity of Peptides. Antibiotics 2023, 12, 725. [Google Scholar] [CrossRef]
- Camoez, M.; Sierra, J.M.; Dominguez, M.A.; Ferrer-Navarro, M.; Vila, J.; Roca, I. Automated categorization of methicillin-resistant Staphylococcus aureus clinical isolates into different clonal complexes by MALDI-TOF mass spectrometry. Clin. Microbiol. Infect. 2016, 22, 161.e161–161.e167. [Google Scholar] [CrossRef]
- Cheah, A.L.Y.; Cheng, A.C.; Spelman, D.; Nation, R.L.; Kong, D.C.M.; McBryde, E.S. Mathematical modelling of vancomycin-resistant enterococci transmission during passive surveillance and active surveillance with contact isolation highlights the need to identify and address the source of acquisition. BMC Infect. Dis. 2018, 18, 511. [Google Scholar] [CrossRef]
- Cherkasov, A.; Hilpert, K.; Jenssen, H.; Fjell, C.D.; Waldbrook, M.; Mullaly, S.C.; Volkmer, R.; Hancock, R.E.W. Use of artificial intelligence in the design of small peptide antibiotics effective against a broad spectrum of highly antibiotic-resistant superbugs. ACS Chem. Biol. 2009, 4, 65–74. [Google Scholar] [CrossRef]
- de Bruin, J.S.; Koller, W.; Zeckl, J.; Blacky, A.; Rappelsberger, A.; Adlassnig, K.P. Arden Syntax MLM Building Blocks for Microbiological Concepts and Their Application in Infection Surveillance. Stud. Health Technol. Inform. 2017, 236, 16–23. [Google Scholar]
- Doan, T.N.; Kong, D.C.M.; Marshall, C.; Kirkpatrick, C.M.J.; McBryde, E.S. Characterising the transmission dynamics of Acinetobacter baumannii in intensive care units using hidden markov models. PLoS ONE 2015, 10, e0132037. [Google Scholar] [CrossRef]
- Feng, C.; Di, J.; Jiang, S.; Li, X.; Hua, F. Machine learning models for prediction of invasion Klebsiella pneumoniae liver abscess syndrome in diabetes mellitus: A singled centered retrospective study. BMC Infect. Dis. 2023, 23, 284. [Google Scholar] [CrossRef]
- Freire, M.P.; Rinaldi, M.; Terrabuio, D.R.B.; Furtado, M.; Pasquini, Z.; Bartoletti, M.; de Oliveira, T.A.; Nunes, N.N.; Lemos, G.T.; Maccaro, A.; et al. Prediction models for carbapenem-resistant Enterobacterales carriage at liver transplantation: A multicenter retrospective study. Transpl. Infect. Dis. 2022, 24, e13920. [Google Scholar] [CrossRef]
- Goodman, K.E.; Simner, P.J.; Klein, E.Y.; Kazmi, A.Q.; Gadala, A.; Toerper, M.F.; Levin, S.; Tamma, P.D.; Rock, C.; Cosgrove, S.E.; et al. Predicting probability of perirectal colonization with carbapenem-resistant Enterobacteriaceae (CRE) and other carbapenem-resistant organisms (CROs) at hospital unit admission. Infect. Control Hosp. Epidemiol. 2019, 40, 541–550. [Google Scholar] [CrossRef]
- Gouareb, R.; Bornet, A.; Proios, D.; Pereira, S.G.; Teodoro, D. Detection of Patients at Risk of Multidrug-Resistant Enterobacteriaceae Infection Using Graph Neural Networks: A Retrospective Study. Health Data Sci. 2023, 3, 0099. [Google Scholar] [CrossRef]
- Hattori, S.; Sekido, R.; Leong, I.W.; Tsutsui, M.; Arima, A.; Tanaka, M.; Yokota, K.; Washio, T.; Kawai, T.; Okochi, M. Machine learning-driven electronic identifications of single pathogenic bacteria. Sci. Rep. 2020, 10, 15525. [Google Scholar] [CrossRef]
- Hsu, C.C.; Lin, Y.E.; Chen, Y.S.; Liu, Y.C.; Muder, R.R. Validation study of artificial neural network models for prediction of methicillin-resistant Staphylococcus aureus carriage. Infect. Control Hosp. Epidemiol. 2008, 29, 607–614. [Google Scholar] [CrossRef]
- Jacques, J.; Martin-Huyghe, H.; Lemtiri-Florek, J.; Taillard, J.; Jourdan, L.; Dhaenens, C.; Delerue, D.; Hansske, A.; Leclercq, V. The detection of hospitalized patients at risk of testing positive to multi-drug resistant bacteria using MOCA-I, a rule-based “white-box” classification algorithm for medical data. Int. J. Med. Inform. 2020, 142, 104242. [Google Scholar] [CrossRef]
- Jakobsen, R.S.; Nielsen, T.D.; Leutscher, P.; Koch, K. A study on the risk stratification for patients within 24 hours of admission for risk of hospital-acquired urinary tract infection using Bayesian network models. Health Informatics J. 2024, 30, 14604582241234232. [Google Scholar] [CrossRef]
- Khaledi, A.; Schniederjans, M.; Pohl, S.; Rainer, R.; Bodenhofer, U.; Xia, B.; Klawonn, F.; Bruchmann, S.; Preusse, M.; Eckweiler, D.; et al. Transcriptome profiling of antimicrobial resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 4722–4733. [Google Scholar] [CrossRef]
- Khaledi, A.; Weimann, A.; Schniederjans, M.; Asgari, E.; Kuo, T.H.; Oliver, A.; Cabot, G.; Kola, A.; Gastmeier, P.; Hogardt, M.; et al. Predicting antimicrobial resistance in Pseudomonas aeruginosa with machine learning-enabled molecular diagnostics. EMBO Mol. Med. 2020, 12, e10264. [Google Scholar] [CrossRef]
- Lapp, Z.; Han, J.H.; Wiens, J.; Goldstein, E.J.C.; Lautenbach, E.; Snitkin, E.S. Patient and microbial genomic factors associated with carbapenem-resistant klebsiella pneumoniae extraintestinal colonization and infection. mSystems 2021, 6, 2. [Google Scholar] [CrossRef]
- Liang, Q.Q.; Ding, S.; Chen, J.; Chen, X.Y.; Xu, Y.S.; Xu, Z.J.; Huang, M. Prediction of carbapenem-resistant gram-negative bacterial bloodstream infection in intensive care unit based on machine learning. BMC Med. Inform. Decis. Mak. 2024, 24, 123. [Google Scholar] [CrossRef]
- Liang, Q.Q.; Zhao, Q.Y.; Xu, X.; Zhou, Y.; Huang, M. Early prediction of carbapenem-resistant Gram-negative bacterial carriage in intensive care units using machine learning. J. Glob. Antimicrob. Resist. 2022, 29, 225–231. [Google Scholar] [CrossRef]
- Lyu, J.W.; Zhang, X.D.; Tang, J.W.; Zhao, Y.H.; Liu, S.L.; Zhao, Y.; Zhang, N.; Wang, D.; Ye, L.; Chen, X.L.; et al. Rapid prediction of multidrug-resistant klebsiella pneumoniae through deep learning analysis of sers spectra. Microbiol. Spectr. 2023, 11, 2. [Google Scholar] [CrossRef]
- Marra, A.R.; Alzunitan, M.; Abosi, O.; Edmond, M.B.; Street, W.N.; Cromwell, J.W.; Salinas, J.L. Modest Clostridiodes difficile infection prediction using machine learning models in a tertiary care hospital. Diagn. Microbiol. Infect. Dis. 2020, 98, 115104. [Google Scholar] [CrossRef]
- Noman, S.M.; Zeeshan, M.; Arshad, J.; Deressa Amentie, M.; Shafiq, M.; Yuan, Y.; Zeng, M.; Li, X.; Xie, Q.; Jiao, X. Machine Learning Techniques for Antimicrobial Resistance Prediction of Pseudomonas Aeruginosa from Whole Genome Sequence Data. Comput. Intell. Neurosci. 2023, 2023, 5236168. [Google Scholar] [CrossRef]
- Panchavati, S.; Zelin, N.S.; Garikipati, A.; Pellegrini, E.; Iqbal, Z.; Barnes, G.; Hoffman, J.; Calvert, J.; Mao, Q.; Das, R. A comparative analysis of machine learning approaches to predict C. difficile infection in hospitalized patients. Am. J. Infect. Control 2022, 50, 250–257. [Google Scholar] [CrossRef]
- Ratzinger, F.; Dedeyan, M.; Rammerstorfer, M.; Perkmann, T.; Burgmann, H.; Makristathis, A.; Dorffner, G.; Loetsch, F.; Blacky, A.; Ramharter, M. Neither Single nor a Combination of Routine Laboratory Parameters can Discriminate between Gram-positive and Gram-negative Bacteremia. Sci. Rep. 2015, 5, 16008. [Google Scholar] [CrossRef]
- Rennert-May, E.; Leal, J.; MacDonald, M.K.; Cannon, K.; Smith, S.; Exner, D.; Larios, O.E.; Bush, K.; Chew, D. Validating administrative data to identify complex surgical site infections following cardiac implantable electronic device implantation: A comparison of traditional methods and machine learning. Antimicrob. Resist. Infect. Control 2022, 11, 138. [Google Scholar] [CrossRef]
- Rhodes, N.J.; Rohani, R.; Yarnold, P.R.; Pawlowski, A.E.; Malczynski, M.; Qi, C.; Sutton, S.H.; Zembower, T.R.; Wunderink, R.G. Machine Learning to Stratify Methicillin-Resistant Staphylococcus aureus Risk among Hospitalized Patients with Community-Acquired Pneumonia. Antimicrob. Agents Chemother. 2023, 67, 1. [Google Scholar] [CrossRef]
- Sambarey, A.; Smith, K.; Chung, C.; Arora, H.S.; Yang, Z.; Agarwal, P.P.; Chandrasekaran, S. Integrative analysis of multimodal patient data identifies personalized predictors of tuberculosis treatment prognosis. iScience 2024, 27, 109025. [Google Scholar] [CrossRef]
- Savin, I.; Ershova, K.; Kurdyumova, N.; Ershova, O.; Khomenko, O.; Danilov, G.; Shifrin, M.; Zelman, V. Healthcare-associated ventriculitis and meningitis in a neuro-ICU: Incidence and risk factors selected by machine learning approach. J. Crit. Care 2018, 45, 95–104. [Google Scholar] [CrossRef]
- Schinkel, M.; Boerman, A.W.; Bennis, F.C.; Minderhoud, T.C.; Lie, M.; Peters-Sengers, H.; Holleman, F.; Schade, R.P.; de Jonge, R.; Wiersinga, W.J.; et al. Diagnostic stewardship for blood cultures in the emergency department: A multicenter validation and prospective evaluation of a machine learning prediction tool. EBioMedicine 2022, 82, 104176. [Google Scholar] [CrossRef]
- Seheult, J.N.; Stram, M.N.; Contis, L.; Pontzer, R.E.; Hardy, S.; Wertz, W.; Baxter, C.M.; Ondras, M.; Kip, P.L.; Snyder, G.M.; et al. Development, Evaluation, and Multisite Deployment of a Machine Learning Decision Tree Algorithm To Optimize Urinalysis Parameters for Predicting Urine Culture Positivity. J. Clin. Microbiol. 2023, 61, e0029123. [Google Scholar] [CrossRef]
- Shohat, N.; Goswami, K.; Tan, T.L.; Yayac, M.; Soriano, A.; Sousa, R.; Wouthuyzen-Bakker, M.; Parvizi, J. 2020 Frank Stinchfield Award: Identifying who will fail following irrigation and debridement for prosthetic joint infection. Bone Joint J. 2020, 102, 11–19. [Google Scholar] [CrossRef]
- Singh, H.; Gonzalez-Juarbe, N.; Pieper, R.; Yu, Y.; Vashee, S. Predictive biomarkers for latent Mycobacterium tuberculosis infection. Tuberculosis 2024, 147, 102399. [Google Scholar] [CrossRef]
- Sundermann, A.J.; Chen, J.; Miller, J.K.; Saul, M.I.; Shutt, K.A.; Griffith, M.P.; Mustapha, M.M.; Ezeonwuka, C.; Waggle, K.; Srinivasa, V.; et al. Outbreak of Pseudomonas aeruginosa Infections from a Contaminated Gastroscope Detected by Whole Genome Sequencing Surveillance. Clin. Infect. Dis. 2021, 73, E638–E642. [Google Scholar] [CrossRef]
- Tacconelli, E.; Górska, A.; De Angelis, G.; Lammens, C.; Restuccia, G.; Schrenzel, J.; Huson, D.H.; Carević, B.; Preoţescu, L.; Carmeli, Y.; et al. Estimating the association between antibiotic exposure and colonization with extended-spectrum β-lactamase-producing Gram-negative bacteria using machine learning methods: A multicentre, prospective cohort study. Clin. Microbiol. Infect. 2020, 26, 87–94. [Google Scholar] [CrossRef]
- Tadesse, B.T.; Khanam, F.; Ahmmed, F.; Liu, X.X.; Islam, M.T.; Kim, D.R.; Kang, S.S.; Im, J.; Chowdhury, F.; Ahmed, T.; et al. Association Among Household Water, Sanitation, and Hygiene (WASH) Status and Typhoid Risk in Urban Slums: Prospective Cohort Study in Bangladesh. JMIR Public Health Surveill. 2023, 9, e41207. [Google Scholar] [CrossRef]
- Tsurumi, A.; Flaherty, P.J.; Que, Y.A.; Ryan, C.M.; Banerjee, A.; Chakraborty, A.; Almpani, M.; Shankar, M.; Goverman, J.; Schulz, J.T.; et al. A preventive tool for predicting bloodstream infections in children with burns. Shock 2023, 59, 393–399. [Google Scholar] [CrossRef]
- Wang, H.Y.; Lee, T.Y.; Tseng, Y.J.; Liu, T.P.; Huang, K.Y.; Chang, Y.T.; Chen, C.H.; Lu, J.J. A new scheme for strain typing of methicillin-resistant Staphylococcus aureus on the basis of matrix-assisted laser desorption ionization time-of-flight mass spectrometry by using machine learning approach. PLoS ONE 2018, 13, e0194289. [Google Scholar] [CrossRef]
- Wang, H.; Lien, F.; Liu, T.; Chen, C.; Chen, C.; Lu, J. Application of a MALDI-TOF analysis platform (ClinProTools) for rapid and preliminary report of MRSA sequence types in Taiwan. PeerJ 2018, 6, e5784. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, G.; Zhao, Y.; Wang, C.; Chen, C.; Ding, Y.; Lin, J.; You, J.; Gao, S.; Pang, X. A deep learning model for predicting multidrug-resistant organism infection in critically ill patients. J. Intensive Care 2023, 11, 49. [Google Scholar] [CrossRef]
- Waterhouse, M.; Morton, A.; Mengersen, K.; Cook, D.; Playford, G. Role of overcrowding in meticillin-resistant Staphylococcus aureus transmission: Bayesian network analysis for a single public hospital. J. Hosp. Infect. 2011, 78, 92–96. [Google Scholar] [CrossRef]
- Wu, G.; Cheligeer, C.; Southern, D.A.; Martin, E.A.; Xu, Y.; Leal, J.; Ellison, J.; Bush, K.; Williamson, T.; Quan, H.; et al. Development of machine learning models for the detection of surgical site infections following total hip and knee arthroplasty: A multicenter cohort study. Antimicrob. Resist. Infect. Control 2023, 12, 88. [Google Scholar] [CrossRef]
- Yan, M.; Yan, M. Monitoring and Early Warning Analysis of the Epidemic Situation of Escherichia coli Based on Big Data Technology and Cloud Computing. J. Healthc. Eng. 2022, 2022, 8739447. [Google Scholar] [CrossRef]
- Zeng, Z.; Wu, J.; Qin, G.; Yu, D.; He, Z.; Zeng, W.; Zhou, H.; Lin, J.; Liu, L.; Qi, C.; et al. Using time-series chest radiographs and laboratory data by machine learning for identifying pulmonary infection and colonization of Acinetobacter baumannii. Respir. Res. 2024, 25, 2. [Google Scholar] [CrossRef]
- Zhong, Z.; Wang, X.; Li, J.; Zhang, B.; Yan, L.; Xu, S.; Chen, G.; Gao, H. A study on the diagnosis of the Helicobacter pylori coccoid form with artificial intelligence technology. Front. Microbiol. 2022, 13, 1008346. [Google Scholar] [CrossRef]
- Zwerwer, L.R.; Luz, C.F.; Soudis, D.; Giudice, N.; Nijsten, M.W.N.; Glasner, C.; Renes, M.H.; Sinha, B. Identifying the need for infection-related consultations in intensive care patients using machine learning models. Sci. Rep. 2024, 14, 2317. [Google Scholar] [CrossRef]
- Roser, M.P.A.; Hasell, J.; Ritchie, H.; Ortiz-Ospina, E.; World Bank Income Groups. World Bank Income Groups; The World Bank: Washington, DC, USA, 2023. [Google Scholar]
- Mustapha, M.M.; Srinivasa, V.R.; Griffith, M.P.; Cho, S.T.; Evans, D.R.; Waggle, K.; Ezeonwuka, C.; Snyder, D.J.; Marsh, J.W.; Harrison, L.H.; et al. Genomic Diversity of Hospital-Acquired Infections Revealed through Prospective Whole-Genome Sequencing-Based Surveillance. mSystems 2022, 7, e0138421. [Google Scholar] [CrossRef]
- Gouareb, R.; Bornet, A.; Proios, D.; Pereira, S.G.; Teodoro, D. Detection of Patients at Risk of Enterobacteriaceae Infection Using Graph Neural Networks: A Retrospective Study. medRxiv 2023. [Google Scholar] [CrossRef]
- Seheult, J.N.; Stram, M.N.; Contis, L.; Pontzer, R.E.; Hardy, S.; Wertz, W.; Baxter, C.M.; Ondras, M.; Kip, P.L.; Snyder, G.M.; et al. Development and Evaluation of a Machine Learning Recursive Partitioning Decision Tree Algorithm to Optimize Urinalysis Parameters to Predict Urine Culture Positivity. bioRxiv 2023. [Google Scholar] [CrossRef]
- Sartelli, M.; Barie, P.S.; Coccolini, F.; Abbas, M.; Abbo, L.M.; Abdukhalilova, G.K.; Abraham, Y.; Abubakar, S.; Abu-Zidan, F.M.; Adebisi, Y.A.; et al. Ten golden rules for optimal antibiotic use in hospital settings: The WARNING call to action. World J. Emerg. Surg. 2023, 18, 35. [Google Scholar] [CrossRef]
- Fitzpatrick, F.; Doherty, A.; Lacey, G. Using artificial intelligence in infection prevention. Curr. Treat. Options Infect. Dis. 2020, 12, 135–144. [Google Scholar] [CrossRef]
- Villanueva, P.; Coffin, S.E.; Mekasha, A.; McMullan, B.; Cotton, M.F.; Bryant, P.A. Comparison of Antimicrobial Stewardship and Infection Prevention and Control Activities and Resources Between Low-/Middle-and High-income Countries. Pediatr. Infect. Dis. J. 2022, 41, S3–S9. [Google Scholar] [CrossRef]
- Vilar-Compte, D.; Camacho-Ortiz, A.; Ponce-de-León, S. Infection Control in Limited Resources Countries: Challenges and Priorities. Curr. Infect. Dis. Rep. 2017, 19, 20. [Google Scholar] [CrossRef]
- Ciecierski-Holmes, T.; Singh, R.; Axt, M.; Brenner, S.; Barteit, S. Artificial intelligence for strengthening healthcare systems in low-and middle-income countries: A systematic scoping review. NPJ Digit. Med. 2022, 5, 162. [Google Scholar] [CrossRef]
- Souza, E.S.; Belei, R.A.; Carrilho, C.M.D.d.M.; Matsuo, T.; Yamada-Ogatta, S.F.; Andrade, G.; Perugini, M.R.E.; Pieri, F.M.; Dessunti, E.M.; Kerbauy, G. Mortality and risks related to healthcare-associated infection. Texto Contexto-Enferm. 2015, 24, 220–228. [Google Scholar] [CrossRef]
- De Oliveira, D.M.; Forde, B.M.; Kidd, T.J.; Harris, P.N.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33, 3. [Google Scholar] [CrossRef]
- Navidinia, M. The clinical importance of emerging ESKAPE pathogens in nosocomial infections. J. Paramed. Sci. 2016, 7, 3. [Google Scholar]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert. Rev. Anti-Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Airlangga, G. Optimizing Machine Learning Models for Urinary Tract Infection Diagnostics: A Comparative Study of Logistic Regression and Random Forest. J. Inform. Ekon. Bisnis 2024, 6, 246–250. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, B.; Haverstick, J.; Ibtehaz, N.; Muszyński, A.; Chen, X.; Chowdhury, M.E.H.; Zughaier, S.M.; Zhao, Y. Differentiation and classification of bacterial endotoxins based on surface enhanced Raman scattering and advanced machine learning. Nanoscale 2022, 14, 8806–8817. [Google Scholar] [CrossRef]
- Ghaffar Nia, N.; Kaplanoglu, E.; Nasab, A. Evaluation of artificial intelligence techniques in disease diagnosis and prediction. Discov. Artif. Intell. 2023, 3, 5. [Google Scholar] [CrossRef]
- Rahman, A.; Debnath, T.; Kundu, D.; Khan, M.S.I.; Aishi, A.A.; Sazzad, S.; Sayduzzaman, M.; Band, S.S. Machine learning and deep learning-based approach in smart healthcare: Recent advances, applications, challenges and opportunities. AIMS Public Health 2024, 11, 58. [Google Scholar] [CrossRef]
- Rahman, A.; Islam, M.J.; Karim, M.R.; Kundu, D.; Kabir, S. An Intelligent Vaccine Distribution Process in COVID-19 Pandemic Through Blockchain-Sdn Framework from Bangladesh Perspective. In Proceedings of the 2021 International Conference on Electronics, Communications and Information Technology (ICECIT), Khulna, Bangladesh, 14–16 September 2021; pp. 1–4. [Google Scholar]
- Rahman, T.; Khandakar, A.; Rahman, A.; Zughaier, S.M.; Al Maslamani, M.; Chowdhury, M.H.; Tahir, A.M.; Hossain, M.S.A.; Chowdhury, M.E.H. TB-CXRNet: Tuberculosis and Drug-Resistant Tuberculosis Detection Technique Using Chest X-ray Images. Cogn. Comput. 2024, 16, 1393–1412. [Google Scholar] [CrossRef]
- Goswami, M.; Mohanty, S.; Pattnaik, P.K. Optimization of Machine Learning Models through Quantization and Data Bit Reduction in Healthcare Datasets. Frankl. Open 2024, 8, 100136. [Google Scholar] [CrossRef]
- Gurevich, E.; El Hassan, B.; El Morr, C. Equity within AI systems: What can health leaders expect? Healthc. Manage. Forum. 2023, 36, 119–124. [Google Scholar] [CrossRef]
- Mondal, H.; Mondal, S. Chapter Thirteen—Ethical and social issues related to AI in healthcare. In Methods in Microbiology; Srivastava, A., Mishra, V., Eds.; Academic Press: Cambridge, MA, USA, 2024; Volume 55, pp. 247–281. [Google Scholar]
- Yadav, N.; Pandey, S.; Gupta, A.; Dudani, P.; Gupta, S.; Rangarajan, K. Data Privacy in Healthcare: In the Era of Artificial Intelligence. Indian. Dermatol. Online J. 2023, 14, 788–792. [Google Scholar] [CrossRef]
- Khalid, N.; Qayyum, A.; Bilal, M.; Al-Fuqaha, A.; Qadir, J. Privacy-preserving artificial intelligence in healthcare: Techniques and applications. Comput. Biol. Med. 2023, 158, 106848. [Google Scholar] [CrossRef]
- Mennella, C.; Maniscalco, U.; De Pietro, G.; Esposito, M. Ethical and regulatory challenges of AI technologies in healthcare: A narrative review. Heliyon 2024, 10, e26297. [Google Scholar] [CrossRef]
- Gerke, S.; Minssen, T.; Cohen, G. Chapter 12—Ethical and legal challenges of artificial intelligence-driven healthcare. In Artificial Intelligence in Healthcare; Bohr, A., Memarzadeh, K., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 295–336. [Google Scholar]
- Bateman, R.M.; Sharpe, M.D.; Jagger, J.E.; Ellis, C.G.; Solé-Violán, J.; López-Rodríguez, M.; Herrera-Ramos, E.; Ruíz-Hernández, J.; Borderías, L.; Horcajada, J.; et al. 36th International Symposium on Intensive Care and Emergency Medicine: Brussels, Belgium, 15–18 March 2016. Crit Care 2016, 20, 94. [Google Scholar] [CrossRef]
- Alie, M.S.; Negesse, Y. Machine learning prediction of adolescent HIV testing services in Ethiopia. Front. Public Health 2024, 12, 1341279. [Google Scholar] [CrossRef]
- Furuno, J.P.; Schweizer, M.L.; McGregor, J.C.; Perencevich, E.N. Economics of infection control surveillance technology: Cost-effective or just cost? Am. J. Infect. Control 2008, 36, S12–S17. [Google Scholar] [CrossRef]
- Li, Y.; Cao, Y.; Wang, M.; Wang, L.; Wu, Y.; Fang, Y.; Zhao, Y.; Fan, Y.; Liu, X.; Liang, H. Development and validation of machine learning models to predict MDRO colonization or infection on ICU admission by using electronic health record data. Antimicrob. Resist. Infect. Control 2024, 13, 74. [Google Scholar] [CrossRef]
- Khan, M.S.; Umer, H.; Faruqe, F. Artificial intelligence for low income countries. Humanit. Soc. Sci. Commun. 2024, 11, 1422. [Google Scholar] [CrossRef]
- Arksey, H.; O’Malley, L. Scoping studies: Towards a methodological framework. Int. J. Social. Res. Methodol. 2005, 8, 19–32. [Google Scholar] [CrossRef]
- Levac, D.; Colquhoun, H.; O’Brien, K.K. Scoping studies: Advancing the methodology. Implement. Sci. 2010, 5, 69. [Google Scholar] [CrossRef]
- Ritchie, J.; Spencer, L. Qualitative Data Analysis for Applied Policy Research. In Analyzing Qualitative Data; Bryman, A.B.B., Ed.; Routledge Member of the Taylor and Francis Group: London, UK, 1994. [Google Scholar]
- Spencer, L.R.J.; O’Connor, W. Analysis: Practices, Principles and Processes. In Qualitative Research Practice; Ritchie, J.L.J., Ed.; SAGE: Washington, DC, USA, 2003; p. 199. [Google Scholar]
- McGowan, J.; Sampson, M.; Salzwedel, D.M.; Cogo, E.; Foerster, V.; Lefebvre, C. PRESS Peer Review of Electronic Search Strategies: 2015 Guideline Statement. J. Clin. Epidemiol. 2016, 75, 40–46. [Google Scholar] [CrossRef]
- Blackwood, D. Peer Review of Electronic Search Strategies (PRESS): “Can you check my systematic review search strategy?”. HLA News 2015, 9–10. [Google Scholar]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.J.; Horsley, T.; Weeks, L.; et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef]
- Foster, E.D.; Deardorff, A. Open science framework (OSF). J. Med. Libr. Assoc. JMLA 2017, 105, 203. [Google Scholar] [CrossRef]
| Author’s Name | Country | Name of AI | Type of AI | Summarized Aim | Scope of Aim | Advantages | Limitations |
|---|---|---|---|---|---|---|---|
| Jeon K, 2022 [21] | Korea | AMRQuest software, v.2.1 | Machine Learning | Presumptive identification of MRSA | Pathogen identification | Enhanced Diagnostic Accuracy | Lack of Real-World Validation |
| Çaǧlayan Ç, 2022 [22] | USA | Logistic regression (LR), random forest (RF), extreme gradient boosting (XGBoost), artificial neural network (ANN), support vector machine (SVM) | Machine Learning | Identify patients likely to be colonized with VRE, CRE, or MRSA upon ICU admission | Pathogen identification | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Ötleş E, 2023 [23] | USA | L2-regularized logistic regression model | Machine Learning | Assess patient risk for hospital-onset CDI and evaluate effectiveness of AI models | Pathogen identification | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Aggarwal S, 2023 [24] | India | Alignment-based methods: BLASTPhage, BLASTHost, CRISPRPred Machine learning models: Random Forest (RF) and Gaussian Naive Bayes (GNB) Hybrid model: Ensemble method: | Machine Learning and Computational Biology | Facilitate researchers in the field of phage therapy | Therapeutic | Predictive Modeling and Risk Assessment | Data Limitation |
| Althomsons S, 2022 [25] | USA | ML/DL techniques after NBH (DT, RF, SVM, Regularized regression, Ensemble methods, GBM, ANNs) | Machine Learning and Deep Learning | Predict excess growth in genotyped tuberculosis clusters, with the goal of early identification of clusters | Pathogen identification | Early Detection and Prevention | Generalizability Issues |
| Aminian M, 2014 [26] | USA, France | Knowledge-based Bayesian network (KBBN) | Machine Learning and Computational Biology | Improve the classification accuracy of Mycobacterium tuberculosis complex (MTBC) clades | Pathogen identification | Enhanced Diagnostic Accuracy | Limited Scope and Applicability |
| Atkinson A, 2023 [27] | Switzerland | Decision trees, and Network graph analysis | Machine Learning | Improve existing outbreak investigation processes | Outbreak investigation and surveillance | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Azé J, 2015 [28] | Netherlands, Pakistan, France | Weka | Machine Learning | Develop a consensual taxonomy for MTC | Pathogen identification | Enhanced Diagnostic Accuracy | Generalizability Issues |
| Bournez C, 2023 [29] | Switzerland | CalcAMP | Machine Learning | Accelerate the discovery of new AMPs as alternatives to antibiotics | Therapeutic | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Camoez M, 2016 [30] | Spain | CLINPROTOOLS, MALDI BIOTYPER | Machine Learning | Automated discrimination of major MRSA lineages and to develop a reliable tool for S. aureus typing | Pathogen identification | Enhanced Diagnostic Accuracy | Limited Scope and Applicability |
| Cheah, A. L. Y, 2018 [31] | Australia | Hidden Markov Model (HMM) in conjunction with Bayesian inference | Machine Learning | Effectively control VRE spread in healthcare settings | Outbreak investigation and surveillance | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Cherkasov A, 2009 [32] | Canada | Artificial Neural Networks (ANNs) | Machine Learning and Deep Learning | Asses the antibacterial, physical, and harmful properties of a variety of small peptide antibiotics | Therapeutic | Improved Treatment Effectiveness | Lack of Real-World Validation |
| de Bruin JS, 2017 [33] | Austria | Rule-based system for processing medical knowledge, which is more related to knowledge representation and reasoning in the field of artificial intelligence. | Knowledge Discovery and Semantic Analysis | Facilitating electronic HAI surveillance | Infection risk assessment | Enhanced Diagnostic Accuracy | Generalizability Issues |
| Doan, T. N, 2015 [34] | Australia | Hidden Markov models (HMMs) | Machine Learning | Characterize the transmission dynamics of Acinetobacter baumannii in ICUs | Pathogen identification | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Feng C, 2023 [35] | China | Artificial Neural Network (ANN), Support Vector Machine (SVM), Logistic Regression, Random Forest, K-Nearest Neighbor, Decision Tree, and XGBoost. | Machine Learning | Predicting invasive Klebsiella pneumoniae liver abscess syndrome (IKPLAS) in diabetes mellitus | Pathogen identification | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Freire, M. P, 2022 [36] | Brazil, Italy | Random Forest Classifier | Machine Learning | Predict CRE colonization | Pathogen identification | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Goodman, K. E, 2019 [37] | USA | Decision trees (DT); classification and regression tree (CART) algorithm, “rpart” package, version 4.1–13, was used in the R statistical package (version 3.0.5) | Machine Learning | Predict the probability of CRO and/or CPO carriage | Pathogen identification | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Gouareb R, 2023 [38] | Switzerland | Graph Neural Networks (GNNs) | Deep Learning | Predict the risk of inpatient colonization by MDR Enterobacteriaceae | Pathogen identification | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Hattori S, 2020 [39] | Japan | Rotation Forest ensembles in Weka | Machine Learning | Early identification of clinically important bacteria | Pathogen identification | Early Detection and Prevention | Lack of Real-World Validation |
| Hsu, C. C, 2008 [40] | USA, Taiwan | Artificial Neural Networks (ANNs) | Machine Learning and Deep Learning | Create a tool that can aid in infection control and potentially reduce the need for active surveillance cultures, which are costly and labor-intensive | Pathogen identification | Enhanced Diagnostic Accuracy | Generalizability Issues |
| Jacques J, 2020 [41] | France | Multi-Objective Classification Algorithm for Imbalanced data (MOCA-I) | Machine Learning | Identify a set of risk factors for MDR pathogen carriage and infection. | Pathogen identification | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Jakobsen, R. S, 2024 [42] | Denmark | Bayesian Network (BN) | Machine Learning | Risk stratification of hospital-acquired urinary tract infections (HA-UTI) | Infection risk assessment | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Khaledi A, 2016 [43] | Germany | Potential Support Vector Machine (P-SVM) | Machine Learning | Genome based ML detection of resistance in Pseudomonas aeruginosa | Antimicrobial resistance and stewardship | Enhanced Diagnostic Accuracy | Limited Scope and Applicability |
| Khaledi A, 2020 [44] | Germany, Spain, Hungry, Romania | SVM | Machine Learning | Predictive models and identified biomarkers of resistance to four commonly administered antimicrobial drugs | Antimicrobial resistance and stewardship | Enhanced Diagnostic Accuracy | Generalizability Issues |
| Lapp Z, 2021 [45] | USA | SVM with a radial basis kernel, L2 regularized logistic regression, Elastic net, Random Forest | Machine Learning | Understand which factors, whether patient-related or microbial genomic, could discriminate between CRKP extraintestinal colonization and infection across multiple healthcare facilities | Pathogen identification | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Liang, Q. Q, 2024 [46] | China | XGBoost, SVM, Random Forest | Machine Learning | Predicting the occurrence of bloodstream infection and associated factors | Pathogen identification | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Liang, Q. Q, 2022 [47] | China | Random forest, XGBoost, Decision tree, Multivariable logistic regression | Machine Learning | Predict the occurrence of CR-GNB carriage in Intensive Care Unit (ICU) patients | Pathogen identification | Early Detection and Prevention | Generalizability Issues |
| Lyu, J. W, 2023 [48] | China | Convolutional Neural Network (CNN) | Deep Learning | Prediction of multidrug-resistant K. pneumoniae | Pathogen identification | Enhanced Diagnostic Accuracy | Lack of Real-World Validation |
| Marra, A. R., 2020 [49] | USA | SVM, decision trees, multilayer perceptron, radial basis function classifiers, K-nearest neighbor, bagging, boosting, logistic regression, random forest, and naïve Bayes models. | Machine Learning | Predict Clostridioides difficile infection in hospitalized patients using routinely available clinical data | Pathogen identification | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Noman, S. M., 2023 [50] | 65 countries | Weka (v3.9.2), Java | Machine Learning | Enhance the accuracy of antimicrobial resistance predictions | Antimicrobial resistance and stewardship | Enhanced Diagnostic Accuracy | Data Limitation |
| Panchavati, S., 2022 [51] | USA | XGBoost, Deep Long Short Term Memory neural network (D-LSTM), and one-dimensional convolutional neural network (1D-CNN) | Machine Learning and Deep Learning | Predict Clostridioides difficile infection (CDI) in hospitalized patients, facilitate enhanced clinical monitoring, earlier diagnosis, and timely implementation of infection control measures | Pathogen identification | Predictive Modeling and Risk Assessment | Lack of Real-World Validation |
| Rabhi, S., 2018 [20] | France | word2vec, Glove | Machine Learning and Deep Learning and Natural Language Processing | Detecting healthcare-associated infections (HAIs), to determine which method provides better accuracy and reliability in classifying HAIs using textual electronic medical records | Infection risk assessment | Enhanced Diagnostic Accuracy | Lack of Real-World Validation (Opacity of CNNs) |
| Ratzinger, F., 2015 [52] | Austria | Weka, R, MDCalc bvba | Machine Learning | Determine whether routine laboratory parameters could be used as surrogate markers to predict the type of bacterial pathogen in bloodstream infections | Infection risk assessment | Predictive Modeling and Risk Assessment | Limited Scope and Applicability |
| Rennert-May, E., 2022 [53] | Canada | Python version 3.9.12 and Scikit-Learn (used to train the logistic regression models) | Machine Learning | Determine the best approach for identifying CIED infections | Infection risk assessment | Enhanced Diagnostic Accuracy | Generalizability Issues |
| Rhodes, N. J., 2023 [54] | USA | Optimal data analysis | Machine Learning | Predict the risk of Methicillin-resistant Staphylococcus aureus (MRSA) in hospitalized patients with community-acquired pneumonia (CAP) early in the course of hospital admission | Pathogen identification | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Sambarey, A., 2024 [55] | Multiple countries | Python v. 3.7.14, Matlab R2021b, R studio v.4.3.0 | Machine Learning and Deep Learning | Improve the prediction of treatment outcomes and guide personalized treatment strategies for TB, particularly in the context of drug-resistant TB | Therapeutic | Predictive Modeling and Risk Assessment | Lack of Real-World Validation |
| Savin, I., 2018 [56] | Russia | RF and XGBoost | Machine Learning | Determine the incidence of healthcare-associated ventriculitis and meningitis (HAVM) in a neuro-ICU | Infection risk assessment | Predictive Modeling and Risk Assessment | Generalizability issues |
| Schinkel, M., 2022 [57] | USA, Netherlands | XGBoost | Machine Learning | Predict blood culture outcomes in the emergency department | Infection risk assessment | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Seheult, J. N., 2023 [58] | USA | Software Represented Using AI: R v 3.4.2 (with the “rpart” package): The machine learning decision tree algorithm (PittUDT) was implemented using the “rpart” package in R. (R v 3.4.2 itself is not AI) | Machine Learning | Optimize urinalysis parameters for predicting urine culture positivity | Infection risk assessment | Cost-Effectiveness and Efficiency | Generalizability Issues |
| Shohat, N., 2020 [59] | USA, Europe | Random Forest (RF) | Machine Learning | Accurately predict the outcome following irrigation and debridement (I&D) surgery for prosthetic joint infection | Infection risk assessment | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Singh, H., 2023 [60] | USA | Weka (version 3.8.6) | Machine Learning | Identify predictive biomarkers for latent Mycobacterium tuberculosis infection | Pathogen identification | Enhanced Diagnostic Accuracy | Generalizability Issues |
| Sundermann, A. J., 2021 [61] | USA | Enhanced Detection System for Healthcare-Associated Transmission (EDS-HAT) | Machine Learning | Enhance outbreak detection in hospitals by combining whole genome sequencing (WGS) surveillance, to identify and trace transmission routes of healthcare-associated infections | Outbreak investigation and surveillance | Early Detection and Prevention | Lack of Real-World Validation |
| Tacconelli, E., 2020 [62] | Italy, Serbia, Romania | Random Forest (RF) algorithm | Machine Learning | Measure the impact of antibiotic exposure on the acquisition of colonization with extended-spectrum β-lactamase-producing Gram-negative bacteria | Therapeutic | Predictive Modeling and Risk Assessment | Technical and Computational Challenges |
| Tadesse, B. T., 2023 [63] | Bangladesh | R Studio analytical software (R Foundation for Statistical Computing) “rpart” for decision tree modeling, “rpart.plot” for tree plotting, “pROC” for ROC curve analysis, “survival” for Cox models, and “dplyr” for data management | Machine Learning | Assess the association between household WASH status and typhoid risk in urban slums | Infection risk assessment | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Tsurumi, A, 2023 [64] | USA | Least Absolute Shrinkage and Selection Operator (LASSO) “machine learning AI algorithm” | Machine Learning | Predicting bloodstream infections in children with burns | Infection risk assessment | Early Detection and Prevention | Lack of Real-World Validation |
| Wang, H. Y. 2018 [65] | Taiwan | Decision tree (DT), Support vector machine (SVM), and k-nearest neighbor (KNN) for predictive modeling | Machine Learning and Deep Learning | Develop a new scheme for strain typing of methicillin-resistant Staphylococcus aureus (MRSA) | Pathogen identification | Enhanced Diagnostic Accuracy | Generalizability Issues |
| Wang, H. Y, 2018 [66] | Taiwan | ClinProTools software version 3.0 | Machine Learning and Deep Learning | Classifying major MLST types of MRSA | Pathogen identification | Enhanced Diagnostic Accuracy | Limited Scope and Applicability |
| Wang, Y, 2023 [67] | China | Backpropagation Neural Network (BPNN) | Deep Learning | Predicting multidrug-resistant organism (MDRO) infection in critically ill patients | Pathogen identification | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Waterhouse, M, 2011 [68] | Australia | Bayesian Networks (implemented in Netica and WinBUGS softwares utilizing AI) | Machine Learning | Understand the complex system of interrelationships between various factors that affect this transmission | Infection risk assessment | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Wu, G, 2023 [69] | Canada | XGBoost | Machine Learning | Automated detection of complex surgical site infections (SSIs) following total hip and knee arthroplasty | Infection risk assessment | Enhanced Diagnostic Accuracy | Generalizability Issues |
| Yan, M, 2022 [70] | China | Markov Model (MM): Machine Learning / Computational Biology Neural Network (NN): Machine Learning Support Vector Machine (SVM): Machine Learning Integrated Promoter Markov Discriminant (IPMD) algorithm: Machine Learning / Computational Biology | Machine Learning and Computational Biology | Establish an early warning system for the epidemic mechanism | Outbreak investigation and surveillance | Early Detection and Prevention | Generalizability Issues |
| Zeng, Z, 2024 [71] | China | Nested Logistic Regression Models (classified under Machine Learning (ML) rather than being standalone AI) | Machine Learning | Accurately classify pulmonary status caused by Acinetobacter baumannii | Pathogen identification | Enhanced Diagnostic Accuracy | Generalizability Issues |
| Zhong, Z, 2022 [72] | China | YOLO v5 | Deep Learning | Diagnostic accuracy of AI models in identifying the Helicobacter pylori | Pathogen identification | Enhanced Diagnostic Accuracy | Lack of Real-World Validation |
| Zwerwer, L. R, 2024 [73] | Netherlands | Long Short-Term Memory (LSTM) neural networks, Gradient Boosting Machines, Random Forest, Logistic Regression | Machine Learning and Deep Learning | Predict the need for infection-related consultations in ICU patients | Therapeutic | Predictive Modeling and Risk Assessment | Generalizability Issues |
| Scope (1) | Countries | Low-Income | Middle-Income | High-Income | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Scope (2) | Aim | Pathogen Identification | Methicillin-Resistant S. aureus | M. tuberculosis | Klebsiella spp. | C. difficile | A. baumannii | Carbapenem-Resistant Gram-Negative | Multidrug-Resistant Organisms | Other |
| Infection Risk Assessment | Healthcare-Associated Infections | Septicemia | Surgical Site Infection | Other Infections | ||||||
| Therapeutic | ||||||||||
| Outbreak Investigation and Surveillance | ||||||||||
| Antimicrobial Resistance Stewardship | ||||||||||
| Scope (3) | Type of AI | Machine Learning | Hybrid | Deep Learning | Computational Biology and Machine Learning | Knowledge Discovery and Semantic Analysis | Machine Learning (ML), Deep Learning (DL), and Natural Language Processing (NLP) | |||
| Scope (4) | Advantages | Enhanced Diagnostic Accuracy | Improved Treatment Effectiveness | Early Detection and Prevention | Cost-Effectiveness and Efficiency | Predictive Modeling and Risk Assessment | ||||
| Scope (5) | Limitations | Generalizability Issues | Lack of Real-World Validation | Limited Scope and Applicability | Technical and Computational Challenges | Data Limitation | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abu-El-Ruz, R.; AbuHaweeleh, M.N.; Hamdan, A.; Rajha, H.E.; Sarah, J.M.; Barakat, K.; Zughaier, S.M. Artificial Intelligence in Bacterial Infections Control: A Scoping Review. Antibiotics 2025, 14, 256. https://doi.org/10.3390/antibiotics14030256
Abu-El-Ruz R, AbuHaweeleh MN, Hamdan A, Rajha HE, Sarah JM, Barakat K, Zughaier SM. Artificial Intelligence in Bacterial Infections Control: A Scoping Review. Antibiotics. 2025; 14(3):256. https://doi.org/10.3390/antibiotics14030256
Chicago/Turabian StyleAbu-El-Ruz, Rasha, Mohannad Natheef AbuHaweeleh, Ahmad Hamdan, Humam Emad Rajha, Jood Mudar Sarah, Kaoutar Barakat, and Susu M. Zughaier. 2025. "Artificial Intelligence in Bacterial Infections Control: A Scoping Review" Antibiotics 14, no. 3: 256. https://doi.org/10.3390/antibiotics14030256
APA StyleAbu-El-Ruz, R., AbuHaweeleh, M. N., Hamdan, A., Rajha, H. E., Sarah, J. M., Barakat, K., & Zughaier, S. M. (2025). Artificial Intelligence in Bacterial Infections Control: A Scoping Review. Antibiotics, 14(3), 256. https://doi.org/10.3390/antibiotics14030256









