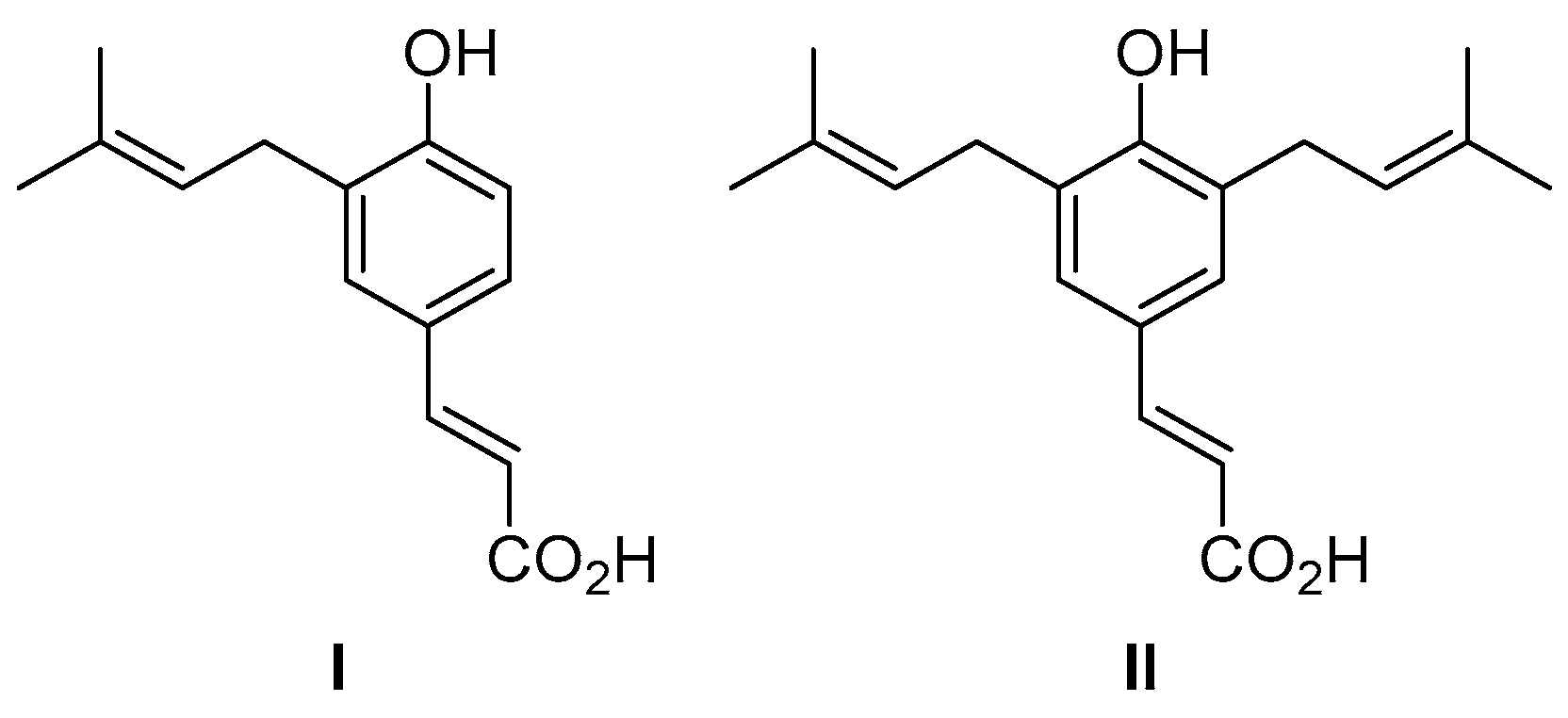
In Vitro and In Silico Studies of the Antimicrobial Activity of Prenylated Phenylpropanoids of Green Propolis and Their Derivatives against Oral Bacteria
Abstract
1. Introduction
2. Results
2.1. In Vitro Antibacterial Activity of Compounds 1–14
2.2. In Silico Studies on the Antibacterial Activity of 2 and 8
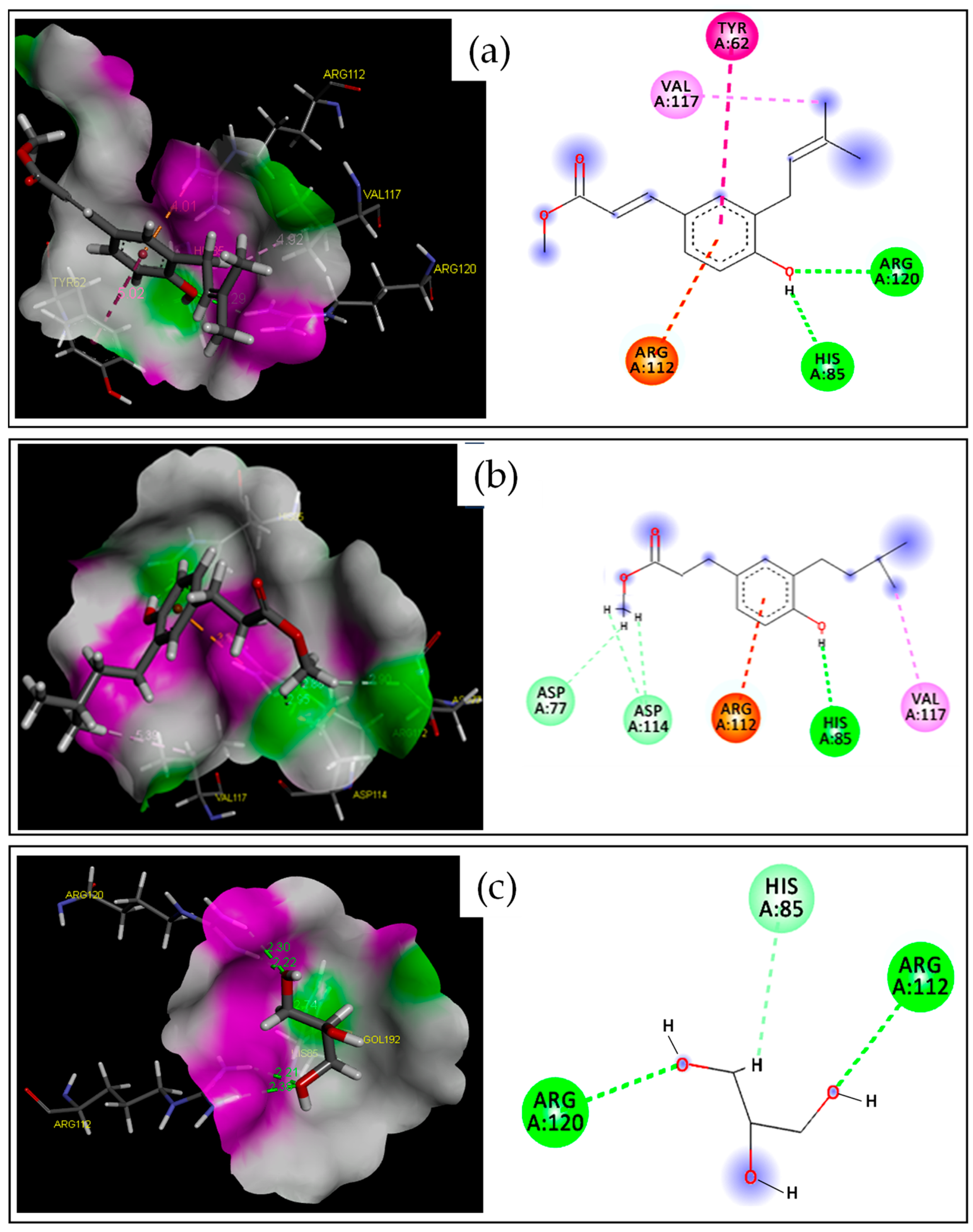
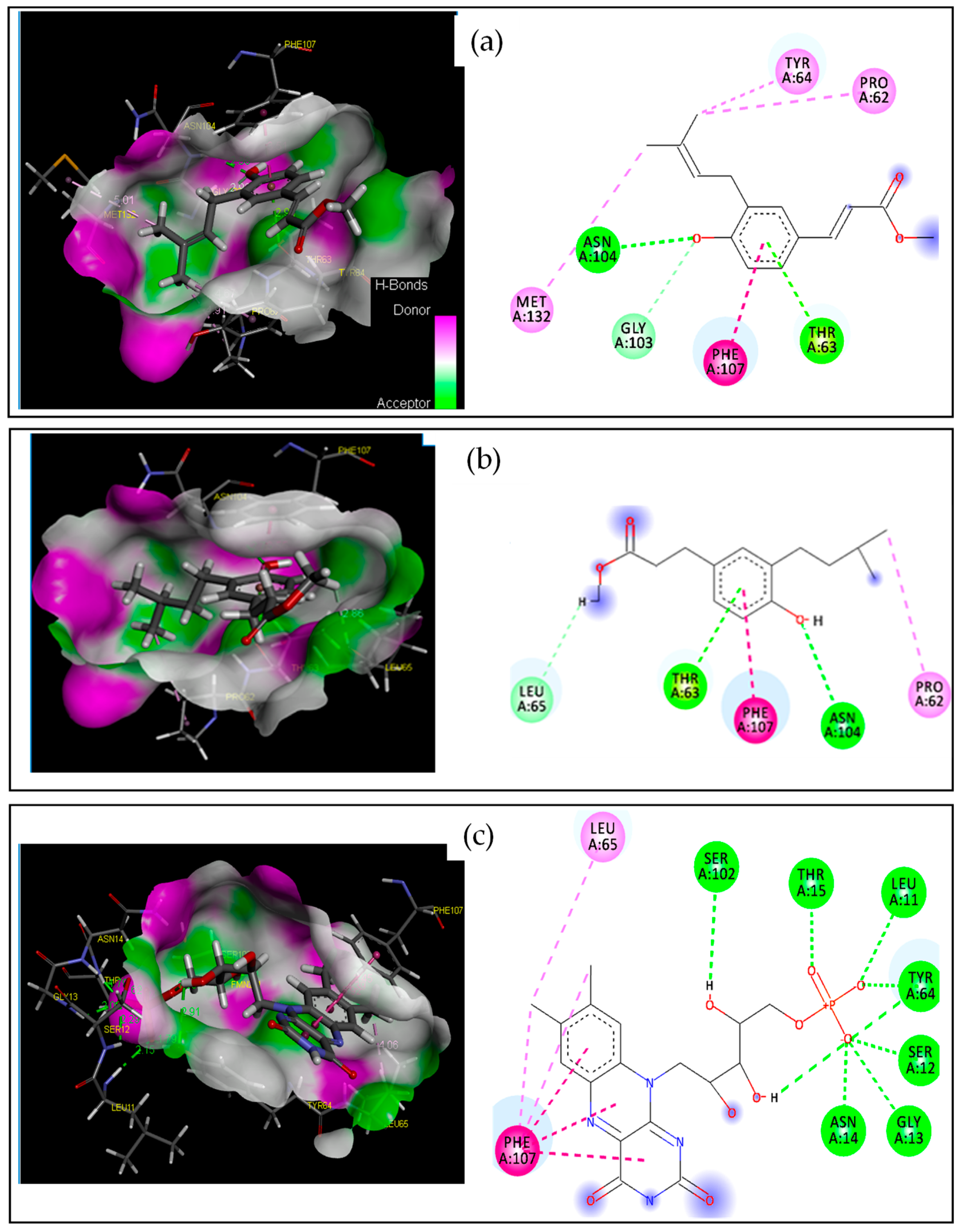
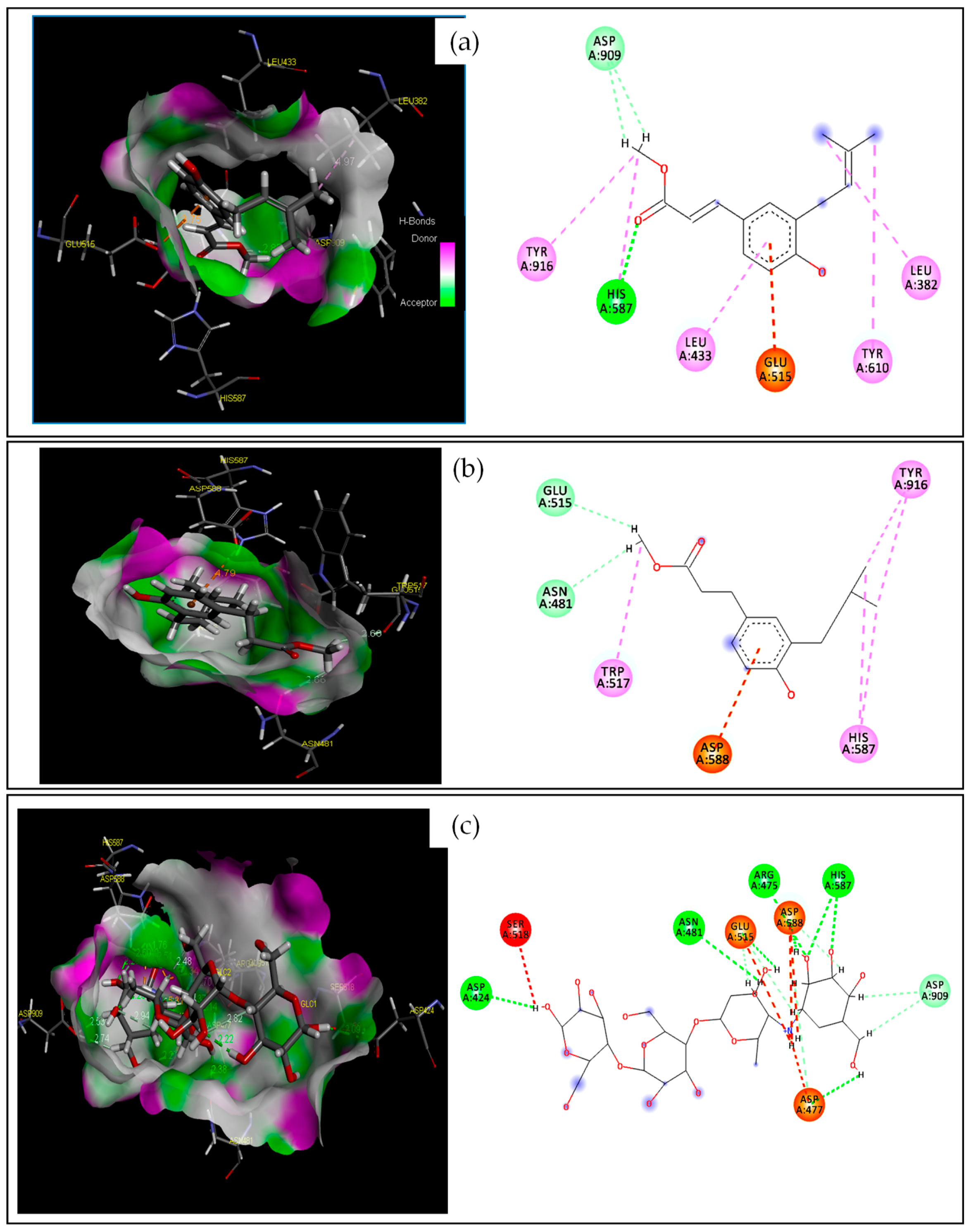
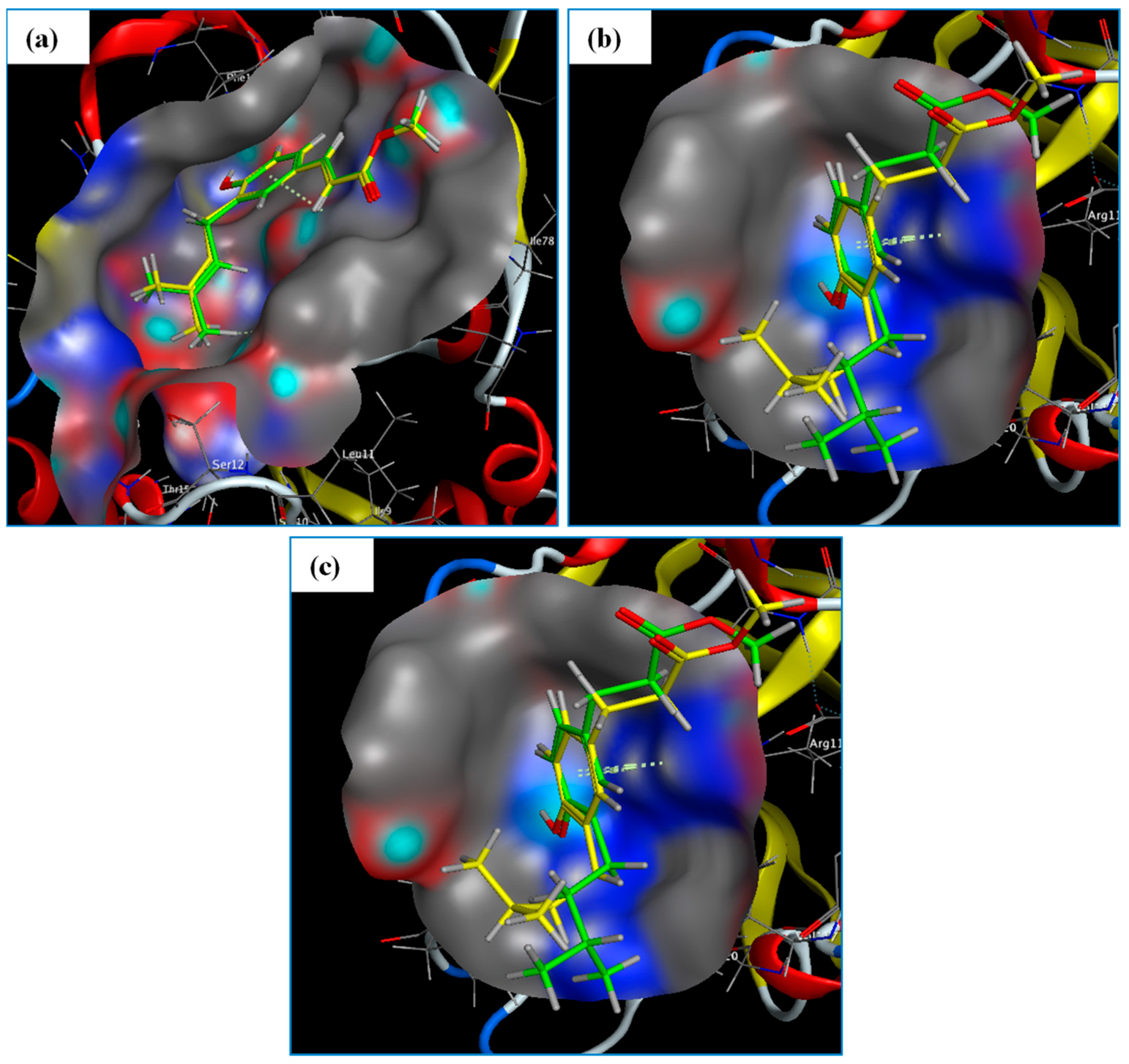
2.2.1. Target–Compound Interactions
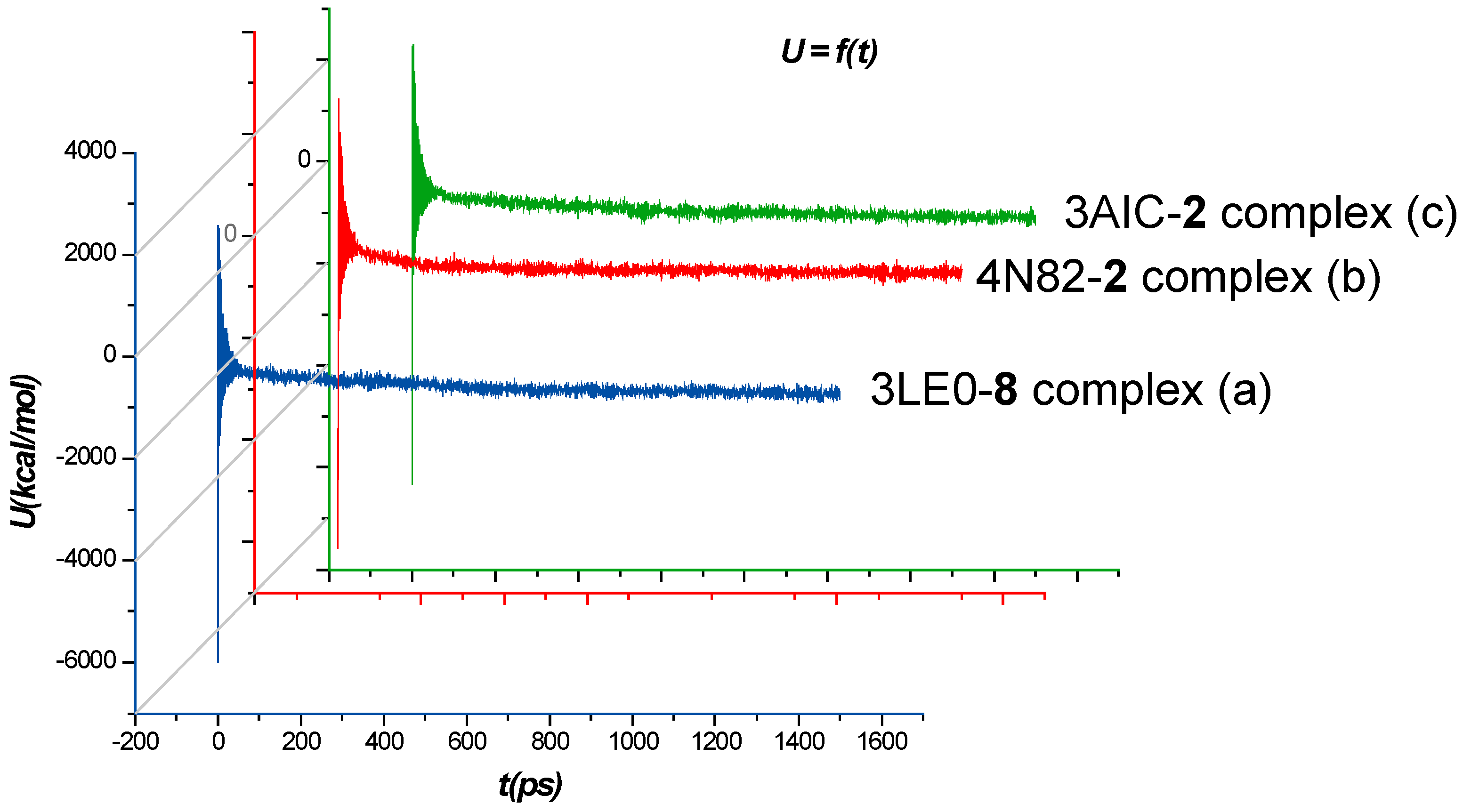
2.2.2. Molecular Dynamics Simulation
2.2.3. Drug-Likeness Prediction and ADMET Properties
Drug-Likeness Evaluation
ADMET Properties
3. Discussion
3.1. In Vitro Antibacterial Activity of Compounds 1–14
3.2. Molecular Docking and Molecular Dynamics
3.3. Drug-Likeness and ADMET Properties
4. Materials and Methods
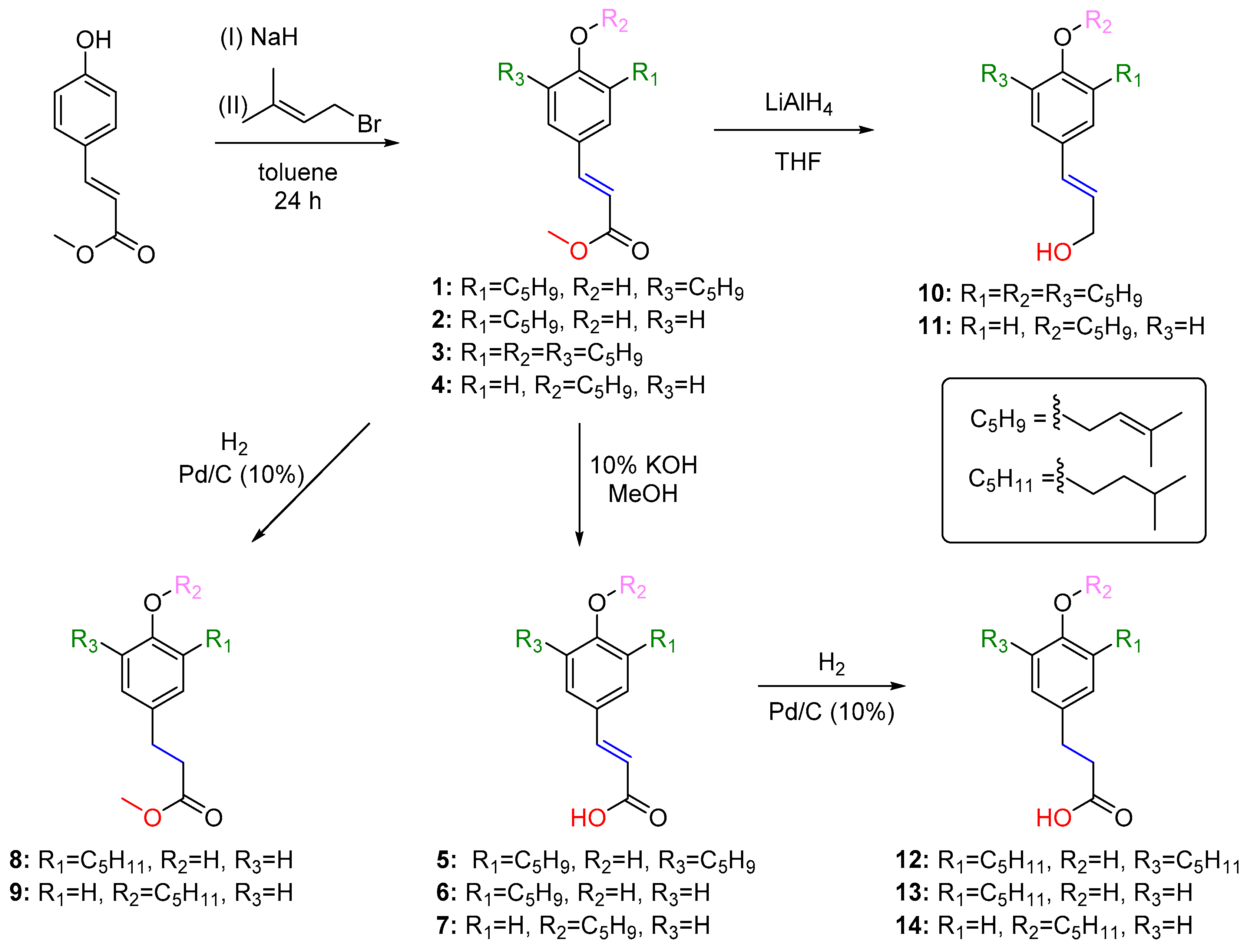
4.1. Synthesis of Compounds 1–14
4.2. Antibacterial Assays
4.3. Computational Methodology
4.3.1. Ligands and Targets Preparation
4.3.2. Docking Method Protocol and Validation
4.3.3. MD Simulations
4.3.4. ADMET Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oliveira, T.A.S.; Santiago, M.B.; Santos, V.H.P.; Silva, E.O.; Martins, C.H.G.; Crotti, A.E.M. Antibacterial activity of essential oils against oral pathogens. Chem. Biodiv. 2022, 19, e202200097. [Google Scholar] [CrossRef]
- Teshome, A.; Muche, A.; Girma, B. Prevalence of dental caries and associated factors in East Africa, 2000–2020: Systematic review and meta-analysis. Front. Public Health 2021, 9, 645091. [Google Scholar] [CrossRef] [PubMed]
- Lendenmann, U.; Grogan, J.; Oppenheim, F. Saliva and dental pellicle-a review. Adv. Dent. Res. 2000, 14, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Chawhuaveang, D.D.; Yu, O.Y.; Yin, I.X.; Lam, W.Y.-H.; Mei, M.L.; Chu, C.-H. Acquired salivary pellicle and oral diseases: A literature review. J. Dent. Sci. 2021, 16, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Reich, M.; Kümmerer, K.; Al-Áhmad, A.; Hannig, C. Fatty acid profile of the initial oral biofilm (pellicle): An in-situ study. Lipids 2013, 48, 929–937. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, W.; Custodio, W.; McDonald, E. New insights into the composition and functions of the acquired enamel pellicle. J. Dent. Res. 2012, 91, 1110–1118. [Google Scholar] [CrossRef]
- Teixeira, E.W.; Napimoga, M.; Carneiro, V. In vitro inhibition of Streptococci binding to enamel acquired pellicle by plant lectins. J. Appl. Microbiol. 2006, 101, 111–116. [Google Scholar] [CrossRef]
- Castro, P.; Tovar, J.A.; Jaramilo, L. Adhesion of Streptococcus mutans to salivary proteins in caries-free and caries-susceptible individuals. Acta Odontol. Latinoam. 2006, 19, 59–66. [Google Scholar]
- Selwitz, R.H.; Ismail, A.I.; Pitts, N.B. Dental caries. Lancet 2007, 369, 51–59. [Google Scholar] [CrossRef]
- Cavalcanti, I.M.; Del Bel Cury, A.A.; Jenkinson, H.F.; Nobbs, A.H. Interactions between Streptococcus oralis, Actinomyces oris, and Candida albicans in the development of multispecies oral microbial biofilms on salivary pellicle. Mol. Oral Microbiol. 2017, 32, 60–73. [Google Scholar] [CrossRef]
- Sahoo, S.R.; Nandini, D.B.; Basandi, P.S.; Selvamani, M.; Donoghue, M. A Comparison of pre- and postbreakfast tooth brushing in caries prevention through the estimation of Streptococcus mutans counts: A prospective clinical and microbiological study. J. Microsc. Ultrastruct. 2022, 10, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Kamal, D.; Hassanein, H.; Akah, M.; Abdelkawy, M.A.; Hamza, H. Caries preventive and antibacterial effects of two natural mouthwashes vs chlorhexidine in high caries-risk patients: A randomized clinical trial. J. Contemp. Dent. Pract. 2020, 21, 1316–1324. [Google Scholar] [CrossRef]
- Fibryanto, E.; Santoso, L. Mouthwashes: A review on its efficacy in preventing dental caries. JKGT 2023, 5, 91–96. [Google Scholar] [CrossRef]
- Masapu, A.; Kumar, S.S.M.; Ashok, K.P.; Anusha, G.; Sangineedy, J.; Ashok, S. Evaluation of the antiplaque efficacy of chlorhexidine mouthwash immediately and 30 min after brushing with fluoride toothpaste—A pilot study. RGUHS J. Dent. Sci. 2019, 11, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Busxer, S. Has resistance to chlorhexidine increased among clinically-relevant bacteria? A systematic review of time course and subpopulation data. PLoS ONE 2021, 16, e0256336. [Google Scholar] [CrossRef]
- Cieplik, F.; Jakubovics, N.S.; Buchalla, W.; Maisch, T.; Hellwig, E.; Al-Ahmad, A. Resistance toward chlorhexidine in oral bacteria—Is there cause for concern? Front. Microbiol. 2019, 10, 587. [Google Scholar] [CrossRef]
- Fahim, A.; Himratul-Aznita, W.H.; Abdul-Rahman, P.S. Allium-sativum and bakuchiol combination: A natural alternative to Chlorhexidine for oral infections? Pakistan J. Med. Sci. 2020, 36, 271–275. [Google Scholar] [CrossRef]
- Soares, K.H.; Firoozi, P.; Souza, G.M.; Martins, B.L.; Falci, S.G.M.; Santos, C.R.R. Efficacy of probiotics compared to chlorhexidine mouthwash in improving periodontal status: A systematic review and meta-analysis. Int. J. Dent. 2023, 2023, 4013004. [Google Scholar] [CrossRef]
- Hossain, R.; Quispe, C.; Khan, R.A.; Saikat, A.S.M.; Ray, P.; Ongalbek, D.; Yeskaliyeva, B.; Jain, D.; Smeriglio, A.; Trombetta, D.; et al. Propolis: An update on its chemistry and pharmacological applications. Chin. Med. 2022, 17, 651. [Google Scholar] [CrossRef]
- Teixeira, E.W.; Negri, G.; Meira, R.M.S.A.; Message, D.; Salatino, A. Plant origin of green propolis: Bee behavior, plant anatomy and chemistry. Evid. Based Complement. Altern. Med. 2005, 2, 85–92. [Google Scholar] [CrossRef]
- Costa, P.; Somensi, L.B.; da Silva, R.; Mariano, L.N.B.; Boeing, T.; Longo, B.; Perfoll, E.; de Souza, P.; Gushiken, L.F.S.; Pellizzon, C.H.; et al. Role of the antioxidant properties in the gastroprotective and gastric healing activity promoted by Brazilian green propolis and the healing efficacy of Artepillin C. Inflammopharmacology 2020, 28, 1009–1025. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, P.F.; Lima, I.M.D.; Munari, C.C.; Bastos, J.K.; da Silva, A.A.; Tavares, D.C. Comparative evaluation of antiproliferative effects of Brazilian green propolis, its main source Baccharis dracunculifolia, and their major constituents artepillin c and baccharin. Planta Med. 2014, 80, 490–492. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.L.; Matsumiya, T.; Hayakari, R.; Shiba, Y.; Kawaguchi, S.; Seya, K.; Ueno, K.; Imaizumi, T. Daily Brazilian green propolis intake elevates blood artepillin C levels in humans. J. Sci. Food Agric. 2021, 101, 4855–4861. [Google Scholar] [CrossRef]
- Kimoto, T.; Koya-Miyata, S.; Hino, K.; Micallef, M.J.; Hanaya, T.; Arai, S.; Ikeda, M.; Kurimoto, M. Pulmonary carcinogenesis induced by ferric nitrilotriacetate in mice and protection from it by Brazilian propolis and artepillin C. Virchows Archiv. Int. J. Phatol. 2001, 438, 259–270. [Google Scholar] [CrossRef]
- Klosek, M.; Sedek, L.; Lewandowska, H.; Czuba, Z.P. The effect of ethanolic extract of Brazilian green propolis and artepillin C on aFGF-1, Eselectin, and CD40L secreted by human gingival fibroblasts. Cent. Eur. J. Immunol. 2021, 46, 438–445. [Google Scholar] [CrossRef]
- Naramoto, K.; Kato, M.; Ichihara, K. Effects of an ethanol extract of Brazilian green propolis on human cytochrome P450 enzyme activities in vitro. J. Agric. Food Chem. 2014, 62, 11296–11302. [Google Scholar] [CrossRef]
- Paulino, N.; Abreu, S.R.L.; Uto, Y.; Koyama, D.; Nagasawa, H.; Hori, H.; Dirsch, V.M.; Vollmar, A.M.; Scremin, A.; Bretz, W.A. Anti-inflammatory effects of a bioavailable compound, Artepillin C, in Brazilian propolis. Eur. J. Pharmacol. 2008, 587, 296–301. [Google Scholar] [CrossRef]
- Ahn, M.R.; Kunimasa, K.; Ohta, T.; Kumazawa, S.; Kamihira, M.; Kaji, K.; Uto, Y.; Hori, H.; Nagasawa, H.; Nakayama, T. Suppression of tumor-induced angiogenesis by Brazilian propolis: Major component artepillin C inhibits in vitro tube formation and endothelial cell proliferation. Cancer Lett. 2007, 252, 235–243. [Google Scholar] [CrossRef]
- Sawaya, A.C.H.F.; Cunha, I.B.S.; Marcucci, M.C. Analytical methods applied to diverse types of Brazilian propolis. Chem. Cent. J. 2011, 5, 27. [Google Scholar] [CrossRef]
- Seibert, J.B.; Bautista-Silva, J.P.; Amparo, T.R.; Petit, A.; Pervier, P.; Almeida, J.C.D.; Azevedo, M.C.; Silveira, B.M.; Brandao, G.C.; de Souza, G.H.B.; et al. Development of propolis nanoemulsion with antioxidant and antimicrobial activity for use as a potential natural preservative. Food Chem. 2019, 287, 61–67. [Google Scholar] [CrossRef]
- Souza, R.M.; de Souza, M.C.; Patitucci, M.L.; Silva, J.F.M. Evaluation of antioxidant and antimicrobial activities and characterization of bioactive components of two Brazilian propolis samples using a pK(a)-guided fractionation. Z. Naturforsch. C 2007, 62, 801–807. [Google Scholar] [CrossRef]
- Veiga, R.S.; Mendonça, S.D.; Mendes, P.B.; Paulino, N.; Mimica, M.J.; Lagareiro Netto, A.A.; Lira, I.S.; López, B.G.C.; Negrão, V.; Marcucci, M.C. Artepillin C and phenolic compounds responsible for antimicrobial and antioxidant activity of green propolis and Baccharis dracunculifolia DC. J. Appl. Microbiol. 2017, 122, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Jorge, R.; Furtado, N.A.J.C.; Sousa, J.P.B.; da Silva, A.A.; Gregorio, L.E.; Martins, C.H.G.; Soares, A.E.E.; Bastos, J.K.; Cunha, W.R.; Silva, M.L.A. Brazilian Propolis: Seasonal variation of the prenylated p-coumaric acids and antimicrobial activity. Pharm. Biol. 2008, 46, 889–893. [Google Scholar] [CrossRef]
- Meto, A.; Colombari, B.; Meto, A.; Boaretto, G.; Pinetti, D.; Marchetti, L.; Benvenuti, S.; Pellati, F.; Blasi, E. Propolis affects Pseudomonas aeruginosa growth, biofilm formation, eDNA release and phenazine production: Potential involvement of polyphenols. Microorganisms 2020, 8, 243. [Google Scholar] [CrossRef] [PubMed]
- Leitão, D.P.S.; Silva Filho, A.A.; Polizello, A.C.M.; Bastos, J.K.; Spadaro, A.C.C. Comparative evaluation of in-vitro effects of Brazilian green propolis and Baccharis dracunculifolia extracts on cariogenic factors of Streptococcus mutans. Biol. Pharm. Bull. 2004, 27, 1834–1839. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, J.G.; Iorio, N.L.P.; Rodrigues, L.F.; Couri, M.L.B.; Farah, A.; Maia, L.C.; Antonio, A.G. Influence of a Brazilian wild green propolis on the enamel mineral loss and Streptococcus mutans’ count in dental biofilm. Arch. Oral Biol. 2016, 65, 77–81. [Google Scholar] [CrossRef]
- Righi, A.A.; Negri, G.; Salatino, A. Comparative chemistry of propolis from eight Brazilian localities. Evid. Based Complement. Altern. Med. 2013, 2013, 267878. [Google Scholar] [CrossRef]
- Salatino, A.; Teixeira, E.W.; Negri, G.; Message, D. Origin and chemical variation of Brazilian propolis. Evid. Based Complement. Altern. Med. 2005, 2, 33–38. [Google Scholar] [CrossRef]
- Shahinozzama, M.; Basak, B.; Emran, R.; Rozario, P.; Obanda, D.N. Artepillin C: A comprehensive review of its chemistry, bioavailability, and pharmacological properties. Fitoterapia 2020, 147, 104775. [Google Scholar] [CrossRef]
- Costa, P.; Almeida, M.O.; Lemos, M.; Arruda, C.; Casoti, R.; Somensi, L.B.; Boeing, T.; Mariott, M.; da Silva, R.; Stein, B.D.; et al. Artepillin C, drupanin, aromadendrin-4′-O-methyl-ether and kaempferide from Brazilian green propolis promote gastroprotective action by diversified mode of action. J. Ethnopharmacol. 2018, 226, 82–89. [Google Scholar] [CrossRef]
- Mishima, S.; Ono, Y.; Araki, Y.; Akao, Y.; Nozawa, Y. Two related cinnamic acid derivatives from Brazilian honey bee propolis, baccharin and drupanin, induce growth inhibition in allografted sarcoma S-180 in mice. Biol. Pharm. Bull. 2005, 28, 1025–1030. [Google Scholar] [CrossRef]
- Antunes, O.A.C.; da Silva, J.F.M.; Estrada, G.O.D. Artepillin C: A review. Lett. Drug Des. Discov. 2008, 5, 88–92. [Google Scholar] [CrossRef]
- Kano, Y.; Horie, N.; Doi, S.; Aramaki, F.; Maeda, H.; Hiragami, F.; Kawamura, K.; Motoda, H.; Koike, Y.; Akiyama, J.; et al. Artepillin C derived from propolis induces neurite outgrowth in PC12m3 cells via ERK and p38 MAPK pathways. Neurochem. Res. 2008, 33, 1795–1803. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.M.; Portapilla, G.B.; Silva, G.M.; Duarte, A.; Rotta, C.G.; Silva, C.; de Albuquerque, S.; Bastos, J.K.; Campo, V.L. Synthesis, antitumor activity and in silico analyses of amino acid derivatives of artepillin C, drupanin and baccharin from green propolis. Bioorg. Med. Chem. 2021, 47, 116372. [Google Scholar] [CrossRef] [PubMed]
- Tiveron, A.P.; Rosalen, P.L.; Franchin, M.; Lacerda, R.C.C.; Bueno-Silva, B.; Benso, B.; Denny, C.; Ikegaki, M.; Alencar, S.M. Chemical characterization and antioxidant, antimicrobial, and anti-inflammatory activities of South Brazilian organic propolis. PLoS ONE 2016, 11, e0165588. [Google Scholar] [CrossRef]
- Feil, S.C.; Lawerence, S.; Mulhern, T.D.; Holien, J.K.; Hotze, E.M.; Farrand, S.; Tweten, R.K.; Parker, M.W. Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity. Structure 2012, 20, 248–258. [Google Scholar] [CrossRef]
- Makhlynets, O.; Boal, A.K.; Rhodes, D.V.; Kitten, T.; Rosenzweig, A.C.; Stubbe, J. Streptococcus sanguinis class Ib ribonucleotide reductase: High activity with both iron and manganese cofactors and structural insights. J. Biol. Chem. 2014, 289, 6259–6272. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Ito, S.; Shimamura, T.; Weyand, S.; Kawarasaki, Y.; Miwaka, T.; Abe, K.; Kobayashi, T.; Cameron, A.D.; Iwata, S. Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans. J. Mol. Biol. 2011, 408, 177–186. [Google Scholar] [CrossRef]
- Nijampatnam, B.; Zhang, H.; Cai, X.; Michalek, S.M.; Wu, H.; Velu, S.E. Inhibition of Streptococcus mutans biofilms by the natural stilbene piceatannol through the inhibition of glucosyltransferases. ACS Omega 2018, 3, 8378–8385. [Google Scholar] [CrossRef]
- Atta, L.; Khalil, R.; Khan, K.M.; Zehra, M.; Saleem, F.; Nur-e-Alam, M.; Ul-Haq, Z. Virtual screening, synthesis and biological evaluation of Streptococcus mutans mediated biofilm inhibitors. Molecules 2022, 27, 1455. [Google Scholar] [CrossRef]
- Liao, Y.; Zhang, M.; Lin, X.; Yan, F. Diaryl urea derivative molecule inhibits cariogenic Streptococcus mutans by affecting exopolysaccharide synthesis, stress response, and nitrogen metabolism. Front. Cell. Infect. Microbiol. 2022, 12, 904488. [Google Scholar] [CrossRef] [PubMed]
- Rios, J.L.; Recio, M.C. Medicinal plants and antimicrobial activity. J. Ethnopharmacol. 2005, 100, 80–84. [Google Scholar] [CrossRef]
- Saleem, M.; Nazir, M.; Ali, M.S.; Hussain, H.; Lee, Y.S.; Riaz, N.; Jabbar, A. Antimicrobial natural products: An update on future antibiotic drug candidates. Nat. Prod. Rep. 2010, 27, 238–254. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, E.M.; Alhatlani, B.Y.; Menezes, R.P.; Martins, C.H.G. Back to nature: Medicinal plants as promising sources for antibacterial drugs in the post-antibiotic era. Plants 2023, 12, 3077. [Google Scholar] [CrossRef] [PubMed]
- Tazawa, S.; Warashina, T.; Noro, T.; Miyase, T. Studies on the constituents of Brazilian Propolis. Chem. Pharm. Bull. 1998, 46, 1477–1479. [Google Scholar] [CrossRef]
- Schmitt, A.; Telikepalli, H.; Mitscher, L.A. Plicatin B, the antimicrobial principle of Psoralea juncea. Phytochemistry 1991, 30, 3569–3570. [Google Scholar] [CrossRef]
- Makade, C.S.; Shenoi, P.R.; Bhongade, B.A.; Shingane, S.A.; Ambulkar, P.C.; Schewale, A.M.; Shewale, A.M. Estimation of MBC: MIC ratio of herbal extracts against common endodontic pathogens. J. Pharm. Bioall. Sci. 2024, 16, S1414–S1416. [Google Scholar] [CrossRef]
- Aga, H.; Shibuya, T.; Sugimoto, T.; Kurimoto, M.; Nakajima, S. Isolation and identification of antimicrobial compounds in Brazilian propolis. Biosci. Biotech. Biochem. 1994, 58, 945–946. [Google Scholar] [CrossRef]
- Liu, X.L.; Xu, Y.J.; Go, M.L. Functionalized chalcones with basic functionalities have antibacterial activity against drug sensitive Staphylococcus aureus. Eur. J. Med. Chem. 2008, 43, 1681–1687. [Google Scholar] [CrossRef]
- Kubo, I.; Muroi, H.; Kubo, A. Structural functions of antimicrobial long-chain alcohols and phenols. Bioorg. Med. Chem. 1995, 3, 873–880. [Google Scholar] [CrossRef]
- Roleira, F.M.F.; Siquet, C.; Orrù, E.; Garrido, E.M.; Garrido, J.; Milhazes, N.; Podda, G.; Paiva-Martins, F.; Reis, S.; Carvalho, R.A. Lipophilic phenolic antioxidants: Correlation between antioxidant profile, partition coefficients and redox properties. Bioorg. Med. Chem. 2010, 18, 5816–5825. [Google Scholar] [CrossRef]
- Oufensou, S.; Casalini, S.; Balmas, V.; Carta, P.; Chtioui, W.; Dettori, M.A.; Fabbri, D.; Migheli, Q.; Delogu, G. Prenylated trans-cinnamic esters and ethers against clinical Fusarium spp.: Repositioning of natural compounds in antimicrobial discovery. Molecules 2021, 26, 658. [Google Scholar] [CrossRef]
- Araya-Cloutier, C.; Vincken, J.-P.; van de Schans, M.G.M.; Hageman, J.; Schafternaar, G.; den Besten, H.M.W.; Gruppen, H. QSAR-based molecular signatures of prenylated (iso)flavonoids underlying antimicrobial potency against and membrane-disruption in Gram positive and Gram negative bacteria. Sci. Rep. 2018, 8, 9267. [Google Scholar] [CrossRef] [PubMed]
- Agu, P.C.; Afiukwa, C.A.; Orji, O.U.; Ezeh, E.M.; Ofoke, I.H.; Ogbu, C.O.; Ugwaja, E.I.; Aja, P.M. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci. Rep. 2023, 13, 13398. [Google Scholar] [CrossRef]
- Yunta, M.J.R. Docking and ligand binding affinity: Uses and pitfalls. Am. J. Model. Optim. 2016, 4, 74–114. [Google Scholar] [CrossRef]
- Bhagat, R.T.; Butle, S.R.; Khobragade, D.S.; Wankhede, S.B.; Prasad, C.C.; Mahure, D.S.; Armarkar, A.V. Molecular Docking in Drug Discovery. J. Pharm. Res. Int. 2021, 33, 46–58. [Google Scholar] [CrossRef]
- Meng, E.C.; Shoichet, B.K.; Kuntz, I.D. Automated docking with grid-based energy evaluation. J. Comput. Chem. 2004, 13, 505–524. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comp. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Feig, M.; Onufriev, A.; Lee, M.S.; Im, W.; Case, D.A.; Brooks, C.L. Performance comparison of generalized born and Poisson methods in the calculation of electrostatic solvation energies for protein structures. J. Comput. Chem. 2004, 25, 265–284. [Google Scholar] [CrossRef]
- Silva, D.R.; Deps, T.D.; Sakaguchi, O.A.S.; Costa, E.M.M.B.; Santos, C.A.O.; Silva, J.P.R.; Silva, B.D.; Ribeiro, F.F.; Mendonça-Júnior, F.J.B.; Silva, A.C.B. Molecular docking of phytochemicals against Streptococcus mutans virulence targets: A proteomic insight into drug plannin. In Oral Health Care—An Important Issue of the Modern Society; Ardelean, L.C., Rusu, L.C., Eds.; IntechOpen: London, UK, 2022; pp. 1–20. [Google Scholar]
- Imberty, A.; Hardman, K.D.; Carver, J.P.; Perez, S. Molecular modelling of protein-carbohydrate interactions. Docking of monosaccharides in the binding site of concanavalin A. Glycobiology 1991, 1, 631–642. [Google Scholar] [CrossRef]
- Wade, R.C.; Goodford, P.J. The role of hydrogen-bonds in drug binding. Progr. Clin. Biol. Res. 1989, 289, 433–444. [Google Scholar]
- Fernandes, L.P.; Silva, J.M.B.; Martins, D.O.S.; Santiago, M.B.; Martins, C.H.G.; Jardim, A.C.G.; Oliveira, G.S.; Pivatto, M.; Souza, R.A.C.; Franca, E.F.; et al. Fragmentation study, dual anti-bactericidal and anti-viral effects and molecular docking of cobalt (III) complexes. Int. J. Mol. Sci. 2020, 21, 8355. [Google Scholar] [CrossRef] [PubMed]
- Velarde, J.J.; Piai, A.; Lichtenstein, I.J.; Lynskey, N.N.; Chou, J.J.; Wessels, M.R. Structure of the Streptococcus pyogenes NAD+ Glycohydrolase translocation domain and its essential role in toxin binding to oropharyngeal keratinocytes. J. Bacteriol. 2022, 204, e0036621. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, S.A.; Dror, R.O. Molecular dynamics simulation for all. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef]
- De Vivo, M.; Masetti, M.; Bottegoni, G.; Cavalli, A. Role of molecular dynamics and related methods in drug discovery. J. Med. Chem. 2016, 59, 4035–4061. [Google Scholar] [CrossRef]
- Salo-Ahen, O.M.; Alanko, I.; Bhadane, R.; Bonvin, A.M.J.J.; Honorato, R.V.; Hossain, S.; Juffer, A.H.; Kabedev, A.; Lahtela-Kakkonen, M.; Larsen, A.S.; et al. Molecular Dynamics simulations in drug discovery and pharmaceutical development. Processes 2021, 9, 71. [Google Scholar] [CrossRef]
- Roy, N.; Ghosh, B.; Roy, D.; Bhaumik, B.; Roy, M.N. Exploring the inclusion complex of a drug (umbelliferone) with α-cyclodextrin optimized by molecular docking and increasing bioavailability with minimizing the doses in human body. ACS Omega 2020, 5, 30243–30251. [Google Scholar] [CrossRef] [PubMed]
- Djebaili, R.; Kenouche, S.; Daoud, I.; Melkemi, N.; Belkadi, A.; Mesli, F. Investigation of [3H]diazepam derivatives as allosteric modulators of GABAA receptor α1β2γ2 subtypes: Combination of molecular docking/dynamic simulations, pharmacokinetics/drug-likeness prediction, and QSAR analysis. Struct. Chem. 2022, 34, 791–823. [Google Scholar] [CrossRef] [PubMed]
- Bickerton, G.R.; Paolini, G.V.; Besnard, J.; Muresan, S.; Hopkins, A.L. Quantifying the chemical beauty of drugs. Nat. Chem. 2012, 4, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug. Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Lipinski, C.A. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 2000, 44, 235–249. [Google Scholar] [CrossRef]
- Pant, S.; Verma, S.; Pathak, R.K.; Singh, D.B. Structure-based drug designing. In Bioinformatics: Methods and Applications; Academic Press: Cambridge, MA, USA, 2022; pp. 219–231. [Google Scholar]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Patra, T.; Bag, S.; Kancherla, R.; Mondal, A.; Dey, A.; Pimparkar, S.; Agasti, S.; Modak, A.; Debabrata Maiti, D. Palladium-catalyzed directed para C-H functionalization of phenols. Angew. Chem. Int. Ed. 2016, 55, 7751–7755. [Google Scholar] [CrossRef]
- Uto, Y.; Hirata, A.; Fujita, T.; Takubo, S.; Nagasawa, H.; Hori, H. First total synthesis of artepillin C established by o,o’-diprenylation of p-halophenols in water. J. Org. Chem. 2002, 67, 2355–2357. [Google Scholar] [CrossRef] [PubMed]
- Kantee, K.; Rukachaisirikul, V.; Tadpetch, K. Synthesis of tetrahydropyranyl diarylheptanoids from Dioscorea villosa. Tetrahedron Lett. 2016, 57, 3505–3509. [Google Scholar] [CrossRef]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; Approved Standard M07; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Duarte, E.A.; Santiago, M.B.; Silva, N.B.S.; Martins, C.H.G.; Gatto, C.C. Crystal design, spectroscopic analyses and antibacterial study of new carbazate ligands and their Cu(II) complexes. Inorg. Chim. Acta 2023, 549, 121421. [Google Scholar] [CrossRef]
- Sarker, S.D.; Nahar, L.; Kumarasamy, Y. Microtitre plate-based antibacterial assay incorporating resazurin as an indicator of cell growth, and its application in the in vitro antibacterial screening of phytochemicals. Methods 2007, 42, 321–324. [Google Scholar] [CrossRef]
- Stewart, J.M.P. Optimization of parameters for semi-empirical methods V: Modification of NDDO approximations and application to 70 elements. J. Mol. Model. 2007, 13, 1173–1213. [Google Scholar] [CrossRef]
- MOE. Molecular Operating Environment (MOE), 2013.08; Chemical Computing Group Inc.: Montreal, QC, Canada, 2014. [Google Scholar]
- Daoud, I.; Melkemi, N.; Salah, T.; Ghalem, S. Combined QSAR, molecular docking and molecular dynamics study on new Acetylcholinesterase and Butyrylcholinesterase inhibitors. Comput. Biol. Chem. 2018, 74, 304–326. [Google Scholar] [CrossRef]
- Daoud, I.; Mesli, F.; Melkemi, N.; Ghalem, S.; Salah, T. Discovery of potential SARS-CoV 3CL protease inhibitors from approved antiviral drugs using: Virtual screening, molecular docking, pharmacophore mapping evaluation and dynamics simulation. J. Biomol. Struct. Dyn. 2021, 40, 12574–12591. [Google Scholar] [CrossRef]
- Bajda, M.; Więckowska, A.; Hebda, M.; Guzior, N.; Sotriffer, C.A.; Malawska, B. Structure-based search for new inhibitors of cholinesterases. Int. J. Mol. Sci. 2013, 14, 5608–5632. [Google Scholar] [CrossRef] [PubMed]
- Bond, S.D.; Leimkuhler, B.J.; Laird, B.B. The Nosé-Poincaré method for constant temperature molecular dynamics. J. Comp. Phys. 1999, 151, 114–134. [Google Scholar] [CrossRef]
- Parikesit, A.A.; Zahroh, H.; Nugroho, A.S.; Hapsari, A.; Tambunan, U.S.F. The computation of cyclic peptide with prolin-prolin bond as fusion inhibitor of DENV envelope protein through molecular docking and molecular dynamics simulation. arXiv-bio 2015, arXiv:1511.01388. [Google Scholar] [CrossRef]
- OriginLab. OriginPro 9.1; OriginLab Corporation: Northampton, MA, USA, 2014. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Pires, D.E.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
| Compound | Enterococcus Faecalis ATCC 4082 | Lactobacillus Paracasei ATCC 11578 | Streptococcus Salivarius ATCC 25975 | Streptococcus Sobrinus ATCC 33478 | Streptococcus Mitis ATCC 49456 | Streptococcus Sanguinis ATCC 10556 | Streptococcus Mutans ATCC 25175 |
|---|---|---|---|---|---|---|---|
| 2 | 500 (2.03) 500 (2.03) | 500 (2.03) 1000 (4.06) | 62.5 (0.25) 250 (1.01) | 62.5 (0.25) 62.5 (0.25) | 31.2 (0.13) 31.2 (0.13) | 31.2 (0.13) 31.2 (0.13) | 31.2 (0.13) 31.2 (0.51) |
| 3 | >2000 (>5.23) >2000 (>5.23) | >2000 (>5.23) >2000 (>5.23) | 2000 (5.23) >2000 (>5.23) | 2000 (5.23) >2000 (>5.23 | 500 (1.31) 1000 (2.61) | 1000 (2.61) 2000 (5.23) | >2000 (5.23) >2000 (5.23) |
| 4 | >2000 (8.12) >2000 (>8.12) | >2000 (8.12) >2000 (>8.12) | >2000 (8.12) >2000 (>8.12) | >2000 (8.12) >2000 (>8.12) | 1000 (4.06) 2000 (8.12) | >2000 (8.12) >2000 (>8.12) | >2000 (8.12) >2000 (>8.12) |
| 5 | >2000 (>6.66) >2000 (>6.66) | 500 (1.66) 1000 (3.33) | 1000 (3.33) 2000 (6.66) | 500 (1.66) 2000 (6.66) | 125 (0.41) 1000 (3.33) | 500 (1.66) 1000 (3.33) | 500 (1.66) 2000 (6.66) |
| 6 | 1000 (3.09) 1000 (3.09) | 500 (1.55) 500 (1.55) | 250 (0.77) 500 (1.55) | 125 (0.39) 125 (0.39) | 250 (0.77) 250 (0.77) | 250 (0.77) 250 (0.77) | 125 (0.39) 250 (0.77) |
| 7 | 500 (2.15) >2000 (>8.61) | 1000 (4.31) 2000 (8.61) | 1000 (4.31) 2000 (8.61) | 1000 (4.31) 1000 (4.31) | 500 (2.15) 500 (2.15) | 500 (2.15) 500 (2.15) | 1000 (4.31) 2000 (8.61) |
| 8 | 125 (0.50) 125 (0.50) | 62.5 (0.25) 250 (1.0) | 62.5 (0.25) 62.5 (0.25) | 125 (0.50) 125 (0.50) | 31.25 (0.12) 31.25 (0.12) | 62.5 (0.25) 62.5 (0.25) | 62.5 (0.25) 62.5 (0.25) |
| 9 | >2000 (8.0) >2000 (8.0) | >2000 (8.0) >2000 (8.0) | 1000 (4.0) 2000 (8.0) | 1000 (4.0) 1000 (4.0) | 500 (2.0) 1000 (4.0) | 1000 (4.0) 1000 (4.0) | 2000 (8.0) 2000 (8.0) |
| 11 | 2000 (9.16) >2000 (>9.1) | 2000 (9.16) >2000 (>9.1) | 62.5 (0.29) 125 (0.57) | 2000 (9.16) 2000 (9.16) | 500 (2.29) 1000 (4.58) | 1000 (4.58) 2000 (9.16) | 1000 (4.58) 2000 (9.16) |
| 12 | 1000 (3.26) 2000 (6.53) | 500 (1.63) 1000 (3.26) | 500 (1.63) 500 (1.63) | 250 (0.82) 500 (1.63) | 62.5 (0.20) 125 (0.41) | 500 (1.63) 1000 (3.26) | 500 (1.63) 500 (1.63) |
| 13 | >2000 (8.46) >2000 (8.46) | >2000 (8.46) >2000 (8.46) | >2000 (8.46) >2000 (8.46) | >2000 (8.46) >2000 (8.46) | 250 (1.06) 1000 (4.23) | 1000 (4.23) 1000 (4.23) | >2000 (8.46) >2000 (8.46) |
| CHD | 3.69 (7.30) 3.69 (7.30) | 1.84 (3.64) 1.84 (3.64) | 0.92 (1.82) 0.92 (1.82) | 1.84 (3.64) 1.84 (3.64) | 3.69 (7.30) 3.69 (7.30) | 3.69 (7.30) 3.69 (7.30) | 0.92 (1.82) 0.92 (1.82) |
| 3LE0 | 4N82 | 3AIC | |
|---|---|---|---|
| Method | X-ray diffraction | X-ray diffraction | X-ray diffraction |
| Microorganism | S. mitis | S. sanguinis | S. mutans |
| Chain | A | A, B, C, D, E, F | A, B, C, D, E |
| Sequence length | 153 | 178 | 488 |
| Resolution (Å) | 1.91 | 1.88 | 3.11 |
| Native ligands | GOL | FMN | ACA |
| S-Score (kcal/mol) | RMSD (Å) | Bonds between Atoms of Compounds and Active Site Residues | |||||
|---|---|---|---|---|---|---|---|
| Atom of Compound | Involved Receptor Atoms | Involved Receptor Residues | Category | Type of Interaction | |||
| 2 | −4.228 | 2.997 | O | HH11 | ARG120(A) | HB | Conventional HB |
| H | NE2 | HIS85(A) | HB | Conventional HB | |||
| / | NH2 | ARG112(A) | Electrostatic | Pi-Cation | |||
| C | / | TYR62(A) | Hydrophobic | Pi-Pi T-shaped | |||
| C | / | VAL117(A) | Hydrophobic | Alkyl | |||
| 8 | −4.476 | 2.932 | H | NE2 | HIS85(A) | HB | Conventional HB |
| H | OD1 | ASP114(A) | HB | Carbon HB | |||
| H | OD2 | ASP77(A) | HB | Carbon HB | |||
| H | OD1 | ASP114(A) | HB | Carbon HB | |||
| / | NH2 | ARG112(A) | Electrostatic | Pi-Cation | |||
| C | / | VAL117(A) | Hydrophobic | Alkyl | |||
| GOL | −3.655 | 2.149 | O1 | HH11 | ARG120(A) | HB | Conventional HB |
| O3 | HH11 | ARG112(A) | HB | Conventional HB | |||
| H12 | NE2 | HIS85(A) | HB | Carbon H-Bond | |||
| Physicochemical Properties | Drug-Likeness Rules | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| TPSA (Å2) | n-ROT | MW (g/mol) | MLogP | n-HA | n-HD | Lipinski | Veber | Egan | |
| WLogP | |||||||||
| (0~140) | (0~11) | (100~500) | (0~5) | (0~12) | (0~7) | ||||
| 2 | 39.42 | 2 | 225.25 | 1.49 | 3 | 0 | Accepted | Accepted | Accepted |
| 2.40 | |||||||||
| 8 | 30.19 | 1 | 223.27 | 2.33 | 2 | 0 | Accepted | Accepted | Accepted |
| 3.01 | |||||||||
| ADMET | Parameters | Compounds | |
|---|---|---|---|
| 2 | 8 | ||
| Absorption | Caco2 (Log Papp × 10−6 cm/s) | 1.178 | 1.417 |
| HIA (%) | 99.207 | 97.682 | |
| Distribution | CNS (logPS) | −1.800 | −1.692 |
| BBB (logBB) | 0.311 | 0.030 | |
| Metabolism | CYP1A2 inhibitor | Yes | Yes |
| CYP2C19 Inhibitor | No | No | |
| CYP2D6 inhibitor | No | No | |
| CYP2D6 substrate | No | No | |
| CYP3A4 substrate | No | No | |
| Excretion | Renal OCT2 substrate | No | No |
| Total clearance (log mL/min/kg) | 0.744 | 0.780 | |
| Toxicity | hERG I and II inhibitors | No | No |
| Hepatotoxicity | No | No | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vieira, T.M.; Barco, J.G.; de Souza, S.L.; Santos, A.L.O.; Daoud, I.; Rahali, S.; Amdouni, N.; Bastos, J.K.; Martins, C.H.G.; Ben Said, R.; et al. In Vitro and In Silico Studies of the Antimicrobial Activity of Prenylated Phenylpropanoids of Green Propolis and Their Derivatives against Oral Bacteria. Antibiotics 2024, 13, 787. https://doi.org/10.3390/antibiotics13080787
Vieira TM, Barco JG, de Souza SL, Santos ALO, Daoud I, Rahali S, Amdouni N, Bastos JK, Martins CHG, Ben Said R, et al. In Vitro and In Silico Studies of the Antimicrobial Activity of Prenylated Phenylpropanoids of Green Propolis and Their Derivatives against Oral Bacteria. Antibiotics. 2024; 13(8):787. https://doi.org/10.3390/antibiotics13080787
Chicago/Turabian StyleVieira, Tatiana M., Julia G. Barco, Sara L. de Souza, Anna L. O. Santos, Ismail Daoud, Seyfeddine Rahali, Noureddine Amdouni, Jairo K. Bastos, Carlos H. G. Martins, Ridha Ben Said, and et al. 2024. "In Vitro and In Silico Studies of the Antimicrobial Activity of Prenylated Phenylpropanoids of Green Propolis and Their Derivatives against Oral Bacteria" Antibiotics 13, no. 8: 787. https://doi.org/10.3390/antibiotics13080787
APA StyleVieira, T. M., Barco, J. G., de Souza, S. L., Santos, A. L. O., Daoud, I., Rahali, S., Amdouni, N., Bastos, J. K., Martins, C. H. G., Ben Said, R., & Crotti, A. E. M. (2024). In Vitro and In Silico Studies of the Antimicrobial Activity of Prenylated Phenylpropanoids of Green Propolis and Their Derivatives against Oral Bacteria. Antibiotics, 13(8), 787. https://doi.org/10.3390/antibiotics13080787







