Characterization of Staphylococcus aureus CC1 and CC1660 of Human and Equine Origin
Abstract
1. Introduction
2. Results and Discussion
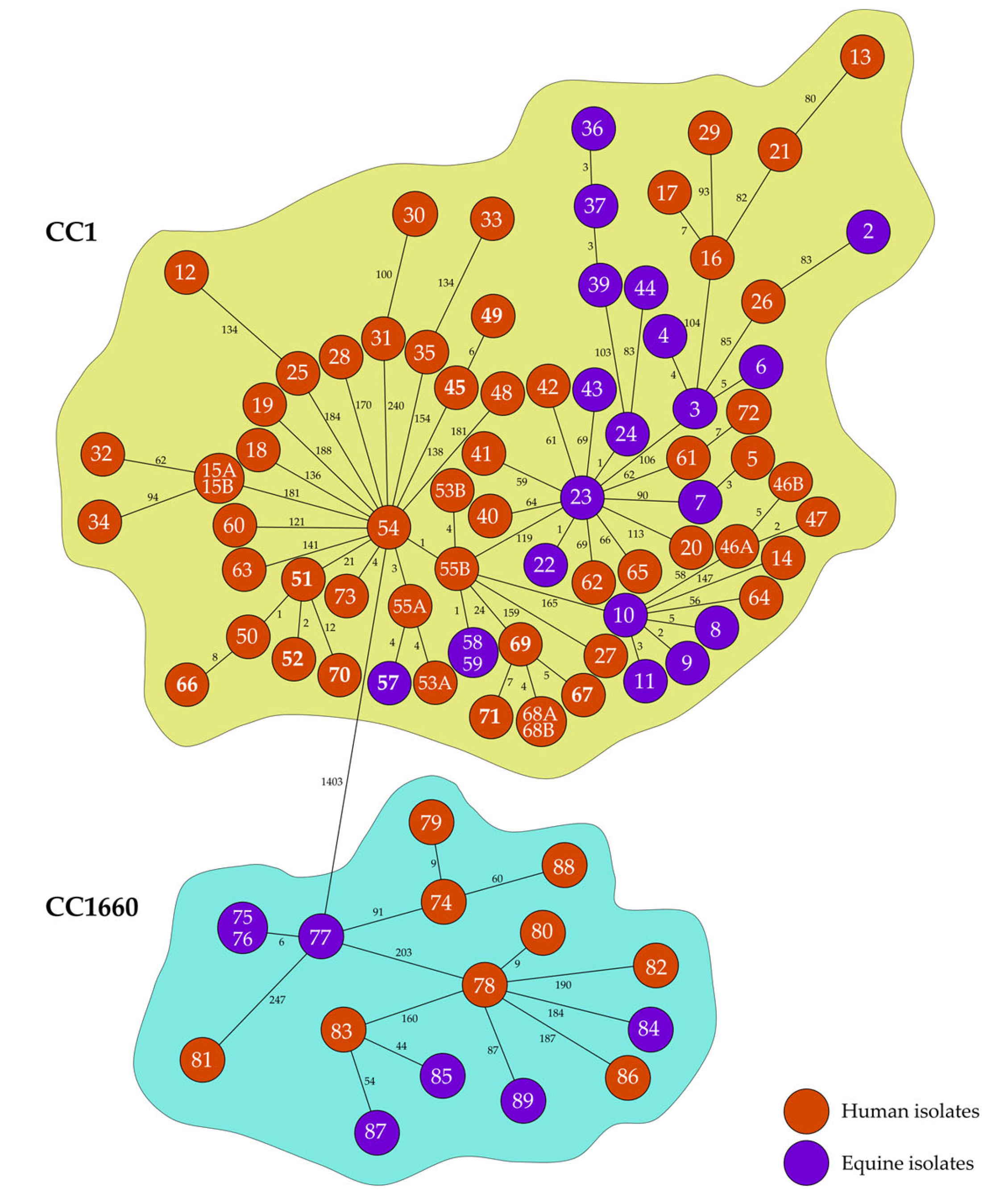
2.1. Genetic Relationships
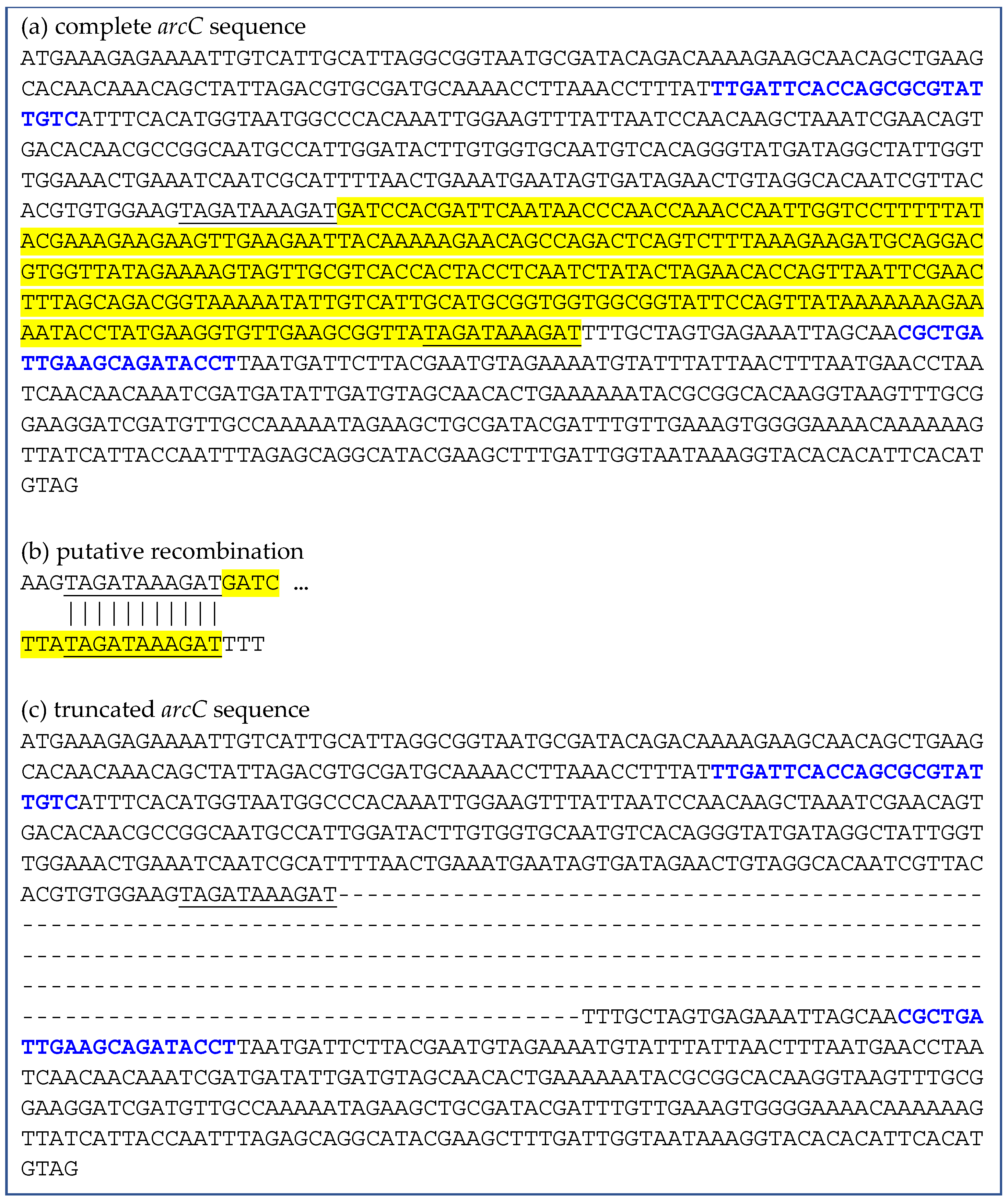
2.2. Deletions Within the arcC Gene
2.3. Antimicrobial Resistance
2.3.1. Antimicrobial Susceptibility Testing and Interpretation of the Results
2.3.2. Correlation of Phenotypic and Genotypic Resistance Properties
2.3.3. SCC Elements
2.4. Clonal Complex-Specific Virulence Factors
2.5. Virulence Factors on Mobile Genetic Elements
3. Materials and Methods
3.1. Isolate Collection
3.2. Whole-Genome Sequencing (WGS) and Sequence Analysis
3.2.1. Whole-Genome Sequencing
3.2.2. Molecular Typing and Phylogenetic Analysis
3.3. Anaerobic Arginine Dehydrogenase Testing
3.4. Antimicrobial Susceptibility Testing
3.5. Detection of Resistance Genes and Resistance-Mediating Mutations
3.6. Determination of Virulence Properties
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BORSA | borderline oxacillin-resistant Staphylococcus aureus |
| bp | base pair |
| CC | clonal complex |
| EC | equine clinic |
| dru | direct repeat unit |
| MIC | minimal inhibitory concentration |
| MLST | multilocus sequence typing |
| MRSA | methicillin-resistant Staphylococcus aureus |
| MSSA | methicillin-susceptible Staphylococcus aureus |
| QRDR | quinolone resistance-determining region |
| S. | Staphylococcus |
| SaPI | staphylococcal pathogenicity island |
| SCC | Staphylococcal Cassette Chromosome |
| SNP | single nucleotide polymorphism |
| spa | staphylococcal protein A |
| ST | sequence type |
| WGS | whole-genome sequencing |
References
- Rasquel-Oliveira, F.S.; Ribeiro, J.M.; Martelossi-Cebinelli, G.; Costa, F.B.; Nakazato, G.; Casagrande, R.; Verri, W.A. Staphylococcus aureus in Inflammation and Pain: Update on Pathologic Mechanisms. Pathogens 2025, 14, 185. [Google Scholar] [CrossRef]
- Holmes, M.A.; Zadoks, R.N. Methicillin resistant S. aureus in human and bovine mastitis. J. Mammary Gland Biol. Neoplasia 2011, 16, 373–382. [Google Scholar] [CrossRef]
- Cuny, C.; Friedrich, A.; Kozytska, S.; Layer, F.; Nübel, U.; Ohlsen, K.; Strommenger, B.; Walther, B.; Wieler, L.; Witte, W. Emergence of methicillin-resistant Staphylococcus aureus (MRSA) in different animal species. Int. J. Med. Microbiol. 2010, 300, 109–117. [Google Scholar] [CrossRef]
- Marshall, K.; Marsella, R. Evolution of the Prevalence of Antibiotic Resistance to Staphylococcus spp. Isolated from Horses in Florida over a 10-Year Period. Vet. Sci. 2023, 10, 71. [Google Scholar] [CrossRef]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef]
- Feil, E.J.; Cooper, J.E.; Grundmann, H.; Robinson, D.A.; Enright, M.C.; Berendt, T.; Peacock, S.J.; Smith, J.M.; Murphy, M.; Spratt, B.G.; et al. How clonal is Staphylococcus aureus? J. Bacteriol. 2003, 185, 3307–3316. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, J.A.; Moore, C.E.; Day, N.P.; Peacock, S.J.; Witney, A.A.; Stabler, R.A.; Husain, S.E.; Butcher, P.D.; Hinds, J. Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J. Bacteriol. 2006, 188, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.; Bray, J.; Maiden, M. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Luedicke, C.; Slickers, P.; Ehricht, R. Molecular epidemiology of Staphylococcus aureus in asymptomatic carriers. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 1159–1165. [Google Scholar] [CrossRef] [PubMed]
- Holtfreter, S.; Grumann, D.; Balau, V.; Barwich, A.; Kolata, J.; Goehler, A.; Weiss, S.; Holtfreter, B.; Bauerfeind, S.S.; Döring, P.; et al. Molecular Epidemiology of Staphylococcus aureus in the General Population in Northeast Germany: Results of the Study of Health in Pomerania (SHIP-TREND-0). J. Clin. Microbiol. 2016, 54, 2774–2785. [Google Scholar] [CrossRef]
- Rao, Q.; Shang, W.; Hu, X.; Rao, X. Staphylococcus aureus ST121: A globally disseminated hypervirulent clone. J. Med. Microbiol. 2015, 64, 1462–1473. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Feßler, A.T.; Burgold-Voigt, S.; Krüger, H.; Mühldorfer, K.; Wibbelt, G.; Liebler-Tenorio, E.M.; Reinicke, M.; Braun, S.D.; Hanke, D.; et al. Staphylococcus aureus isolates from Eurasian Beavers (Castor fiber) carry a novel phage-borne bicomponent leukocidin related to the Panton-Valentine leukocidin. Sci. Rep. 2021, 11, 24394. [Google Scholar] [CrossRef] [PubMed]
- Moon, D.C.; Kim, B.Y.; Tamang, M.D.; Nam, H.M.; Jang, G.C.; Jung, S.C.; Lee, H.S.; Park, Y.H.; Lim, S.K. Genome Sequence of a Unique t2247-ST692-III Livestock-Associated Methicillin-Resistant Staphylococcus aureus Strain from Chicken Carcass. Genome Announc. 2016, 4, e00026-16. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Gavier-Widén, D.; Hotzel, H.; Peters, M.; Guenther, S.; Lazaris, A.; Loncaric, I.; Müller, E.; Reissig, A.; Ruppelt-Lorz, A.; et al. Diversity of Staphylococcus aureus Isolates in European Wildlife. PLoS ONE 2016, 11, e0168433. [Google Scholar] [CrossRef]
- García-Álvarez, L.; Holden, M.T.; Lindsay, H.; Webb, C.R.; Brown, D.F.; Curran, M.D.; Walpole, E.; Brooks, K.; Pickard, D.J.; Teale, C.; et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: A descriptive study. Lancet Infect. Dis. 2011, 11, 595–603. [Google Scholar] [CrossRef]
- Vancraeynest, D.; Haesebrouck, F.; Deplano, A.; Denis, O.; Godard, C.; Wildemauwe, C.; Hermans, K. International dissemination of a high virulence rabbit Staphylococcus aureus clone. J. Vet. Med. B Infect. Dis. Vet. Public Health 2006, 53, 418–422. [Google Scholar] [CrossRef]
- Burgold-Voigt, S.; Monecke, S.; Busch, A.; Bocklisch, H.; Braun, S.D.; Diezel, C.; Hotzel, H.; Liebler-Tenorio, E.M.; Müller, E.; Reinicke, M.; et al. Characterisation of a Staphylococcus aureus Isolate Carrying Phage-Borne Enterotoxin E from a European Badger (Meles meles). Pathogens 2023, 12, 704. [Google Scholar] [CrossRef]
- Bar-Gal, G.K.; Blum, S.E.; Hadas, L.; Ehricht, R.; Monecke, S.; Leitner, G. Host-specificity of Staphylococcus aureus causing intramammary infections in dairy animals assessed by genotyping and virulence genes. Vet. Microbiol. 2015, 176, 143–154. [Google Scholar] [CrossRef]
- Smyth, D.S.; Feil, E.J.; Meaney, W.J.; Hartigan, P.J.; Tollersrud, T.; Fitzgerald, J.R.; Enright, M.C.; Smyth, C.J. Molecular genetic typing reveals further insights into the diversity of animal-associated Staphylococcus aureus. J. Med. Microbiol. 2009, 58, 1343–1353. [Google Scholar] [CrossRef]
- Schlotter, K.; Ehricht, R.; Hotzel, H.; Monecke, S.; Pfeffer, M.; Donat, K. Leukocidin genes lukF-P83 and lukM are associated with Staphylococcus aureus clonal complexes 151, 479 and 133 isolated from bovine udder infections in Thuringia, Germany. Vet. Res. 2012, 43, 42. [Google Scholar] [CrossRef]
- Richardson, E.J.; Bacigalupe, R.; Harrison, E.M.; Weinert, L.A.; Lycett, S.; Vrieling, M.; Robb, K.; Hoskisson, P.A.; Holden, M.T.G.; Feil, E.J.; et al. Gene exchange drives the ecological success of a multi-host bacterial pathogen. Nat. Ecol. Evol. 2018, 2, 1468–1478. [Google Scholar] [CrossRef]
- Voss, A.; Loeffen, F.; Bakker, J.; Klaassen, C.; Wulf, M. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 2005, 11, 1965–1966. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Kuhnert, P.; Hotzel, H.; Slickers, P.; Ehricht, R. Microarray based study on virulence-associated genes and resistance determinants of Staphylococcus aureus isolates from cattle. Vet. Microbiol. 2007, 125, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Witte, W.; Strommenger, B.; Stanek, C.; Cuny, C. Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, Central Europe. Emerg. Infect. Dis. 2007, 13, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Nemati, M.; Hermans, K.; Lipinska, U.; Denis, O.; Deplano, A.; Struelens, M.; Devriese, L.A.; Pasmans, F.; Haesebrouck, F. Antimicrobial resistance of old and recent Staphylococcus aureus isolates from poultry: First detection of livestock-associated methicillin-resistant strain ST398. Antimicrob. Agents Chemother. 2008, 52, 3817–3819. [Google Scholar] [CrossRef]
- Cuny, C.; Abdelbary, M.M.H.; Köck, R.; Layer, F.; Scheidemann, W.; Werner, G.; Witte, W. Methicillin-resistant Staphylococcus aureus from infections in horses in Germany are frequent colonizers of veterinarians but rare among MRSA from infections in humans. One Health 2016, 2, 11–17. [Google Scholar] [CrossRef]
- Murphy, R.J.T.; Ramsay, J.P.; Lee, Y.T.; Pang, S.; O’Dea, M.A.; Pearson, J.C.; Axon, J.E.; Raby, E.; Abdulgader, S.M.; Whitelaw, A.; et al. Multiple introductions of methicillin-resistant Staphylococcus aureus ST612 into Western Australia associated both with human and equine reservoirs. Int. J. Antimicrob. Agents 2019, 54, 681–685. [Google Scholar] [CrossRef]
- Cuny, C.; Witte, W. MRSA in equine hospitals and its significance for infections in humans. Vet. Microbiol. 2017, 200, 59–64. [Google Scholar] [CrossRef]
- Albrecht, N.; Jatzwauk, L.; Slickers, P.; Ehricht, R.; Monecke, S. Clonal replacement of epidemic methicillin-resistant Staphylococcus aureus strains in a German university hospital over a period of eleven years. PLoS ONE 2011, 6, e28189. [Google Scholar] [CrossRef]
- Merz, A.; Stephan, R.; Johler, S. Staphylococcus aureus Isolates from Goat and Sheep Milk Seem to Be Closely Related and Differ from Isolates Detected from Bovine Milk. Front. Microbiol. 2016, 7, 319. [Google Scholar] [CrossRef]
- Azara, E.; Piras, M.G.; Parisi, A.; Tola, S. Antimicrobial susceptibility and genotyping of Staphylococcus aureus isolates collected between 1986 and 2015 from ovine mastitis. Vet. Microbiol. 2017, 205, 53–56. [Google Scholar] [CrossRef]
- Carfora, V.; Giacinti, G.; Sagrafoli, D.; Marri, N.; Giangolini, G.; Alba, P.; Feltrin, F.; Sorbara, L.; Amoruso, R.; Caprioli, A.; et al. Methicillin-resistant and methicillin-susceptible Staphylococcus aureus in dairy sheep and in-contact humans: An intra-farm study. J. Dairy Sci. 2016, 99, 4251–4258. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Burgold-Voigt, S.; Feßler, A.T.; Krapf, M.; Loncaric, I.; Liebler-Tenorio, E.M.; Braun, S.D.; Diezel, C.; Müller, E.; Reinicke, M.; et al. Characterisation of Staphylococcus aureus Strains and Their Prophages That Carry Horse-Specific Leukocidin Genes lukP/Q. Toxins 2025, 17, 20. [Google Scholar] [CrossRef] [PubMed]
- Sieber, S.; Gerber, V.; Jandova, V.; Rossano, A.; Evison, J.M.; Perreten, V. Evolution of multidrug-resistant Staphylococcus aureus infections in horses and colonized personnel in an equine clinic between 2005 and 2010. Microb. Drug Resist. 2011, 17, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.Z.; Espinosa-Gongora, C.; Damborg, P.; Sieber, R.N.; Munk, R.; Husted, L.; Moodley, A.; Skov, R.; Larsen, J.; Guardabassi, L. Horses in Denmark Are a Reservoir of Diverse Clones of Methicillin-Resistant and -Susceptible Staphylococcus aureus. Front. Microbiol. 2017, 8, 543. [Google Scholar] [CrossRef]
- Sung, J.M.; Lloyd, D.H.; Lindsay, J.A. Staphylococcus aureus host specificity: Comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology 2008, 154, 1949–1959. [Google Scholar] [CrossRef]
- Monecke, S.; Coombs, G.; Shore, A.C.; Coleman, D.C.; Akpaka, P.; Borg, M.; Chow, H.; Ip, M.; Jatzwauk, L.; Jonas, D.; et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE 2011, 6, e17936. [Google Scholar] [CrossRef]
- Lawal, O.U.; Ayobami, O.; Abouelfetouh, A.; Mourabit, N.; Kaba, M.; Egyir, B.; Abdulgader, S.M.; Shittu, A.O. A 6-Year Update on the Diversity of Methicillin-Resistant Staphylococcus aureus Clones in Africa: A Systematic Review. Front. Microbiol. 2022, 13, 860436. [Google Scholar] [CrossRef]
- Williamson, D.A.; Monecke, S.; Heffernan, H.; Ritchie, S.R.; Roberts, S.A.; Upton, A.; Thomas, M.G.; Fraser, J.D. High usage of topical fusidic acid and rapid clonal expansion of fusidic acid-resistant Staphylococcus aureus: A cautionary tale. Clin. Infect. Dis. 2014, 59, 1451–1454. [Google Scholar] [CrossRef]
- Alba, P.; Feltrin, F.; Cordaro, G.; Porrero, M.C.; Kraushaar, B.; Argudín, M.A.; Nykäsenoja, S.; Monaco, M.; Stegger, M.; Aarestrup, F.M.; et al. Livestock-Associated Methicillin Resistant and Methicillin Susceptible Staphylococcus aureus Sequence Type (CC)1 in European Farmed Animals: High Genetic Relatedness of Isolates from Italian Cattle Herds and Humans. PLoS ONE 2015, 10, e0137143. [Google Scholar] [CrossRef]
- Leopold, S.R.; Goering, R.V.; Witten, A.; Harmsen, D.; Mellmann, A. Bacterial whole-genome sequencing revisited: Portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J. Clin. Microbiol. 2014, 52, 2365–2370. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, S.A.; Chia, N.; Jeraldo, P.R.; Quest, D.J.; Johnson, J.A.; Boxrud, D.J.; Taylor, A.J.; Chen, J.; Jenkins, G.D.; Drucker, T.M.; et al. Comparison of Whole-Genome Sequencing Methods for Analysis of Three Methicillin-Resistant Staphylococcus aureus Outbreaks. J. Clin. Microbiol. 2017, 55, 1946–1953. [Google Scholar] [CrossRef] [PubMed]
- Graveland, H.; Wagenaar, J.A.; Bergs, K.; Heesterbeek, H.; Heederik, D. Persistence of livestock associated MRSA CC398 in humans is dependent on intensity of animal contact. PLoS ONE 2011, 6, e16830. [Google Scholar] [CrossRef] [PubMed]
- Ramón-Maiques, S.; Marina, A.; Guinot, A.; Gil-Ortiz, F.; Uriarte, M.; Fita, I.; Rubio, V. Substrate binding and catalysis in carbamate kinase ascertained by crystallographic and site-directed mutagenesis studies: Movements and significance of a unique globular subdomain of this key enzyme for fermentative ATP production in bacteria. J. Mol. Biol. 2010, 397, 1261–1275. [Google Scholar] [CrossRef]
- Zhang, L.; Thomas, J.C.; Didelot, X.; Robinson, D.A. Molecular signatures identify a candidate target of balancing selection in an arcD-like gene of Staphylococcus epidermidis. J. Mol. Evol. 2012, 75, 43–54. [Google Scholar] [CrossRef]
- Lodder, G.; Werckenthin, C.; Schwarz, S.; Dyke, K. Molecular analysis of naturally occuring ermC-encoding plasmids in staphylococci isolated from animals with and without previous contact with macrolide/lincosamide antibiotics. FEMS Immunol. Med. Microbiol. 1997, 18, 7–15. [Google Scholar] [CrossRef]
- EMA (European Medicines Agency). Categorisation of Antibiotics in the European Union; EMA: Amsterdam, The Netherlands, 2019. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 7th ed.; CLSI Supplement VET01S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2024. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 35th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2025. [Google Scholar]
- Guignard, B.; Entenza, J.M.; Moreillon, P. Beta-lactams against methicillin-resistant Staphylococcus aureus. Curr. Opin. Pharmacol. 2005, 5, 479–489. [Google Scholar] [CrossRef]
- Scholtzek, A.D.; Hanke, D.; Walther, B.; Eichhorn, I.; Stöckle, S.D.; Klein, K.S.; Gehlen, H.; Lübke-Becker, A.; Schwarz, S.; Feßler, A.T. Molecular Characterization of Equine Staphylococcus aureus Isolates Exhibiting Reduced Oxacillin Susceptibility. Toxins 2019, 11, 535. [Google Scholar] [CrossRef]
- Feßler, A.; Scott, C.; Kadlec, K.; Ehricht, R.; Monecke, S.; Schwarz, S. Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 2010, 65, 619–625. [Google Scholar] [CrossRef]
- Brennan, G.I.; Abbott, Y.; Burns, A.; Leonard, F.; McManus, B.A.; O’Connell, B.; Coleman, D.C.; Shore, A.C. The Emergence and Spread of Multiple Livestock-Associated Clonal Complex 398 Methicillin-Resistant and Methicillin-Susceptible Staphylococcus aureus Strains among Animals and Humans in the Republic of Ireland, 2010–2014. PLoS ONE 2016, 11, e0149396. [Google Scholar] [CrossRef]
- Scholtzek, A.D.; Hanke, D.; Eichhorn, I.; Walther, B.; Lübke-Becker, A.; van Duijkeren, E.; Köck, R.; Schwarz, S.; Feßler, A.T. Heterogeneity of antimicrobial susceptibility testing results for sulfamethoxazole/trimethoprim obtained from clinical equine Staphylococcus aureus isolates using different methods. Vet. Microbiol. 2020, 242, 108600. [Google Scholar] [CrossRef] [PubMed]
- Kehrenberg, C.; Schwarz, S. fexA, a novel Staphylococcus lentus gene encoding resistance to florfenicol and chloramphenicol. Antimicrob. Agents Chemother. 2004, 48, 615–618. [Google Scholar] [CrossRef] [PubMed]
- Goering, R.V.; Morrison, D.; Al-Doori, Z.; Edwards, G.F.; Gemmell, C.G. Usefulness of mec-associated direct repeat unit (dru) typing in the epidemiological analysis of highly clonal methicillin-resistant Staphylococcus aureus in Scotland. Clin. Microbiol. Infect. 2008, 14, 964–969. [Google Scholar] [CrossRef]
- Monecke, S.; König, E.; Earls, M.R.; Leitner, E.; Müller, E.; Wagner, G.E.; Poitz, D.M.; Jatzwauk, L.; Vremerǎ, T.; Dorneanu, O.S.; et al. An epidemic CC1-MRSA-IV clone yields false-negative test results in molecular MRSA identification assays: A note of caution, Austria, Germany, Ireland, 2020. Euro Surveill. 2020, 25, 2000929. [Google Scholar] [CrossRef] [PubMed]
- Cuny, C.; Strommenger, B.; Witte, W.; Stanek, C. Clusters of infections in horses with MRSA ST1, ST254, and ST398 in a veterinary hospital. Microb. Drug Resist. 2008, 14, 307–310. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Seemann, T. mlst. Github. Available online: https://github.com/tseemann/mlst (accessed on 1 September 2025).
- Bartels, M.D.; Petersen, A.; Worning, P.; Nielsen, J.B.; Larner-Svensson, H.; Johansen, H.K.; Andersen, L.P.; Jarløv, J.O.; Boye, K.; Larsen, A.R.; et al. Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2014, 52, 4305–4308. [Google Scholar] [CrossRef]
- Silva, M.; Machado, M.P.; Silva, D.N.; Rossi, M.; Moran-Gilad, J.; Santos, S.; Ramirez, M.; Carriço, J.A. chewBBACA: A complete suite for gene-by-gene schema creation and strain identification. Microb. Genom. 2018, 4, e000166. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 6th ed.; CLSI Standard VET01; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2024. [Google Scholar]
- BVL. BVL-Report 14.6 Bericht zur Resistenzmonitoringstudie 2018 [BVL-Report 14.6 Report on the Resistance Monitoring Study 2018]; BVL: Berlin, Germany, 2020. [Google Scholar]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.U. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2024, 53, D609–D617. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef]
- Monecke, S.; Gavier-Widen, D.; Mattsson, R.; Rangstrup-Christensen, L.; Lazaris, A.; Coleman, D.C.; Shore, A.C.; Ehricht, R. Detection of mecC-positive Staphylococcus aureus (CC130-MRSA-XI) in diseased European hedgehogs (Erinaceus europaeus) in Sweden. PLoS ONE 2013, 8, e66166. [Google Scholar] [CrossRef]
- Monecke, S.; Jatzwauk, L.; Müller, E.; Nitschke, H.; Pfohl, K.; Slickers, P.; Reissig, A.; Ruppelt-Lorz, A.; Ehricht, R. Diversity of SCCmec Elements in Staphylococcus aureus as Observed in South-Eastern Germany. PLoS ONE 2016, 11, e0162654. [Google Scholar] [CrossRef]
- Coleman, D.; Knights, J.; Russell, R.; Shanley, D.; Birkbeck, T.H.; Dougan, G.; Charles, I. Insertional inactivation of the Staphylococcus aureus beta-toxin by bacteriophage phi 13 occurs by site- and orientation-specific integration of the phi 13 genome. Mol. Microbiol. 1991, 5, 933–939. [Google Scholar] [CrossRef]
- Coleman, D.C.; Sullivan, D.J.; Russell, R.J.; Arbuthnott, J.P.; Carey, B.F.; Pomeroy, H.M. Staphylococcus aureus Bacteriophages Mediating the Simultaneous Lysogenic Conversion of β-Lysin, Staphylokinase and Enterotoxin A: Molecular Mechanism of Triple Conversion. J. Gen. Microbiol. 1989, 135, 1679–1697. [Google Scholar] [CrossRef]
| Antimicrobial Agents | MIC in (µg/mL) | S | I | R | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.008 | 0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | 1024 | (µg/mL) | |||
| Oxacillin (total) | - | - | - | 2 | 14 | 46 | 17 | 2 | 3 | - | 7 | ||||||||||
| Equine | - | - | - | - | 1 | 10 + 3 | 4 + 1 | 1 | 1 + 2 | - | 4 | ≤2 | - | ≥4 | |||||||
| Human | - | - | - | 1 + 1 | 12 + 1 | 29 + 4 | 9 + 3 | 1 | - | - | 3 | ≤2 | - | ≥4 | |||||||
| Penicillin (total) | 3 | 14 | 2 | - | - | - | 1 | 10 | 17 | 8 | 24 | 8 | 4 | ||||||||
| Equine | 1 | 2 | - | - | - | - | - | 2 | 3 + 1 | 3 + 1 | 4 + 1 | 5 | 3 + 1 | ≤0.5 | 1 | ≥2 | |||||
| Human | 1 + 1 | 7 + 5 | 2 | - | - | - | 1 | 8 | 10 + 3 | 4 | 19 | 3 | ≤0.12 | - | ≥0.25 | ||||||
| Ampicillin (total) | - | 2 | 16 | 1 | 1 | - | 16 | 12 | 22 | 8 | 11 | 2 | - | ||||||||
| Equine | - | 2 | 1 | - | - | - | 3 | 2 | 5 + 2 | 2 + 1 | 7 | 1 + 1 | - | ≤0.25 | 0.5 | ≥1 | |||||
| Human | - | - | 9 + 6 | 1 | 1 | - | 13 | 7 + 3 | 15 | 5 | 4 | - | - | ||||||||
| Amoxicillin/Clavulanic Acid (total) | - | - | 2 | 17 | 19 | 35 | 11 | - | - | 6 | 1 | - | - | ||||||||
| Equine | - | - | 1 | 2 | 3 | 10 + 2 | 3 + 2 | - | - | 3 | 1 | ||||||||||
| Human | - | - | 1 | 10 + 5 | 16 | 22 + 1 | 4 + 2 | - | - | 3 | - | - | - | ||||||||
| Imipenem (total) | 31 | 53 | - | - | - | - | 3 | - | 1 | 3 | - | - | - | ||||||||
| Equine | 7 + 2 | 9 + 5 | - | - | - | - | - | - | 1 | 3 | - | - | - | ||||||||
| Human | 17 + 5 | 35 + 4 | - | - | - | - | 3 | - | - | - | - | - | - | ||||||||
| Ceftiofur (total) | - | - | - | - | - | 58 | 25 | 1 | - | - | - | 3 | 4 | ||||||||
| Equine | - | - | - | - | - | 14 + 4 | 2 + 3 | - | - | - | - | - | 4 | ||||||||
| Human | - | - | - | - | - | 33 + 7 | 18 + 2 | 1 | - | - | - | 3 | - | ||||||||
| Cefquinome (total) | - | - | - | 1 | - | 34 | 47 | 2 | - | 1 | 5 | 1 | - | ||||||||
| Equine | - | - | - | 1 | - | 8 + 3 | 7 + 3 | 1 | - | - | 3 | 1 | - | ||||||||
| Human | - | - | - | - | - | 19 + 4 | 32 + 5 | 1 | - | 1 | 2 | - | - | ||||||||
| Cefalothin (total) | 1 | 2 | 18 | 56 | 7 | - | - | - | 3 | 3 | 1 | - | - | ||||||||
| Equine | 1 | 1 | 1 | 13 + 3 | 2 + 2 | - | - | - | - | 3 | 1 | - | - | ||||||||
| Human | - | 1 | 12 + 5 | 36 + 4 | 3 | - | - | - | 3 | - | - | - | - | ||||||||
| Cefotaxime (total) | - | - | - | - | - | - | - | 33 | 50 | 1 | - | - | 7 | ||||||||
| Equine | - | - | - | - | - | - | - | 4 + 2 | 11 + 5 | 1 | 4 | ||||||||||
| Human | - | - | - | - | - | - | - | 23 + 4 | 29 + 5 | - | - | 3 | |||||||||
| Cefoperazone (total) | - | - | - | - | - | 21 | 52 | 11 | - | - | 7 | ||||||||||
| Equine | - | - | - | - | - | 1 + 3 | 12 + 1 | 3 + 3 | - | - | 4 | ||||||||||
| Human | - | - | - | - | - | 12 + 5 | 35 + 4 | 5 | - | - | 3 | ||||||||||
| Erythromycin (total) | - | - | - | - | 9 | 70 | 3 | - | - | - | - | - | 9 | ||||||||
| Equine | - | - | - | - | 5 + 3 | 11 + 4 | - | - | - | - | - | - | 4 | ≤0.5 | 1–4 | ≥8 | |||||
| Human | - | - | - | - | 1 | 47 + 8 | 3 | - | - | - | - | - | 5 | ≤0.5 | 1–4 | ≥8 | |||||
| Tylosin tartrate (total) | - | - | - | - | 5 | 67 | 19 | - | - | - | - | - | - | ||||||||
| Equine | - | - | - | - | 3 + 2 | 15 + 5 | 2 | - | - | - | - | - | - | ||||||||
| Human | - | - | - | - | - | 38 + 9 | 17 | - | - | - | - | - | - | ||||||||
| Tulathromycin (total) | - | - | - | - | - | 1 | 11 | 56 | 17 | 4 | 2 | ||||||||||
| Equine | - | - | - | - | - | 1 | 2 + 4 | 12 + 3 | 1 | 4 | - | ||||||||||
| Human | - | - | - | - | - | - | 1 + 4 | 36 + 5 | 16 | - | 2 | ||||||||||
| Tilmicosin (total) | - | - | 1 | 5 | 48 | 37 | - | - | - | - | - | - | - | ||||||||
| Equine | - | - | 1 | 3 + 2 | 7 + 5 | 9 | - | - | - | - | - | - | |||||||||
| Human | - | - | - | - | 28 + 8 | 27 + 1 | - | - | - | - | - | - | |||||||||
| Clindamycin (total) | - | 2 | 22 | 66 | 1 | - | - | - | - | - | - | - | - | ||||||||
| Equine | - | 1 + 1 | 6 + 2 | 12 + 4 | 1 | - | - | - | - | - | - | - | - | ||||||||
| Human | - | - | 14 | 41 + 9 | - | - | - | - | - | - | - | - | - | ≤0.5 | 1–2 | ≥4 | |||||
| Pirlimycin (total) | - | - | - | 2 | 22 | 66 | 1 | - | - | - | - | - | - | ||||||||
| Equine | - | - | - | 2 | 9 + 3 | 9 + 4 | - | - | - | - | - | - | - | ||||||||
| Human | - | - | - | - | 10 | 45 + 8 | 1 | - | - | - | - | - | - | ||||||||
| Tiamulin (total) | - | - | 1 | 1 | 13 | 74 | 2 | - | - | - | - | - | - | ||||||||
| Equine | - | - | 1 | 1 | 2 + 3 | 17 + 3 | - | - | - | - | - | - | - | ||||||||
| Human | - | - | - | - | 8 | 45 + 9 | 2 | - | - | - | - | - | - | ||||||||
| Ciprofloxacin (total) | - | - | 1 | 3 | 37 | 24 | 8 | 18 | - | - | - | - | - | ||||||||
| Equine | - | - | 1 | 1 | 9 + 4 | 7 + 2 | - | 3 | - | - | - | - | - | ||||||||
| Human | - | - | - | 1 + 1 | 18 + 6 | 14 + 1 | 7 + 1 | 15 | - | - | - | - | - | ≤1 | 2 | ≥4 | |||||
| Enrofloxacin (total) | - | - | 3 | 40 | 24 | 7 | 17 | - | - | - | - | - | - | ||||||||
| Equine | - | - | 1 + 1 | 9 + 6 | 7 | - | 3 | - | - | - | - | - | - | ≤0.12 | 0.25 | ≥0.5 | |||||
| Human | - | - | 1 | 19 + 6 | 14 + 3 | 7 | 14 | - | - | - | - | - | - | ||||||||
| Marbofloxacin (total) | - | - | - | - | 15 | 48 | 18 | 10 | - | - | - | - | - | ||||||||
| Equine | - | - | - | - | 5 + 1 | 8 + 6 | 5 | 2 | - | - | - | - | - | ||||||||
| Human | - | - | - | - | 8 + 1 | 27 + 7 | 12 + 1 | 8 | - | - | - | - | - | ||||||||
| Gentamicin (total) | 1 | 27 | 25 | 5 | 1 | - | 1 | 2 | 13 | 14 | 2 | - | - | ||||||||
| Equine | - | 4 + 2 | 4 + 1 | 1 | 1 | - | - | - | 2 + 3 | 6 | 2 | - | - | ≤4 | 8 | ≥16 | |||||
| Human | 1 | 18 + 3 | 16 + 4 | 4 | - | - | 1 | 2 | 8 | 7 + 1 | - | - | - | ≤4 | 8 | ≥16 | |||||
| Streptomycin (total) | - | - | - | - | 12 | 56 | 11 | 1 | - | 1 | 10 | - | - | ||||||||
| Equine | - | - | - | - | 2 + 3 | 10 + 3 | 4 | - | - | 1 | 4 | - | - | ||||||||
| Human | - | - | - | - | 6 + 1 | 36 + 7 | 7 | 1 | - | - | 6 | - | - | ||||||||
| Neomycin (total) | - | 4 | 40 | 12 | 5 | 7 | 8 | 6 | 7 | 2 | - | ||||||||||
| Equine | - | 1 | 8 + 5 | 1 | 1 | 2 | 1 + 1 | 2 | 4 | 1 | - | ||||||||||
| Human | - | 1 + 2 | 23 + 4 | 9 + 2 | 4 | 5 | 5 + 1 | 4 | 3 | 1 | - | ||||||||||
| Tetracycline (total) | 2 | 43 | 16 | - | - | - | - | 2 | 21 | 7 | - | - | - | ||||||||
| Equine | 1 | 6 + 5 | 3 + 1 | - | - | - | - | 2 | 8 + 1 | - | - | - | - | ≤4 | 8 | ≥16 | |||||
| Human | 1 | 26 + 6 | 10 + 2 | - | - | - | - | - | 11 + 1 | 7 | - | - | - | ≤4 | 8 | ≥16 | |||||
| Doxycycline (total) | 3 | 33 | 19 | 6 | - | 10 | 19 | 1 | - | - | - | - | - | ||||||||
| Equine | 1 + 1 | 7 + 3 | 2 + 2 | - | - | 6 + 1 | 4 | - | - | - | - | - | - | ≤0.12 | 0.25 | ≥0.5 | |||||
| Human | 1 | 19 + 4 | 13 + 2 | 5 + 1 | - | 3 | 14 + 1 | 1 | - | - | - | - | - | ≤4 | 8 | ≥16 | |||||
| Sulfamethoxazole/trimethoprim (total) | - | 3 | 48 | 9 | 7 | 1 | 3 | 1 | 6 | 12 | 1 | - | - | ||||||||
| Equine | - | 2 | 9 + 1 | 1 + 3 | 4 | - | 2 + 1 | 1 | - | 3 | - | - | - | ≤2/38 | - | ≥4/76 | |||||
| Human | - | 1 | 31 + 7 | 4 + 1 | 3 | 1 | - | - | 6 | 9 | 1 | - | - | ≤2/38 | - | ≥4/76 | |||||
| Florfenicol (total) | - | - | - | - | 2 | 69 | 19 | - | - | 1 | - | - | - | ||||||||
| Equine | - | - | - | - | 1 + 1 | 17 + 6 | 2 | - | - | - | - | - | - | ||||||||
| Human | - | - | - | - | - | 39 + 7 | 15 + 2 | - | - | 1 | - | - | - | ||||||||
| Linezolid (total) | - | - | - | - | - | 8 | 62 | 21 | - | - | - | - | - | ||||||||
| Equine | - | - | - | - | - | 2 + 3 | 17 + 4 | 1 | - | - | - | - | - | ≤4 | - | ≥8 | |||||
| Human | - | - | - | - | - | 3 | 36 + 5 | 16 + 4 | - | - | - | - | - | ≤4 | - | ≥8 | |||||
| Vancomycin (total) | - | - | - | - | - | 6 | 83 | 2 | - | - | - | - | - | ||||||||
| Equine | - | - | - | - | - | 3 | 20 + 4 | - | - | - | - | - | - | ||||||||
| Human | - | - | - | - | - | 1 + 2 | 52 + 7 | 2 | - | - | - | - | - | ≤2 | 4–8 | ≥16 | |||||
| Quinupristin/Dalfopristin (total) | - | - | - | - | 18 | 70 | 3 | - | - | - | - | - | - | ||||||||
| Equine | - | - | - | - | 4 + 4 | 16 + 3 | - | - | - | - | - | - | - | ||||||||
| Human | - | - | - | - | 6 + 4 | 46 + 5 | 3 | - | - | - | - | - | - | ≤1 | 2 | ≥4 | |||||
| Isolates Phenotype Total | Isolates Genotype Total | CC1 Horses | CC1 Humans | CC1660 Horses | CC1660 Humans | PEN | OXA | TET | GEN | ERY | SXT | FQ | NEO | STR | FFN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18 | 16 | - | 8 | 3 | 5 | ||||||||||
| 2 | 2 | - | - | folP (F17L) | |||||||||||
| 31 | 32 | 9 | 19 | - | 3 | blaZ | |||||||||
| 1 | 1 | - | - | - | blaZ | ||||||||||
| 1 | 1 | - | 1 | - | - | blaZ | fexA | ||||||||
| 2 | 2 | - | 2 | - | - | blaZ | dfrG + folP (F17L and A184V) | ||||||||
| 2 | 2 | - | 2 | - | - | blaZ | erm(C) | str | |||||||
| 3 | 1 | - | 1 | - | - | blaZ | aacA-aphD | dfrS1 | grlA (S80Y) | ||||||
| 4 | - | 1 | 1 | - | blaZ | aacA-aphD | dfrS1 | ||||||||
| 1 | - | - | 1 | - | blaZ | aacA-aphD | dfrS1 | ||||||||
| 1 | - | - | 1 | - | blaZ | aacA-aphD | dfrS1 | ||||||||
| 1 | 1 | - | 1 | - | - | blaZ | aacA-aphD | dfrS1/dfrG + folP (F17L and A184V) | |||||||
| 1 | 1 | - | 1 | - | - | blaZ | tet(K) | aphA3 | aad(E) | ||||||
| 1 | 1 | - | - | - | 1 | tet(L) | aacA-aphD | dfrS1 | aadD | ||||||
| 4 | 3 | 3 | - | - | - | blaZ | tet(L) | aacA-aphD | dfrK | aadD | |||||
| 1 | - | 1 | - | - | blaZ | tet(L) | aacA-aphD | dfrG | aadD | ||||||
| 1 | 1 | - | - | 1 | - | blaZ | tet(L) | aacA-aphD | dfrK | aadD | str | ||||
| 3 | 3 | - | 3 | - | - | blaZ | mecA | tet(K) | erm(C) | aphA3 | aad(E) | ||||
| 13 | 2 | - | 2 | - | - | blaZ | tet(L) | aacA-aphD | dfrG + folP (F17L and A184V) | aadD | |||||
| 1 | - | 1 | - | - | blaZ | tet(L) | aacA-aphD | dfrS1/dfrG + folP (F17L and A184V) | grlA (S80Y) | aadD | |||||
| 13 | - | 10 | - | - | blaZ | tet(L) | aacA-aphD | dfrG + folP (F17L and A184V) | grlA (S80Y) | aadD | |||||
| 3 | 3 | - | - | - | blaZ | tet(L) | aacA-aphD | dfrG + folP (F17L and A184V) | grlA (S80Y) | aadD | |||||
| 4 | 4 | 4 | - | - | - | blaZ | mecA | tet(K) | aacA-aphD | erm(C) | dfrS1 | aphA3 | aad(E) |
| Virulence Genes | Localization | In CC1 Isolates (n = 75) | In CC1660 Isolates (n = 16) | In Human Isolates (n = 67) | In Equine Isolates (n = 24) |
|---|---|---|---|---|---|
| tst-1 + sec + sel | SaPI | 0 | 3 (18.75%) | 3 (4.48%) | 0 |
| seb | SaPI | 4 (5.33%) | 0 | 4 (5.97%) | 0 |
| seb + sek + seq | SaPI | 4 (5.33%) | 0 | 4 (5.97%) | 0 |
| sek + seq | SaPI | 1 (1.33%) | 0 | 1 (1.49%) | 0 |
| scn2 + vwb3 | SaPI | 49 (65.33%) | 16 (100%) | 42 (62.69%) | 23 (95.83%) |
| sea + sak + scn | prophage | 7 (9.33%) | 0 | 7 (10.45%) | 0 |
| sak + scn | prophage | 8 (10.67%) | 0 | 4 (5.97%) | 4 (16.67%) |
| lukP + lukQ + scn-eq | prophage | 46 (61.33%) | 14 (87.50%) | 38 (56.72%) | 22 (91.67%) |
| sak_phi-42e | prophage | 46 (61.33%) | 13 (81.25%) | 41 (61.19%) | 18 (75%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jahnen, J.; Cuny, C.; Witte, W.; Ehricht, R.; Monecke, S.; Hanke, D.; Ahrens, T.; Leal, M.; Costa, S.S.; Couto, I.; et al. Characterization of Staphylococcus aureus CC1 and CC1660 of Human and Equine Origin. Antibiotics 2025, 14, 1082. https://doi.org/10.3390/antibiotics14111082
Jahnen J, Cuny C, Witte W, Ehricht R, Monecke S, Hanke D, Ahrens T, Leal M, Costa SS, Couto I, et al. Characterization of Staphylococcus aureus CC1 and CC1660 of Human and Equine Origin. Antibiotics. 2025; 14(11):1082. https://doi.org/10.3390/antibiotics14111082
Chicago/Turabian StyleJahnen, Johanna, Christiane Cuny, Wolfgang Witte, Ralf Ehricht, Stefan Monecke, Dennis Hanke, Tanja Ahrens, Marta Leal, Sofia S. Costa, Isabel Couto, and et al. 2025. "Characterization of Staphylococcus aureus CC1 and CC1660 of Human and Equine Origin" Antibiotics 14, no. 11: 1082. https://doi.org/10.3390/antibiotics14111082
APA StyleJahnen, J., Cuny, C., Witte, W., Ehricht, R., Monecke, S., Hanke, D., Ahrens, T., Leal, M., Costa, S. S., Couto, I., Schwarz, S., & Feßler, A. T. (2025). Characterization of Staphylococcus aureus CC1 and CC1660 of Human and Equine Origin. Antibiotics, 14(11), 1082. https://doi.org/10.3390/antibiotics14111082







