Evaluating the Diagnostic Performance of Long-Read Metagenomic Sequencing Compared to Culture and Antimicrobial Susceptibility Testing for Detection of Bovine Respiratory Bacteria and Indicators of Antimicrobial Resistance
Abstract
1. Introduction
2. Results
2.1. Detection of Target Bacteria via Long-Read Metagenomic Sequencing
2.2. Detection of Antimicrobial Resistance Genes in Bacterial Reads
2.3. Phenotypic Non-Susceptibility
2.4. Bayesian Latent Class Models for Detection of BRD Bacteria
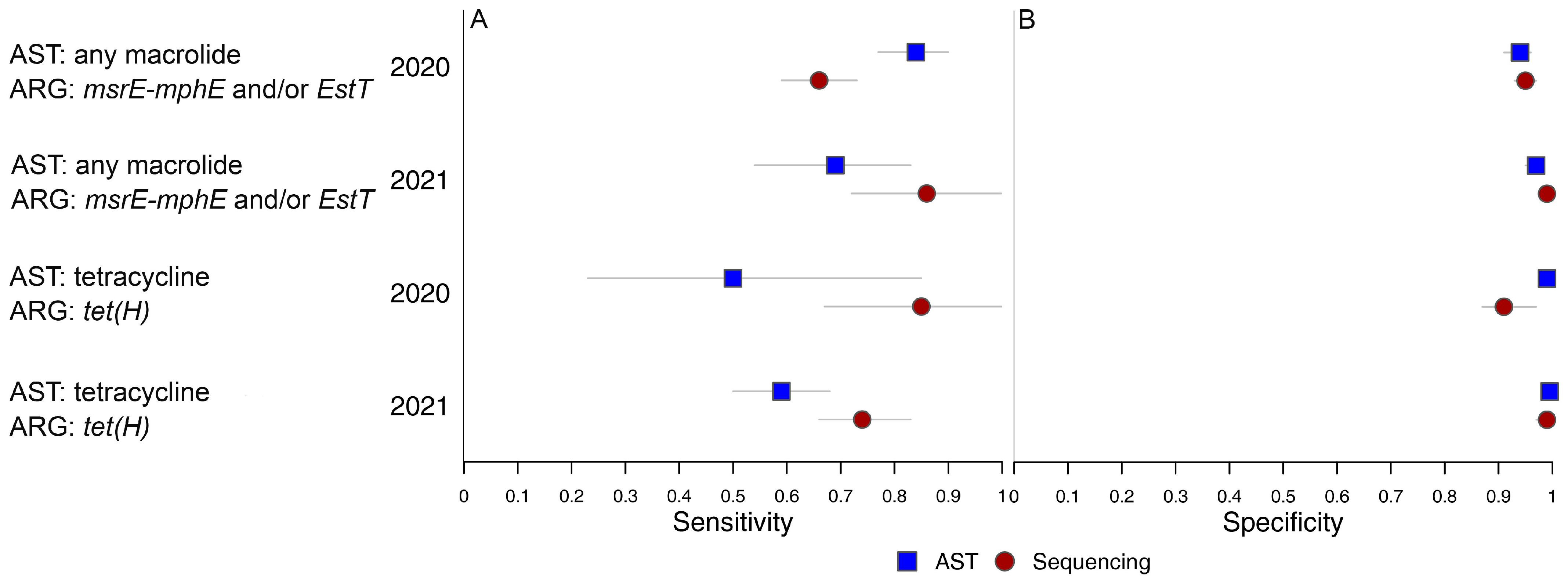
2.5. Bayesian Latent Class Models for Detection of Antimicrobial Non-Susceptibility in BRD Bacteria
2.6. Sensitivity Analyses Examining the Assumptions of the BLCM
2.7. Impacts of Se and Sp on Positive and Negative Predictive Value Based on Long-Read Metagenomic Sequencing
2.8. Differences in Detection of BRD-Associated Bacteria over Time Based on Long-Read Metagenomic Sequencing
2.9. Differences in Detection of ARGs in BRD-Associated Bacteria over Time
3. Discussion
4. Methods
4.1. Ethics Statement
4.2. Animals and Sample Population
4.3. Bacteriology and AST Protocols
4.4. Metagenomic Sequencing Sample Preparation Protocols
4.5. Sample Processing Protocol
4.6. Library Preparation and Metagenomic Sequencing Protocol
4.7. Preprocessing/Quality Control (QC)
4.8. Read Classification and Host Filtering
4.9. Antimicrobial Resistance Gene Detection
4.10. Data Management and Statistical Analyses
4.11. Bayesian Latent Class Models
4.12. Statistical Analysis of Differences in Bacterial ARG Detection Among Time Points
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial Resistance |
| AMU | Antimicrobial Use |
| ARG | Antimicrobial Resistance Gene |
| AST | Antimicrobial Susceptibility Testing |
| BHI | Brain Heart Infusion Broth |
| BLCM | Bayesian Latent Class Model |
| BRD | Bovine Respiratory Disease |
| BW | Body Weight |
| CARD | Comprehensive Antimicrobial Resistance Database |
| CCAC | Canadian Council of Animal Care |
| CLSI | Clinical and Laboratory Standards Institute |
| CrI | Credible Intervals |
| C/S | Culture and Antimicrobial Susceptibility Testing |
| DHPR | Dihydrofolate Reductase |
| DNPS | Deep Nasopharyngeal Swab |
| DOF | Days on Feed |
| MIC | Minimum Inhibitory Concentration |
| NPV | Negative Predictive Value |
| OIE | World Organization for Animal Health |
| ONT | Oxford Nanopore Technologies |
| PBS | Phosphate-Buffered Saline |
| PCR | Polymerase Chain Reaction |
| PPV | Positive Predictive Value |
| Se | Sensitivity |
| Sp | Specificity |
References
- Jorgensen, J.H. Laboratory issues in the detection and reporting of antibacterial resistance. Infect. Dis. Clin. N. Am. 1997, 11, 785–802. [Google Scholar] [CrossRef]
- Galhano, B.S.; Ferrari, R.G.; Panzenhagen, P.; de Jesus, A.C.S.; Conte-Junior, C.A. Antimicrobial Resistance Gene Detection Methods for Bacteria in Animal-Based Foods: A Brief Review of Highlights and Advantages. Microorganisms 2021, 9, 923. [Google Scholar] [CrossRef]
- Boolchandani, M.; D’Souza, A.W.; Dantas, G. Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet. 2019, 20, 356–370. [Google Scholar] [CrossRef]
- Aidara-Kane, A.; Angulo, F.J.; Conly, J.M.; Minato, Y.; Silbergeld, E.K.; McEwen, S.A.; Collignon, P.J.; Balkhy, H.; Collignon, P.; Conly, J.; et al. World Health Organization (WHO) guidelines on use of medically important antimicrobials in food-producing animals. Antimicrob. Resist. Infect. Control 2018, 7, 7. [Google Scholar] [CrossRef]
- Public Health Agency of Canada. Pan-Canadian Action Plan on Antimicrobial Resistance. 2023. Available online: https://www.canada.ca/en/public-health/services/publications/drugs-health-products/pan-canadian-action-plan-antimicrobial-resistance.html (accessed on 29 September 2025).
- Canadian Academy of Health Sciences. Antimicrobial Resistance/Antimicrobial Use in Food-Producing Animals in Canada: Strategic Interventions to Strengthen Antimicrobial Stewardship. 2025. Available online: https://cahs-acss.ca/wp-content/uploads/2025/03/CAHS-AMU-Report_EN_Final-1.pdf (accessed on 29 September 2025).
- Herman, E.K.; Lacoste, S.R.; Freeman, C.N.; Otto, S.J.G.; McCarthy, E.L.; Links, M.G.; Stothard, P.; Waldner, C.L. Bacterial enrichment prior to third-generation metagenomic sequencing improves detection of BRD pathogens and genetic determinants of antimicrobial resistance in feedlot cattle. Front. Microbiol. 2024, 15, 1386319. [Google Scholar] [CrossRef]
- United States Department of Agriculture; Animal and Plant Health Inspection Service; Veterinary Services. National Animal Health Monitoring System Feedlot 2011 Part IV: Health and Health Management on U.S. Feedlots with a Capacity of 1000 or More Head. 2013. Available online: https://www.aphis.usda.gov/sites/default/files/feed11_dr_partiv.pdf (accessed on 29 September 2025).
- Brault, S.A.; Hannon, S.J.; Gow, S.P.; Warr, B.N.; Withell, J.; Song, J.; Williams, C.M.; Otto, S.J.G.; Booker, C.W.; Morley, P.S. Antimicrobial Use on 36 Beef Feedlots in Western Canada: 2008–2012. Front. Vet. Sci. 2019, 6, 329. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Antimicrobial Resistance Surveillance System (GLASS): Molecular Methods for Antimicrobial Resistance (AMR) Diagnostics to Enhance the Global Antimicrobial Resistance Surveillance System; World Health Organization: Geneva, Switzerland, 2019; Available online: https://www.who.int/publications/i/item/WHO-WSI-AMR-2019.1 (accessed on 29 September 2025).
- Beef Cattle Research Council. Research Priorities. 2025. Available online: https://www.beefresearch.ca/research/research-priorities/ (accessed on 29 September 2025).
- McLain, J.E.; Cytryn, E.; Durso, L.M.; Young, S. Culture-based Methods for Detection of Antibiotic Resistance in Agroecosystems: Advantages, Challenges, and Gaps in Knowledge. J. Environ. Qual. 2016, 45, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Owen, J.R.; Noyes, N.; Young, A.E.; Prince, D.J.; Blanchard, P.C.; Lehenbauer, T.W.; Aly, S.S.; Davis, J.H.; O’Rourke, S.M.; Abdo, Z.; et al. Whole-Genome Sequencing and Concordance Between Antimicrobial Susceptibility Genotypes and Phenotypes of Bacterial Isolates Associated with Bovine Respiratory Disease. G3 Genes Genomes Genet. 2017, 7, 3059–3071. [Google Scholar] [CrossRef] [PubMed]
- Snyder, E.R.; Savitske, B.J.; Credille, B.C. Concordance of disk diffusion, broth microdilution, and whole-genome sequencing for determination of in vitro antimicrobial susceptibility of Mannheimia haemolytica. J. Vet. Intern. Med. 2020, 34, 2158–2168. [Google Scholar] [CrossRef]
- Freeman, C.N.; Herman, E.K.; Abi Younes, J.; Ramsay, D.E.; Erikson, N.; Stothard, P.; Links, M.G.; Otto, S.J.G.; Waldner, C. Evaluating the potential of third generation metagenomic sequencing for the detection of BRD pathogens and genetic determinants of antimicrobial resistance in chronically ill feedlot cattle. BMC Vet. Res. 2022, 18, 211. [Google Scholar] [CrossRef]
- Burbick, C.R.; Fajt, V.R.; Frey, E.; Fritz, H.; Goodman, L.B.; Lorenz, C.; Lubbers, B.V.; Marshall, E.; Rankin, S.C.; Silva, M. Benefits and challenges of creating veterinary antibiograms for empiric antimicrobial selection in support of antimicrobial stewardship and advancement of one-health goals. Am. J. Vet. Res. 2023, 84. [Google Scholar] [CrossRef] [PubMed]
- Dutta, E.; Loy, J.D.; Deal, C.A.; Wynn, E.L.; Clawson, M.L.; Clarke, J.; Wang, B. Development of a Multiplex Real-Time PCR Assay for Predicting Macrolide and Tetracycline Resistance Associated with Bacterial Pathogens of Bovine Respiratory Disease. Pathogens 2021, 10, 64. [Google Scholar] [CrossRef]
- Klima, C.L.; Zaheer, R.; Cook, S.R.; Booker, C.W.; Hendrick, S.; Alexander, T.W.; McAllister, T.A. Pathogens of Bovine Respiratory Disease in North American Feedlots Conferring Multidrug Resistance via Integrative Conjugative Elements. J. Clin. Microbiol. 2014, 52, 438–448. [Google Scholar] [CrossRef]
- Hirsch, C.; Timsit, E.; Uddin, M.S.; Guan, L.L.; Alexander, T.W. Comparison of pathogenic bacteria in the upper and lower respiratory tracts of cattle either directly transported to a feedlot or co-mingled at auction markets prior to feedlot placement. Front. Vet. Sci. 2023, 9, 1026470. [Google Scholar] [CrossRef] [PubMed]
- Woolums, A.; Karisch, B.; Frye, J.; Epperson, W.; Smith, D.; Blanton, J.; Austin, F.; Kaplan, R.; Hiott, L.; Woodley, T.; et al. Multidrug resistant Mannheimia haemolytica isolated from high-risk beef stocker cattle after antimicrobial metaphylaxis and treatment for bovine respiratory disease. Vet. Microbiol. 2018, 221, 143. [Google Scholar] [CrossRef]
- Ramsay, D.; McDonald, W.; Thompson, M.; Erickson, N.; Gow, S.; Osgood, N.D.; Waldner, C. Contagious acquisition of antimicrobial resistance is critical for explaining emergence in western Canadian feedlots—Insights from an agent-based modelling tool. Front. Vet. Sci. 2025, 11, 2024. [Google Scholar] [CrossRef]
- Johnson, W.O.; Jones, G.; Gardner, I.A. Gold standards are out and Bayes is in: Implementing the cure for imperfect reference tests in diagnostic accuracy studies. Prev. Vet. Med. 2019, 167, 113–127. [Google Scholar] [CrossRef]
- Collins, J.; Huynh, M. Estimation of diagnostic test accuracy without full verification: A review of latent class methods. Stat. Med. 2014, 33, 4141–4169. [Google Scholar] [CrossRef]
- Enøe, C.; Georgiadis, M.P.; Johnson, W.O. Estimation of sensitivity and specificity of diagnostic tests and disease prevalence when the true disease state is unknown. Prev. Vet. Med. 2000, 45, 61–81. [Google Scholar] [CrossRef]
- Cheung, A.; Dufour, S.; Jones, G.; Kostoulas, P.; Stevenson, M.A.; Singanallur, N.B.; Firestone, S.M. Bayesian latent class analysis when the reference test is imperfect. Rev. Sci. Tech. 2021, 40, 271–286. [Google Scholar] [CrossRef] [PubMed]
- Lewis, F.I.; Torgerson, P.R. A tutorial in estimating the prevalence of disease in humans and animals in the absence of a gold standard diagnostic. Emerg. Themes Epidemiol. 2012, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- World Organization for Animal Health. Principles and methods of validation of diagnostic assays for infectious diseases. Man. Diagn. Tests Vaccines Terr. Anim. 2023. [Google Scholar]
- Rattanapanadda, P.; Ramsay, D.; Butters, A.; Booker, C.W.; Hannon, S.J.; Hendrick, S.; Van Donkersgoed, J.; Warr, B.N.; Gow, S.P.; Morley, P.S. The prevalence and antimicrobial resistance of respiratory pathogens isolated from feedlot cattle in Canada. Front. Microbiol. 2025, 16, 2025. [Google Scholar] [CrossRef]
- Abi Younes, J.N.; Campbell, J.R.; Otto, S.J.G.; Gow, S.P.; Woolums, A.R.; Jelinski, M.; Lacoste, S.; Waldner, C.L. Variation in pen-level prevalence of BRD bacterial pathogens and antimicrobial resistance following feedlot arrival in beef calves. Antibiotics 2024, 13, 322. [Google Scholar] [CrossRef]
- Holman, D.B.; Klima, C.L.; Ralston, B.J.; Niu, Y.D.; Stanford, K.; Alexander, T.W.; McAllister, T.A. Metagenomic Sequencing of Bronchoalveolar Lavage Samples from Feedlot Cattle Mortalities Associated with Bovine Respiratory Disease. Genome Announc. 2017, 5. [Google Scholar] [CrossRef]
- Gaeta, N.C.; Lima, S.F.; Teixeira, A.G.V.; Ganda, E.; Oikonomou, G.; Gregory, L.; Bicalho, R.C. Deciphering upper respiratory tract microbiota complexity in healthy calves and calves that develop respiratory disease using shotgun metagenomics. J. Dairy Sci. 2017, 100, 1445–1458. [Google Scholar] [CrossRef] [PubMed]
- Neal, K.; Amachawadi, R.G.; White, B.J.; Shippy, T.D.; Theurer, M.E.; Larson, R.L.; Lubbers, B.V.; Kleinhenz, M. Nasopharyngeal Bacterial Prevalence and Microbial Diversity at First Treatment for Bovine Respiratory Disease (BRD) and Its Associations with Health and Mortality Outcomes in Feedyard Cattle. Microorganisms 2024, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Van Driessche, L.; De Neve, C.; Haesebrouck, F.; van Leenen, K.; Boyen, F.; Pardon, B. Storage time and temperature affect the isolation rate of Mannheimia haemolytica and Pasteurella multocida from bovine bronchoalveolar lavage samples. BMC Vet. Res. 2020, 16, 238. [Google Scholar] [CrossRef]
- Stanford, K.; Zaheer, R.; Klima, C.; McAllister, T.; Peters, D.; Niu, Y.D.; Ralston, B. Antimicrobial resistance in members of the bacterial bovine respiratory disease complex isolated from lung tissue of cattle mortalities managed with or without the use of antimicrobials. Microorganisms 2020, 8, 288. [Google Scholar] [CrossRef]
- Smith, E.M.; Monaghan, E.M.; Huntley, S.J.; Green, L.E. Short communication: Preliminary investigation into the effect of freezing and a cryopreservant on the recovery of mastitis pathogens from ewe milk. J. Dairy Sci. 2011, 94, 4850–4855. [Google Scholar] [CrossRef]
- Simione, F.P.; Brown, E.M.; Buck, C. American Type Culture Collection. ATCC Preservation Methods: Freezing and Freeze-Drying, 2nd ed.; Simione, F.P., Brown, E.M., Buck, C., Eds.; American Type Culture Collection: Rockville, MD, USA, 1991. [Google Scholar]
- Simione, F.P., Jr. Key issues relating to the genetic stability and preservation of cells and cell banks. J. Parenter. Sci. Technol. 1992, 46, 226–232. [Google Scholar]
- Pérez-Burillo, S.; Hinojosa-Nogueira, D.; Navajas-Porras, B.; Blasco, T.; Balzerani, F.; Lerma-Aguilera, A.; León, D.; Pastoriza, S.; Apaolaza, I.; Planes, F.J.; et al. Effect of Freezing on Gut Microbiota Composition and Functionality for In Vitro Fermentation Experiments. Nutrients 2021, 13, 2207. [Google Scholar] [CrossRef]
- Corbett, C.S.; De Buck, J.; Barkema, H.W. Effects of freezing on ability to detect Mycobacterium avium subsp. paratuberculosis from bovine tissues following culture. J. Vet. Diagn. Investig. 2018, 30, 743–746. [Google Scholar] [CrossRef]
- Garzon, A.; Hoyos-Jaramillo, A.; Hustad, S.; Byrne, B.A.; Fritz, H.M.; Lehenbauer, T.W.; Aly, S.; Pereira, R. In vitro evaluation of the effect of transport medium, temperature, and time on the recovery of Mannheimia haemolytica and Pasteurella multocida. JDS Commun. 2023, 4, 214–218. [Google Scholar] [CrossRef]
- Dohoo, I.R. Veterinary Epidemiologic Research, 2nd ed.; VER, Inc.: Charlottetown, PEI, Canada, 2014. [Google Scholar]
- Su, M.; Satola, S.W.; Read, T.D. Genome-Based Prediction of Bacterial Antibiotic Resistance. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Bonin, N.; Doster, E.; Worley, H.; Pinnell, L.J.; Bravo, J.E.; Ferm, P.; Marini, S.; Prosperi, M.; Noyes, N.; Morley, P.S.; et al. MEGARes and AMR++, v3.0: An updated comprehensive database of antimicrobial resistance determinants and an improved software pipeline for classification using high-throughput sequencing. Nucleic Acids Res. 2023, 51, D744–D752. [Google Scholar] [CrossRef] [PubMed]
- Dhindwal, P.; Thompson, C.; Kos, D.; Planedin, K.; Jain, R.; Jelinski, M.; Ruzzini, A. A neglected and emerging antimicrobial resistance gene encodes for a serine-dependent macrolide esterase. Proc. Natl. Acad. Sci. USA 2023, 120, e2219827120. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.S.; Warrass, R.; Douthwaite, S. Macrolide resistance conferred by rRNA mutations in field isolates of Mannheimia haemolytica and Pasteurella multocida. J. Antimicrob. Chemother. 2014, 70, 420–423. [Google Scholar] [CrossRef]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020, 21, 30. [Google Scholar] [CrossRef]
- Griffin, D.; Chengappa, M.M.; Kuszak, J.; McVey, D.S. Bacterial pathogens of the bovine respiratory disease complex. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 381–394. [Google Scholar] [CrossRef]
- Confer, A.W. Update on bacterial pathogenesis in BRD. Anim. Health Res. Rev. 2009, 10, 145–148. [Google Scholar] [CrossRef]
- Credille, B. Antimicrobial resistance in Mannheimia haemolytica: Prevalence and impact. Anim. Health Res. Rev. 2020, 21, 196–199. [Google Scholar] [CrossRef]
- Klima, C.L.; Holman, D.B.; Cook, S.R.; Conrad, C.C.; Ralston, B.J.; Allan, N.; Anholt, R.M.; Niu, Y.D.; Stanford, K.; Hannon, S.J.; et al. Multidrug resistance in Pasteurellaceae associated with bovine respiratory disease mortalities in North America from 2011 to 2016. Front. Microbiol. 2020, 11, 606438. [Google Scholar] [CrossRef]
- O’Connor, A.M.; Hu, D.; Totton, S.C.; Scott, N.; Winder, C.B.; Wang, B.; Wang, C.; Glanville, J.; Wood, H.; White, B.; et al. A systematic review and network meta-analysis of injectable antibiotic options for the control of bovine respiratory disease in the first 45 days post arrival at the feedlot. Anim. Health Res. Rev. 2019, 20, 163–181. [Google Scholar] [CrossRef]
- DeDonder, K.D.; Apley, M.D. A literature review of antimicrobial resistance in Pathogens associated with bovine respiratory disease. Anim. Health Res. Rev. 2015, 16, 125–134. [Google Scholar] [CrossRef]
- Andrés-Lasheras, S.; Ha, R.; Zaheer, R.; Lee, C.; Booker, C.W.; Dorin, C.; Van Donkersgoed, J.; Deardon, R.; Gow, S.; Hannon, S.J.; et al. Prevalence and Risk Factors Associated with Antimicrobial Resistance in Bacteria Related to Bovine Respiratory Disease—A Broad Cross-Sectional Study of Beef Cattle at Entry Into Canadian Feedlots. Front. Vet. Sci. 2021, 8, 692646. [Google Scholar] [CrossRef] [PubMed]
- Kos, D.; Jelinski, M.; Ruzzini, A. Retrospective analysis of antimicrobial resistance associated with bovine respiratory disease. Appl. Environ. Microbiol. 2025, 91, e0190924. [Google Scholar] [CrossRef] [PubMed]
- Klima, C.L.; Alexander, T.W.; Read, R.R.; Gow, S.P.; Booker, C.W.; Hannon, S.; Sheedy, C.; McAllister, T.A.; Selinger, L.B. Genetic characterization and antimicrobial susceptibility of Mannheimia haemolytica isolated from the nasopharynx of feedlot cattle. Vet. Microbiol. 2011, 149, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Desmolaize, B.; Rose, S.; Warrass, R.; Douthwaite, S. A novel Erm monomethyltransferase in antibiotic-resistant isolates of Mannheimia haemolytica and Pasteurella multocida. Mol. Microbiol. 2011, 80, 184–194. [Google Scholar] [CrossRef]
- Michael, G.B.; Eidam, C.; Kadlec, K.; Meyer, K.; Sweeney, M.T.; Murray, R.W.; Watts, J.L.; Schwarz, S. Increased MICs of gamithromycin and tildipirosin in the presence of the genes erm(42) and msr(E)-mph(E) for bovine Pasteurella multocida and Mannheimia haemolytica. J. Antimicrob. Chemother. 2012, 67, 1555–1557. [Google Scholar] [CrossRef]
- Kadlec, K.; Brenner Michael, G.; Sweeney, M.T.; Brzuszkiewicz, E.; Liesegang, H.; Daniel, R.; Watts, J.L.; Schwarz, S. Molecular basis of macrolide, triamilide, and lincosamide resistance in Pasteurella multocida from bovine respiratory disease. Antimicrob. Agents Chemother. 2011, 55, 2475–2477. [Google Scholar] [CrossRef]
- Chaslus-Dancla, E.; Lesage-Descauses, M.-C.; Leroy-Sétrin, S.; Martel, J.-L.; Lafont, J.-P. Tetracycline resistance determinants, Tet B and Tet M, detected in Pasteurella haemolytica and Pasteurella multocida from bovine herds. J. Antimicrob. Chemother. 1995, 36, 815–819. [Google Scholar] [CrossRef]
- Benedict, K.M.; Gow, S.P.; Reid-Smith, R.J.; Booker, C.W.; McAllister, T.A.; Morley, P.S. Latent class comparison of test accuracy when evaluating antimicrobial susceptibility using disk diffusion and broth microdilution to test E. coli and Mannheimia haemolytica isolates recovered from beef feedlot cattle. Epidemiol. Infect. 2014, 142, 2314–2325. [Google Scholar] [CrossRef]
- D’Amours, G.H.; Ward, T.I.; Mulvey, M.R.; Read, R.R.; Morck, D.W. Genetic diversity and tetracycline resistance genes of Histophilus somni. Vet. Microbiol. 2011, 150, 362–372. [Google Scholar] [CrossRef]
- Desmolaize, B.; Rose, S.; Wilhelm, C.; Warrass, R.; Douthwaite, S. Combinations of macrolide resistance determinants in field isolates of Mannheimia haemolytica and Pasteurella multocida. Antimicrob. Agents Chemother. 2011, 55, 4128–4133. [Google Scholar] [CrossRef]
- Rose, S.; Desmolaize, B.; Jaju, P.; Wilhelm, C.; Warrass, R.; Douthwaite, S. Multiplex PCR to identify macrolide resistance determinants in Mannheimia haemolytica and Pasteurella multocida. Antimicrob. Agents Chemother. 2012, 56, 3664–3669. [Google Scholar] [CrossRef]
- Snyder, E.R.; Alvarez-Narvaez, S.; Credille, B.C. Genetic characterization of susceptible and multi-drug resistant Mannheimia haemolytica isolated from high-risk stocker calves prior to and after antimicrobial metaphylaxis. Vet. Microbiol. 2019, 235, 110–117. [Google Scholar] [CrossRef]
- Deschner, D.; Voordouw, M.J.; Fernando, C.; Campbell, J.; Waldner, C.L.; Hill, J.E. Identification of genetic markers of resistance to macrolide class antibiotics in Mannheimia haemolytica isolates from a Saskatchewan feedlot. Appl. Environ. Microbiol. 2024, 90, e0050224. [Google Scholar] [CrossRef]
- Abi Younes, J.; Ramsay, D.E.; Lacoste, S.; Deschner, D.; Hill, J.E.; Campbell, J.; Waldner, C.L. Changes in the phenotypic susceptibility of Mannheimia haemolytica isolates to macrolide antimicrobials during the early feeding period following metaphylactic tulathromycin use in western Canadian feedlot calves. Can. Vet. J. 2022, 63, 920–928. [Google Scholar]
- Donlon, J.D.; McAloon, C.G.; Mee, J.F. Performance of various interpretations of clinical scoring systems for diagnosis of respiratory disease in dairy calves in a temperate climate using Bayesian latent class analysis. J. Dairy Sci. 2024, 107, 7138–7152. [Google Scholar] [CrossRef]
- Berman, J.; Francoz, D.; Abdallah, A.; Dufour, S.; Buczinski, S. Development and validation of a clinical respiratory disease scoring system for guiding treatment decisions in veal calves using a Bayesian framework. J. Dairy Sci. 2022, 105, 9917–9933. [Google Scholar] [CrossRef]
- Timsit, E.; Dendukuri, N.; Schiller, I.; Buczinski, S. Diagnostic accuracy of clinical illness for bovine respiratory disease (BRD) diagnosis in beef cattle placed in feedlots: A systematic literature review and hierarchical Bayesian latent-class meta-analysis. Prev. Vet. Med. 2016, 135, 67–73. [Google Scholar] [CrossRef]
- White, B.J.; Renter, D.G. Bayesian estimation of the performance of using clinical observations and harvest lung lesions for diagnosing bovine respiratory disease in post-weaned beef calves. J. Vet. Diagn. Investig. 2009, 21, 446. [Google Scholar] [CrossRef] [PubMed]
- Hesp, A.; Veldman, K.; Brouwer, M.S.M.; Wagenaar, J.A.; Mevius, D.; van Schaik, G. Latent class analysis to assess whole-genome sequencing versus broth microdilution for monitoring antimicrobial resistance in livestock. Prev. Vet. Med. 2021, 193, 105406. [Google Scholar] [CrossRef]
- Bokma, J.; Vereecke, N.; Pas, M.L.; Chantillon, L.; Vahl, M.; Weesendorp, E.; Deurenberg, R.H.; Nauwynck, H.; Haesebrouck, F.; Theuns, S.; et al. Evaluation of Nanopore Sequencing as a Diagnostic Tool for the Rapid Identification of Mycoplasma bovis from Individual and Pooled Respiratory Tract Samples. J. Clin. Microbiol. 2021, 59, e0111021. [Google Scholar] [CrossRef]
- Burckhardt, I.; Zimmermann, S. Susceptibility Testing of Bacteria Using Maldi-Tof Mass Spectrometry. Front. Microbiol. 2018, 9, 1744. [Google Scholar] [CrossRef]
- Bayot, M.L.; Bragg, B.N. Antimicrobial Susceptibility Testing. In StatPearls; StatPearls Publishing Copyright © 2025; StatPearls Publishing LLC: Treasure Island, FL, USA, 2025. [Google Scholar]
- Giguère, S.; Prescott, J.F.; Dowling, P.M. Antimicrobial Therapy in Veterinary Medicine, 5th ed.; Wiley Blackwell: Ames, IA, USA, 2013. [Google Scholar]
- Rubin, J.E.; Damborg, P. Antimicrobial Susceptibility Testing Methods and Interpretation of Results. In Antimicrobial Therapy in Veterinary Medicine; Wiley: Hoboken, NJ, USA, 2024; pp. 13–28. [Google Scholar]
- Abi Younes, J.N.; Campbell, J.R.; Gow, S.P.; Woolums, A.R.; Waldner, C.L. Association between respiratory disease pathogens in calves near feedlot arrival with treatment for bovine respiratory disease and subsequent antimicrobial resistance status. Front. Vet. Sci. 2024, 11. [Google Scholar] [CrossRef] [PubMed]
- Gardner, I.A.; Stryhn, H.; Lind, P.; Collins, M.T. Conditional dependence between tests affects the diagnosis and surveillance of animal diseases. Prev. Vet. Med. 2000, 45, 107–122. [Google Scholar] [CrossRef] [PubMed]
- Georgiadis, M.P.; Johnson, W.O.; Gardner, I.A.; Singh, R. Correlation-adjusted estimation of sensitivity and specificity of two diagnostic tests. J. R. Stat. Soc. Ser. C (Appl. Stat.) 2003, 52, 63–76. [Google Scholar] [CrossRef]
- Nissly, R.H.; Zaman, N.; Ibrahim, P.A.S.; McDaniel, K.; Lim, L.; Kiser, J.N.; Bird, I.; Chothe, S.K.; Bhushan, G.L.; Vandegrift, K.; et al. Influenza C and D viral load in cattle correlates with bovine respiratory disease (BRD): Emerging role of orthomyxoviruses in the pathogenesis of BRD. Virology 2020, 551, 10–15. [Google Scholar] [CrossRef]
- Kudirkiene, E.; Aagaard, A.K.; Schmidt, L.M.B.; Pansri, P.; Krogh, K.M.; Olsen, J.E. Occurrence of major and minor pathogens in calves diagnosed with bovine respiratory disease. Vet. Microbiol. 2021, 259, 109135. [Google Scholar] [CrossRef]
- Holman, D.B.; Timsit, E.; Booker, C.W.; Alexander, T.W. Injectable antimicrobials in commercial feedlot cattle and their effect on the nasopharyngeal microbiota and antimicrobial resistance. Vet. Microbiol. 2018, 214, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Holman, D.B.; Yang, W.; Alexander, T.W. Antibiotic treatment in feedlot cattle: A longitudinal study of the effect of oxytetracycline and tulathromycin on the fecal and nasopharyngeal microbiota. Microbiome 2019, 7, 86. [Google Scholar] [CrossRef]
- Guo, Y.; McMullen, C.; Timsit, E.; Hallewell, J.; Orsel, K.; van der Meer, F.; Yan, S.; Alexander, T.W. Genetic relatedness and antimicrobial resistance in respiratory bacteria from beef calves sampled from spring processing to 40 days after feedlot entry. Vet. Microbiol. 2020, 240, 108478. [Google Scholar] [CrossRef] [PubMed]
- Crosby, W.B.; Karisch, B.B.; Hiott, L.M.; Pinnell, L.J.; Pittman, A.; Frye, J.G.; Jackson, C.R.; Loy, J.D.; Epperson, W.B.; Blanton, J.; et al. Tulathromycin metaphylaxis increases nasopharyngeal isolation of multidrug resistant Mannheimia haemolytica in stocker heifers. Front. Vet. Sci. 2023, 10. [Google Scholar] [CrossRef] [PubMed]
- Snyder, E.; Credille, B.; Berghaus, R.; Giguère, S. Prevalence of multi drug antimicrobial resistance in isolated from high-risk stocker cattle at arrival and two weeks after processing. J. Anim. Sci. 2017, 95, 1124–1131. [Google Scholar] [CrossRef]
- Olfert, E.D.; Cross, B.M.; McWilliam, A.A. Guide to the Care and Use of Experimental Animals, 2nd ed.; The Council: Ottawa, ON, USA, 1993.
- National Research Council. Nutrient Requirements of Beef Cattle: Seventh Revised Edition: Update 2000; The National Academies Press: Washington, DC, USA, 2000; p. 248. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacterial Isolated from Animals, 6th ed.; CLSI Supplement VET01S ed.; The Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2023. [Google Scholar]
- Wick, R.R.; Judd, L.M.; Holt, K.E. Deepbinner: Demultiplexing barcoded Oxford Nanopore reads with deep convolutional neural networks. PLoS Comput. Biol. 2018, 14, e1006583. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 2017, 3, e000132. [Google Scholar] [CrossRef]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, T.; Kodali, V.K.; Pujar, S.; Brover, V.; Robbertse, B.; Farrell, C.M.; Oh, D.H.; Astashyn, A.; Ermolaeva, O.; Haddad, D.; et al. NCBI RefSeq: Reference sequence standards through 25 years of curation and annotation. Nucleic Acids Res. 2025, 53, D243–D257. [Google Scholar] [CrossRef]
- Hayes, B.J.; Daetwyler, H.D. 1000 Bull Genomes Project to Map Simple and Complex Genetic Traits in Cattle: Applications and Outcomes. Annu. Rev. Anim. Biosci. 2019, 7, 89–102. [Google Scholar] [CrossRef]
- Lu, J.; Rincon, N.; Wood, D.E.; Breitwieser, F.P.; Pockrandt, C.; Langmead, B.; Salzberg, S.L.; Steinegger, M. Metagenome analysis using the Kraken software suite. Nat. Protoc. 2022, 17, 2815–2839. [Google Scholar] [CrossRef]
- Bushnell, B. BBTools. Available online: https://sourceforge.net/projects/bbmap/ (accessed on 29 September 2025).
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating species abundance in metagenomics data. PeerJ Comput. Sci. 2017, 3, e104. [Google Scholar] [CrossRef]
- Li, H. Seqtk. Available online: https://github.com/lh3/seqtk (accessed on 29 September 2025).
- Seeman, T. Abricate (1.0.1). Available online: https://github.com/tseemann/abricate (accessed on 29 September 2025).
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.-H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2022, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed]
- Plummer, M. JAGS: A Program for Analysis of Bayesian Graphical Models using Gibbs Sampling. In Proceedings of the 3rd International Workshop on Distributed Statistical Computing (DSC 2003), Vienna, Austria, 20–22 March 2003. [Google Scholar]
- Denwood, M.J. runjags: An R Package Providing Interface Utilities, Model Templates, Parallel Computing Methods and Additional Distributions for MCMC Models in JAGS. J. Stat. Softw. 2016, 71, 1–25. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available online: https://www.r-project.org/ (accessed on 29 September 2025).
- Sergeant, E.S.G. Epitools Epidemiological Calculators. Available online: http://epitools.ausvet.com.au/ (accessed on 29 September 2025).

| Year | Bacteria | Median Total Base Pairs (bp) | Median of Per Sample Median Read Lengths (bp) | Median Theoretical Coverage (×Genome Size) 1 | Median Number of Reads | Percent (%) of Samples with ≥1 Read |
|---|---|---|---|---|---|---|
| 2020 (n = 909 samples) | M. haemolytica | 5,161,422 | 1321 | 1.8 | 2304 | 100% (909) |
| P. multocida | 1,088,197 | 1371 | 0.47 | 525 | 99.7% (906) | |
| H. somni | 11,787 | 1099 | 0.01 | 8 | 89.5% (814) | |
| 2021 (n = 1079 samples) | M. haemolytica | 3,979,840 | 3858 | 1.4 | 775 | 100% (1079) |
| P. multocida | 228,236 | 3351 | 0.10 | 46 | 99.3% (1071) | |
| H. somni | 13,170 | 2591 | 0.01 | 4 | 78.3% (845) |
| Number (%) of Samples in Which Gene Was Detected | ||||
|---|---|---|---|---|
| Gene 1 | Resistance class | Total (n = 1985) | 2020 (n = 909) | 2021 (n = 1076) 2 |
| sul2 | sulfonamides | 327 (16%) | 234 (26%) | 93 (8.6%) |
| tet(H) | tetracyclines | 313 (16%) | 138 (15%) | 175 (16%) |
| mphE | macrolides | 181 (9.1%) | 178 (20%) | 3 (0.3%) |
| msrE | macrolides | 177 (8.9%) | 174 (19%) | 3 (0.3%) |
| APH(3″)-Ib | aminoglycosides | 165 (8.3%) | 85 (9.4%) | 80 (7.4%) |
| APH(3′)-Ia | aminoglycosides | 160 (8.1%) | 84 (9.2%) | 76 (7.1%) |
| APH(6)-Id | aminoglycosides | 157 (7.9%) | 81 (8.9%) | 76 (7.1%) |
| EstT | macrolides | 131 (6.6%) | 60 (6.6%) | 71 (6.6%) |
| 2020 (n = 909) | 2021 (n = 1076) 1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tulathromycin Metaphylaxis | Oxytetracycline Metaphylaxis (n = 559) | Tulathromycin Metaphylaxis (n = 517) | |||||||||||
| DOF | n | msrE-mphE | EstT | tet(H) | n | msrE-mphE | EstT | tet(H) | n | msrE-mphE | EstT | tet(H) | |
| M. haemolytica | Arrival processing | 426 | 10 (2.3%) | 5 (1.2%) | 52 (12%) | 214 | 0 | 2 (0.9%) | 5 (2.3%) | 198 | 0 | 0 | 7 (3.5%) |
| 13 | 404 | 134 (33%) | 33 (8.2%) | 57 (14%) | 215 | 0 | 8 (3.7%) | 45 (21%) | 200 | 1 (0.5%) | 27 (14%) | 36 (18%) | |
| 36 | 79 | 40 (51%) | 11 (14%) | 22 (28%) | 130 | 0 | 2 (1.5%) | 32 (25%) | 119 | 1 (0.8%) | 28 (24%) | 34 (29%) | |
| P. multocida | Arrival processing | 426 | 2 (0.5%) | 1 (0.2%) | 22 (5.2%) | 214 | 0 | 0 | 2 (0.9%) | 198 | 0 | 0 | 5 (2.5%) |
| 13 | 404 | 9 (2.2%) | 3 (0.7%) | 15 (3.7%) | 215 | 0 | 0 | 25 (12.0%) | 200 | 0 | 0 | 8 (4.0%) | |
| 36 | 79 | 0 | 0 | 1 (1.3%) | 130 | 0 | 0 | 19 (15%) | 119 | 0 | 0 | 6 (5.0%) | |
| H. somni | Arrival processing | 426 | 1 (0.2%) | 6 (1.4%) | 20 (4.7%) | 214 | 0 | 0 | 0 | 198 | 0 | 0 | 0 |
| 13 | 404 | 59 (15%) | 30 (7.4%) | 10 (2.5%) | 215 | 0 | 4 (1.9%) | 11 (5.1%) | 200 | 0 | 15 (7.5%) | 8 (4.0%) | |
| 36 | 79 | 17 (22%) | 6 (7.6%) | 3 (3.8%) | 130 | 0 | 1 (0.8%) | 15 (12%) | 119 | 2 (1.7%) | 16 (13%) | 12 (10%) | |
| Any of: M. haemolytica, P. multocida, or H. somni | Arrival processing 2 | 426 | 13 (3.1%) | 9 (2.1%) | 58 (14%) | 214 | 0 | 2 (0.9%) | 5 (2.3%) | 198 | 0 | 0 | 9 (4.5%) |
| 13 | 404 | 138 (34%) | 39 (10%) | 58 (14%) | 215 | 0 | 8 (3.7%) | 52 (24%) | 200 | 1 (0.5%) | 30 (15%) | 36 (18%) | |
| 36 | 79 | 40 (51%) | 12 (15%) | 22 (28%) | 130 | 0 | 2 (1.5%) | 37 (28%) | 119 | 2 (1.7%) | 29 (24%) | 36 (30%) | |
| 2020 (n = 909) | 2021 (n = 1076) 3 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tulathromycin Metaphylaxis | Oxytetracycline Metaphylaxis (n = 559) | Tulathromycin Metaphylaxis (n = 517) | ||||||||||||||
| DOF | n | Any Macrolide | GAM or TULA | TILD or TILM 4 | TET | n | Any Macrolide | GAM or TULA | TILD or TILM 4 | TET | n | Any Macrolide | GAM or TULA | TILD or TILM 4 | TET | |
| M. haemolytica | Arrival processing | 426 | 5 (1.2%) | 1 (0.2%) | 5 (1.2%) | 5 (1.2%) | 214 | 8 (3.7%) | 0 | 8 (3.7%) | 0 | 198 | 5 (2.5%) | 0 | 5 (2.5%) | 0 |
| 13 | 404 | 196 (49%) | 196 (49%) | 135 (33%) | 14 (3.5%) | 215 | 4 (1.9%) | 1 (0.5%) | 4 (1.9%) | 1 (0.5%) | 200 | 22 (11%) | 21 (11%) | 22 (11%) | 21 (11%) | |
| 36 | 79 | 44 (56%) | 43 (54%) | 33 (42%) | 8 (10%) | 130 | 5 (3.8%) | 0 | 5 (3.8%) | 0 | 119 | 27 (23%) | 10 (8.4%) | 27 (23%) | 26 (22%) | |
| P. multocida | Arrival processing | 426 | 0 | 0 | 0 | 13 (3.1%) | 214 | 0 | 0 | 0 | 4 (1.9%) | 198 | 0 | 0 | 0 | 4 (2.0%) |
| 13 | 404 | 1 (0.2%) | 1 (0.2%) | 0 | 5 (1.2%) | 215 | 0 | 0 | 0 | 29 (13%) | 200 | 0 | 0 | 0 | 0 | |
| 36 | 79 | 1 (1.3%) | 1 (1.3%) | 0 | 3 (3.8%) | 130 | 0 | 0 | 0 | 19 (15%) | 119 | 1 (0.8%) | 1 (0.8%) | 0 | 0 | |
| H. somni | Arrival processing | 426 | 22 (5.2%) | 22 (5.2%) | 16 (3.8%) | 0 | 214 | 2 (0.9%) | 2 (0.9%) | 0 | 1 (0.5%) | 198 | 0 | 0 | 0 | 0 |
| 13 | 404 | 8 (2.0%) | 6 (1.5%) | 2 (0.5%) | 0 | 215 | 1 (0.5%) | 0 | 1 (0.5%) | 11 (5.1%) | 200 | 1 (0.5%) | 1 (0.5%) | 0 | 0 | |
| 36 | 79 | 12 (15%) | 12 (15%) | 5 (6.3%) | 0 | 130 | 2 (1.5%) | 2 (1.5%) | 0 | 12 (9.2%) | 119 | 2 (1.7%) | 2 (1.7%) | 0 | 6 (5.0%) | |
| Any of: M. haemolytica, P. multocida, or H. somni | Arrival processing | 426 | 27 (6.3%) | 23 (5.4%) | 21 (4.9%) | 18 (4.2%) | 214 | 10 (4.7%) | 2 (0.9%) | 8 (3.7%) | 5 (2.3%) | 198 | 5 (2.5%) | 0 | 5 (2.5%) | 4 (2.0%) |
| 13 | 404 | 201 (50%) | 200 (50%) | 136 (34%) | 19 (4.7%) | 215 | 5 (2.3%) | 1 (0.5%) | 5 (2.3%) | 40 (19%) | 200 | 23 (12%) | 22 (11%) | 22 (11%) | 21 (11%) | |
| 36 | 79 | 53 (67%) | 52 (66%) | 36 (46%) | 11 (14%) | 130 | 7 (5.4%) | 2 (1.5%) | 5 (3.8%) | 31 (24%) | 119 | 30 (25%) | 13 (11%) | 27 (23%) | 32 (27%) | |
| 2020 (n = 909) | 2021 (n = 1079) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tulathromycin Metaphylaxis | Oxytetracycline Metaphylaxis | Tulathromycin Metaphylaxis | ||||||||
| Theoretical Coverage Cutoff | DOF | n | Samples Above Cutoff | Theoretical Coverage Cutoff | DOF | n | Samples Above Cutoff | n | Samples Above Cutoff | |
| M. haemolytica | >5.1× | Arrival processing | 426 | 140 (33%) | >1.7× | Arrival processing | 215 | 100 (47%) | 199 | 98 (49%) |
| 13 | 404 | 138 (34%) | 13 | 215 | 120 (56%) | 200 | 74 (37%) | |||
| 36 | 79 | 56 (71%) | 36 | 130 | 66 (51%) | 120 | 72 (60%) | |||
| P. multocida | >1.2× | Arrival processing | 426 | 258 (61%) | >0.26× | Arrival processing | 215 | 92 (43%) | 199 | 90 (45%) |
| 13 | 404 | 31 (7.7%) | 13 | 215 | 86 (40%) | 200 | 32 (16%) | |||
| 36 | 79 | 17 (22%) | 36 | 130 | 73 (56%) | 120 | 39 (33%) | |||
| H. somni | >0.09× | Arrival processing | 426 | 50 (12%) | >0.05× | Arrival processing | 215 | 13 (6.1%) | 199 | 28 (14%) |
| 13 | 404 | 47 (12%) | 13 | 215 | 34 (16%) | 200 | 22 (11%) | |||
| 36 | 79 | 27 (34%) | 36 | 130 | 94 (72%) | 120 | 66 (55%) | |||
| 2020 (n = 909) | 2021 (n = 1079) | |||||
|---|---|---|---|---|---|---|
| Bacteria | Metric | Method | Median | 95% CrI | Median | 95% CrI |
| M. haemolytica | Sensitivity | Culture | 0.99 | 0.96, 0.999 | 0.90 | 0.81, 0.996 |
| Sequencing 1 | 0.71 | 0.65, 0.78 | 0.91 | 0.87, 0.96 | ||
| Specificity | Culture | 0.97 | 0.91, 0.999 | 0.99 | 0.95, 0.999 | |
| Sequencing 1 | 0.92 | 0.89, 0.95 | 0.90 | 0.83, 0.996 | ||
| P. multocida | Sensitivity | Culture | 0.86 | 0.81, 0.90 | 0.77 | 0.70, 0.84 |
| Sequencing 1 | 0.96 | 0.92, 0.999 | 0.89 | 0.84, 0.94 | ||
| Specificity | Culture | 0.94 | 0.91, 0.96 | 0.99 | 0.96, 0.999 | |
| Sequencing 1 | 0.98 | 0.96, 0.999 | 0.97 | 0.93, 0.999 | ||
| H. somni | Sensitivity | Culture | 0.84 | 0.65, 0.999 | 0.79 | 0.73, 0.86 |
| Sequencing 1 | 0.52 | 0.36, 0.67 | 0.86 | 0.81, 0.91 | ||
| Specificity | Culture | 0.97 | 0.95, 0.99 | 0.99 | 0.98, 0.999 | |
| Sequencing 1 | 0.90 | 0.88, 0.92 | 0.97 | 0.95, 0.99 | ||
| 2020 (n = 909) | 2021 (n = 1076) | |||||
|---|---|---|---|---|---|---|
| Model | Metric | Method | Median | 95% CrI | Median | 95% CrI |
| AST: any macrolide ARG: msrE-mphE | Se | AST | 0.86 | 0.80, 0.92 | n/a | |
| Seq | 0.61 | 0.55, 0.68 | ||||
| Sp | AST | 0.94 | 0.91, 0.96 | n/a | ||
| Seq | 0.97 | 0.95, 0.99 | ||||
| AST: GAM or TULA 1 ARG: msrE-mphE | Se | AST | 0.85 | 0.79, 0.91 | n/a | |
| Seq | 0.60 | 0.54, 0.67 | ||||
| Sp | AST | 0.95 | 0.92, 0.97 | n/a | ||
| Seq | 0.97 | 0.95, 0.98 | ||||
| AST: TILD or TILM 2 ARG: msrE-mphE | Se | AST | 0.58 | 0.50, 0.66 | n/a | |
| Seq | 0.62 | 0.53, 0.70 | ||||
| Sp | AST | 0.95 | 0.93, 0.97 | n/a | ||
| Seq | 0.97 | 0.95, 0.99 | ||||
| AST: any macrolide ARG: EstT | Se | AST | 0.67 | 0.55, 0.78 | 0.69 | 0.54, 0.84 |
| Seq | 0.13 | 0.09, 0.17 | 0.86 | 0.72, 0.999 | ||
| Sp | AST | 0.96 | 0.93, 0.999 | 0.97 | 0.95, 0.979 | |
| Seq | 0.98 | 0.96, 0.99 | 0.99 | 0.978, 0.998 | ||
| AST: GAM or TULA 1 ARG: EstT | Se | AST | 0.65 | 0.54, 0.77 | 0.38 | 0.26, 0.509 |
| Seq | 0.12 | 0.09, 0.16 | 0.69 | 0.514, 0.85 | ||
| Sp | AST | 0.96 | 0.93, 0.99 | 0.995 | 0.99, 0.999 | |
| Seq | 0.98 | 0.96, 0.99 | 0.99 | 0.99, 0.999 | ||
| AST: TILD or TILM 2 ARG: EstT | Se | AST | 0.52 | 0.39, 0.67 | 0.69 | 0.54, 0.83 |
| Seq | 0.15 | 0.10, 0.20 | 0.92 | 0.79, 0.999 | ||
| Sp | AST | 0.97 | 0.95, 0.999 | 0.97 | 0.96, 0.983 | |
| Seq | 0.98 | 0.97, 0.998 | 0.99 | 0.978, 0.998 | ||
| AST: any macrolide ARG: msrE-mphE and/or EstT | Se | AST | 0.84 | 0.77, 0.90 | 0.69 | 0.54, 0.83 |
| Seq | 0.66 | 0.59, 0.73 | 0.86 | 0.72, 0.998 | ||
| Sp | AST | 0.94 | 0.91, 0.96 | 0.97 | 0.95, 0.979 | |
| Seq | 0.95 | 0.93, 0.97 | 0.99 | 0.978, 0.998 | ||
| AST: TET ARG: tet(H) | Se | AST | 0.50 | 0.23, 0.85 | 0.59 | 0.50, 0.68 |
| Seq | 0.85 | 0.67, 0.999 | 0.74 | 0.66, 0.83 | ||
| Sp | AST | 0.99 | 0.98, 0.999 | 0.995 | 0.99, 0.999 | |
| Seq | 0.91 | 0.87, 0.97 | 0.99 | 0.97, 0.999 | ||
| Bacteria | Year/ Metaphylaxis | DOF Comparison | Odds Ratio | 95% CI Lower | 95% CI Upper | p-Value |
|---|---|---|---|---|---|---|
| M. haemolytica | 2020/Tulathromycin | 13 DOF vs. 1 DOF | 1.1 | 0.80 | 1.4 | 0.65 |
| 36 DOF vs. 1 DOF | 5.5 | 3.2 | 9.4 | ≤0.001 | ||
| 36 DOF vs. 13 DOF | 5.1 | 3.0 | 8.8 | ≤0.001 | ||
| P. multocida | 2020/Tulathromycin | 13 DOF vs. 1 DOF | 0.03 | 0.015 | 0.06 | ≤0.001 |
| 36 DOF vs. 1 DOF | 0.12 | 0.06 | 0.25 | ≤0.001 | ||
| 36 DOF vs. 13 DOF | 4.1 | 1.9 | 8.6 | ≤0.001 | ||
| H. somni | 2020/Tulathromycin | 13 DOF vs. 1 DOF | 0.98 | 0.64 | 1.5 | 0.94 |
| 36 DOF vs. 1 DOF | 3.9 | 2.3 | 6.9 | ≤0.001 | ||
| 36 DOF vs. 13 DOF | 4.0 | 2.3 | 7.0 | ≤0.001 | ||
| M. haemolytica | 2021/Tulathromycin | 13 DOF vs. 1 DOF | 0.59 | 0.39 | 0.89 | 0.012 |
| 36 DOF vs. 1 DOF | 1.6 | 0.99 | 2.6 | 0.055 | ||
| 36 DOF vs. 13 DOF | 2.7 | 1.7 | 4.5 | ≤0.001 | ||
| P. multocida | 2021/Tulathromycin | 13 DOF vs. 1 DOF | 0.15 | 0.08 | 0.28 | ≤0.001 |
| 36 DOF vs. 1 DOF | 0.53 | 0.30 | 0.92 | 0.025 | ||
| 36 DOF vs. 13 DOF | 3.5 | 1.8 | 6.6 | ≤0.001 | ||
| H. somni | 2021/Tulathromycin | 13 DOF vs. 1 DOF | 0.75 | 0.41 | 1.4 | 0.36 |
| 36 DOF vs. 1 DOF | 8.4 | 4.8 | 15 | ≤0.001 | ||
| 36 DOF vs. 13 DOF | 11 | 6.2 | 20 | ≤0.001 | ||
| M. haemolytica | 2021/Oxytetracycline | 13 DOF vs. 1 DOF | 1.5 | 1.002 | 2.2 | 0.049 |
| 36 DOF vs. 1 DOF | 1.04 | 0.6 | 1.7 | 0.88 | ||
| 36 DOF vs. 13 DOF | 0.7 | 0.4 | 1.1 | 0.14 | ||
| P. multocida | 2021/Oxytetracycline | 13 DOF vs. 1 DOF | 0.9 | 0.5 | 1.3 | 0.49 |
| 36 DOF vs. 1 DOF | 2.2 | 1.2 | 3.8 | 0.006 | ||
| 36 DOF vs. 13 DOF | 2.6 | 1.4 | 4.5 | 0.001 | ||
| H. somni | 2021/Oxytetracycline | 13 DOF vs. 1 DOF | 2.9 | 1.5 | 5.7 | 0.002 |
| 36 DOF vs. 1 DOF | 46 | 22 | 94 | ≤0.001 | ||
| 36 DOF vs. 13 DOF | 16 | 8.8 | 28 | ≤0.001 |
| ARG | Year/Metaphylaxis | DOF Comparison | Odds Ratio | 95% CI Lower | 95% CI Upper | p-Value |
|---|---|---|---|---|---|---|
| mphE-msrE | 2020/ Tulathromycin | 13 DOF vs. 1 DOF | 20 | 11 | 36 | ≤0.001 |
| 36 DOF vs. 1 DOF | 42 | 20 | 88 | ≤0.001 | ||
| 36 DOF vs. 13 DOF | 2.1 | 1.3 | 3.6 | 0.004 | ||
| EstT | 2020/ Tulathromycin | 13 DOF vs. 1 DOF | 5.7 | 2.5 | 13 | ≤0.001 |
| 36 DOF vs. 1 DOF | 11 | 3.7 | 33 | ≤0.001 | ||
| 36 DOF vs. 13 DOF | 1.9 | 0.85 | 4.4 | 0.12 | ||
| tet(H) | 2020/ Tulathromycin | 13 DOF vs. 1 DOF | 1.1 | 0.71 | 1.6 | 0.78 |
| 36 DOF vs. 1 DOF | 2.5 | 1.4 | 4.6 | 0.002 | ||
| 36 DOF vs. 13 DOF | 2.4 | 1.3 | 4.3 | 0.004 | ||
| mphE-msrE | 2021/ Tulathromycin 1 | 13 DOF vs. 1 DOF | 0.995 | 0.03 | ∞ | 0.99 |
| 36 DOF vs. 1 DOF | 6.5 | 0.69 | ∞ | 0.11 | ||
| 36 DOF vs. 13 DOF | 5.1 | 0.40 | 269 | 0.30 | ||
| EstT | 2021/ Tulathromycin 1 | 13 DOF vs. 1 DOF | 50 | 8.7 | ∞ | ≤0.001 |
| 36 DOF vs. 1 DOF | 93 | 16 | ∞ | ≤0.001 | ||
| 36 DOF vs. 13 DOF | 1.9 | 1.03 | 3.5 | 0.04 | ||
| tet(H) | 2021/ Tulathromycin | 13 DOF vs. 1 DOF | 5.0 | 2.3 | 11 | ≤0.001 |
| 36 DOF vs. 1 DOF | 11 | 4.7 | 25 | ≤0.001 | ||
| 36 DOF vs. 13 DOF | 2.2 | 1.2 | 3.9 | 0.008 | ||
| mphE-msrE | 2021/ Oxytetracycline | 13 DOF vs. 1 DOF | n/a | |||
| 36 DOF vs. 1 DOF | n/a | |||||
| 36 DOF vs. 13 DOF | n/a | |||||
| EstT | 2021/ Oxytetracycline | 13 DOF vs. 1 DOF | 5.5 | 0.94 | 32 | 0.058 |
| 36 DOF vs. 1 DOF | 2.4 | 0.27 | 21 | 0.43 | ||
| 36 DOF vs. 13 DOF | 0.44 | 0.07 | 2.6 | 0.37 | ||
| tet(H) | 2021/ Oxytetracycline | 13 DOF vs. 1 DOF | 20 | 6.8 | 57 | ≤0.001 |
| 36 DOF vs. 1 DOF | 32 | 9.9 | 100 | ≤0.001 | ||
| 36 DOF vs. 13 DOF | 1.6 | 0.88 | 2.9 | 0.12 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abi Younes, J.N.; McLeod, L.; Otto, S.J.G.; Chai, Z.; Lacoste, S.; McCarthy, E.L.; Links, M.G.; Herman, E.K.; Stothard, P.; Gow, S.P.; et al. Evaluating the Diagnostic Performance of Long-Read Metagenomic Sequencing Compared to Culture and Antimicrobial Susceptibility Testing for Detection of Bovine Respiratory Bacteria and Indicators of Antimicrobial Resistance. Antibiotics 2025, 14, 1114. https://doi.org/10.3390/antibiotics14111114
Abi Younes JN, McLeod L, Otto SJG, Chai Z, Lacoste S, McCarthy EL, Links MG, Herman EK, Stothard P, Gow SP, et al. Evaluating the Diagnostic Performance of Long-Read Metagenomic Sequencing Compared to Culture and Antimicrobial Susceptibility Testing for Detection of Bovine Respiratory Bacteria and Indicators of Antimicrobial Resistance. Antibiotics. 2025; 14(11):1114. https://doi.org/10.3390/antibiotics14111114
Chicago/Turabian StyleAbi Younes, Jennifer N., Lianne McLeod, Simon J. G. Otto, Zhijian Chai, Stacey Lacoste, E. Luke McCarthy, Matthew G. Links, Emily K. Herman, Paul Stothard, Sheryl P. Gow, and et al. 2025. "Evaluating the Diagnostic Performance of Long-Read Metagenomic Sequencing Compared to Culture and Antimicrobial Susceptibility Testing for Detection of Bovine Respiratory Bacteria and Indicators of Antimicrobial Resistance" Antibiotics 14, no. 11: 1114. https://doi.org/10.3390/antibiotics14111114
APA StyleAbi Younes, J. N., McLeod, L., Otto, S. J. G., Chai, Z., Lacoste, S., McCarthy, E. L., Links, M. G., Herman, E. K., Stothard, P., Gow, S. P., Campbell, J. R., & Waldner, C. L. (2025). Evaluating the Diagnostic Performance of Long-Read Metagenomic Sequencing Compared to Culture and Antimicrobial Susceptibility Testing for Detection of Bovine Respiratory Bacteria and Indicators of Antimicrobial Resistance. Antibiotics, 14(11), 1114. https://doi.org/10.3390/antibiotics14111114








