Bacterial and Genetic Features of Raw Retail Pork Meat: Integrative Analysis of Antibiotic Susceptibility, Whole-Genome Sequencing, and Metagenomics
Abstract
1. Introduction
2. Results
2.1. Isolation of Indicator Bacteria
2.2. Antibiotic Susceptibility Testing (AST) and Whole-Genome Sequencing (WGS) of Isolated Indicator Bacteria
2.3. Antibiotic Residue Testing
2.4. Metagenomics
2.4.1. Read Statistics
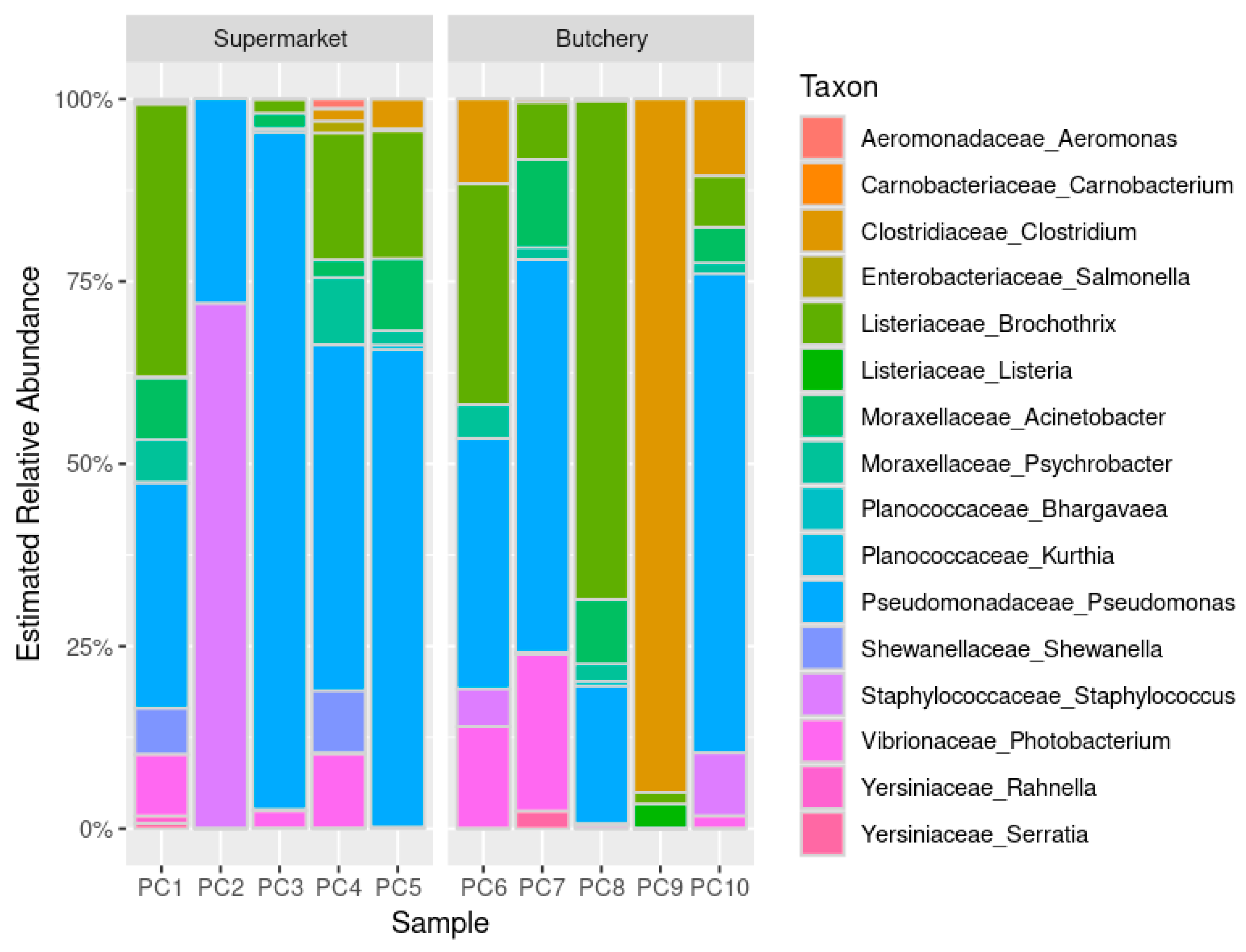
2.4.2. Estimated Relative Abundance
2.4.3. Resistome Prediction
Antibiotic Resistance Gene Prediction
Virulence Factor and Toxin Gene Prediction
3. Materials and Methods
3.1. Ethical Clearance and Study Definitions
3.2. Study Setting and Sampling
3.3. AST and WGS of Isolated Indicator Bacteria from Raw Meat Samples
3.4. Metagenomics
3.5. Resistome Gene Abundance Estimates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Department of Agriculture Land Reform and Rural Development. A Profile of the South African Pork Market Value Chain. Available online: http://webapps1.daff.gov.za/AmisAdmin/upload/Pork%20Market%20Value%20Chain%20Profile%202021.pdf (accessed on 26 October 2023).
- Manyi-Loh, C.E.; Lues, R. A South African perspective on the microbiological and chemical quality of meat: Plausible public health implications. Microorganisms 2023, 11, 2484. [Google Scholar] [CrossRef] [PubMed]
- Eckstrom, K.; Barlow, J.W. Resistome metagenomics from plate to farm: The resistome and microbial composition during food waste feeding and composting on a Vermont poultry farm. PLoS ONE 2019, 14, e0219807. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cao, W.; Liang, S.; Yamasaki, S.; Chen, X.; Shi, L.; Ye, L. Metagenomic characterization of bacterial community and antibiotic resistance genes in representative ready-to-eat food in southern China. Sci. Rep. 2020, 10, 15175. [Google Scholar] [CrossRef]
- Samtiya, M.; Matthews, K.R.; Dhewa, T.; Puniya, A.K. Antimicrobial resistance in the food chain: Trends, mechanisms, pathways, and possible regulation strategies. Foods 2022, 11, 2966. [Google Scholar] [CrossRef] [PubMed]
- Bezanson, G.S.; MacInnis, R.; Potter, G.; Hughes, T. Presence and potential for horizontal transfer of antibiotic resistance in oxidase-positive bacteria populating raw salad vegetables. Int. J. Food Microbiol. 2008, 127, 37–42. [Google Scholar] [CrossRef]
- Sultana, F.; Kamrunnahar; Afroz, H.; Jahan, A.; Fakruddin, M.; Datta, S. Multi-antibiotic resistant bacteria in frozen food (ready to cook food) of animal origin sold in Dhaka, Bangladesh. Asian Pac. J. Trop. Biomed. 2014, 4, S268–S271. [Google Scholar] [CrossRef]
- Patel, S.J.; Wellington, M.; Shah, R.M.; Ferreira, M.J. Antibiotic stewardship in food-producing animals: Challenges, progress, and opportunities. Clin. Ther. 2020, 42, 1649–1658. [Google Scholar] [CrossRef]
- Urban-Chmiel, R.; Marek, A.; Stepien-Pysniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic resistance in bacteria-A review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef]
- Ramatla, T.; Ngoma, L.; Adetunji, M.; Mwanza, M. Evaluation of antibiotic residues in raw meat using different analytical methods. Antibiotics 2017, 6, 34. [Google Scholar] [CrossRef]
- World Health Organization. WHO Guidelines on Use of Medically Important Antimicrobials in Food-Producing Animals. Available online: https://www.who.int/publications/i/item/9789241550130 (accessed on 31 October 2023).
- Food and Agriculture Organization of the United Nations; World Health Organization. CX/MRL 2-2021: Maximum Residue Limits (MRLs) and Risk Management Recommendations (RMRs) for Residues of Veterinary Drugs in Foods. Available online: https://www.fao.org/fao-who-codexalimentarius/codex-texts/maximum-residue-limits/en/ (accessed on 20 July 2023).
- Foodstuffs Cosmetics and Disinfectants Amendment Act 39 of 2007. Available online: https://www.gov.za/sites/default/files/gcis_document/201409/a39-07.pdf (accessed on 26 October 2023).
- Meat Safety Act 40 of 2000. Available online: https://www.gov.za/sites/default/files/gcis_document/201409/a40-000.pdf (accessed on 26 October 2023).
- EUCAST. EUCAST Breakpoint Tables for Interpretation of MICs and Zone Diameters (v. 13.1). 2023. Available online: https://www.eucast.org/eucast_news/news_singleview?tx_ttnews%5Btt_news%5D=518&cHash=2509b0db92646dffba041406dcc9f20c (accessed on 1 February 2023).
- Stellato, G.; La Storia, A.; De Filippis, F.; Borriello, G.; Villani, F.; Ercolini, D. Overlap of spoilage-associated microbiota between meat and the meat processing environment in small-scale and large-scale retail distributions. Appl. Environ. Microbiol. 2016, 82, 4045–4054. [Google Scholar] [CrossRef]
- Stellato, G.; Utter, D.R.; Voorhis, A.; De Angelis, M.; Eren, A.M.; Ercolini, D. A few Pseudomonas oligotypes dominate in the meat and dairy processing environment. Front. Microbiol. 2017, 8, 264. [Google Scholar] [CrossRef] [PubMed]
- Fuertes-Perez, S.; Hauschild, P.; Hilgarth, M.; Vogel, R.F. Biodiversity of Photobacterium spp. isolated from meats. Front. Microbiol. 2019, 10, 2399. [Google Scholar] [CrossRef] [PubMed]
- Lulietto, M.F.; Sechi, P.; Borgogni, E.; Cenci-Goga, B.T. Meat spoilage: A critical review of a neglected alteration due to ropy slime producing bacteria. Ital. J. Anim. Sci. 2016, 14, 4011. [Google Scholar] [CrossRef]
- Cha, M.H.; Kim, S.H.; Kim, S.; Lee, W.; Kwak, H.S.; Chi, Y.M.; Woo, G.J. Antimicrobial resistance profile of Acinetobacter spp. isolates from retail meat samples under Campylobacter-selective conditions. J. Microbiol. Biotechnol. 2021, 31, 733–739. [Google Scholar] [CrossRef]
- Schulz, L. Consumers Respond to Meat Price Differences. Available online: https://www.extension.iastate.edu/agdm/articles/schulz/SchApr22.html (accessed on 31 October 2023).
- Kwenda, S.; Allam, M.; Khumualo, Z.T.H.; Mtshali, S.; Mnyameni, F.; Ismail, A. Jekesa: An Automated Easy-to-Use Pipeline for Bacterial Whole Genome Typing. 2020. Available online: https://github.com/stanikae/jekesa (accessed on 3 April 2023).
- Krueger, F.; James, F.; Ewels, P.; Afyounian, E.; Schuster-Boeckler, B. FelixKrueger/TrimGalore: v0.6.7—DOI via Zenodo (0.6.7). Zenodo. 2021. Available online: https://zenodo.org/records/5127899 (accessed on 1 February 2023).
- Souvorov, A.; Agarwala, R.; Lipman, D.J. SKESA: Strategic k-mer extension for scrupulous assemblies. Genome Biol. 2018, 19, 153. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Jolley, K.A.; Maiden, M.C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010, 11, 595. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Allesoe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Lu, J.; Rincon, N.; Wood, D.E.; Breitwieser, F.P.; Pockrandt, C.; Langmead, B.; Salzberg, S.L.; Steinegger, M. Metagenome analysis using the Kraken software suite. Nat. Protoc. 2022, 17, 2815–2839. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- de Nies, L.; Lopes, S.; Busi, S.B.; Galata, V.; Heintz-Buschart, A.; Laczny, C.C.; May, P.; Wilmes, P. PathoFact: A pipeline for the prediction of virulence factors and antimicrobial resistance genes in metagenomic data. Microbiome 2021, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, J.; Gibson, M.K.; Franzosa, E.A.; Segata, N.; Dantas, G.; Huttenhower, C. High-specificity targeted functional profiling in microbial communities with ShortBRED. PLoS Comput. Biol. 2015, 11, e1004557. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.; Diarra, M.S.; Checkley, S.; Bohaychuk, V.; Masson, L. Characterization of antimicrobial resistance and virulence genes in Enterococcus spp. isolated from retail meats in Alberta, Canada. Int. J. Food Microbiol. 2012, 156, 222–230. [Google Scholar] [CrossRef]
- Tyson, G.H.; Nyirabahizi, E.; Crarey, E.; Kabera, C.; Lam, C.; Rice-Trujillo, C.; McDermott, P.F.; Tate, H. Prevalence and antimicrobial resistance of Enterococci isolated from retail meats in the United States, 2002 to 2014. Appl. Environ. Microbiol. 2018, 84, e01902-17. [Google Scholar] [CrossRef]
- de Jong, A.; Simjee, S.; Garch, F.E.; Moyaert, H.; Rose, M.; Youala, M.; Dry, M.; Group, E.S. Antimicrobial susceptibility of enterococci recovered from healthy cattle, pigs and chickens in nine EU countries (EASSA Study) to critically important antibiotics. Vet. Microbiol. 2018, 216, 168–175. [Google Scholar] [CrossRef]
- Amuasi, G.R.; Dsani, E.; Owusu-Nyantakyi, C.; Owusu, F.A.; Mohktar, Q.; Nilsson, P.; Adu, B.; Hendriksen, R.S.; Egyir, B. Enterococcus species: Insights into antimicrobial resistance and whole-genome features of isolates recovered from livestock and raw meat in Ghana. Front. Microbiol. 2023, 14, 1254896. [Google Scholar] [CrossRef] [PubMed]
- Novais, C.; Freitas, A.R.; Silveira, E.; Antunes, P.; Silva, R.; Coque, T.M.; Peixe, L. Spread of multidrug-resistant Enterococcus to animals and humans: An underestimated role for the pig farm environment. J. Antimicrob. Chemother. 2013, 68, 2746–2754. [Google Scholar] [CrossRef] [PubMed]
- Hart, W.S.; Heuzenroeder, M.W.; Barton, M.D. Antimicrobial resistance in Campylobacter spp., Escherichia coli and Enterococci associated with pigs in Australia. J. Vet. Med. B Infect. Dis. Vet. Public Health 2004, 51, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Strasheim, W.; Etter, E.M.C.; Lowe, M.; Perovic, O. Method to Assess Farm-Level Vaccine and Antibiotic Usage Utilizing Financial Documentation: A Pilot Study in a Commercial Pig Farm in South Africa From 2016 to 2018. Front. Vet. Sci. 2022, 9, 856729. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Klumper, U.; Shi, L.; Ye, L.; Li, M. From pig breeding environment to subsequently produced pork: Comparative analysis of antibiotic resistance denes and cacterial community composition. Front. Microbiol. 2019, 10, 43. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Hu, Z.; Li, Z.; Zhang, X.; Jia, C.; Li, T.; Dai, M.; Tan, C.; Xu, Z.; Wu, B.; et al. Antimicrobial resistance and population genomics of multidrug-resistant Escherichia coli in pig farms in mainland China. Nat. Commun. 2022, 13, 1116. [Google Scholar] [CrossRef] [PubMed]
- Bertels, F.; Silander, O.K.; Pachkov, M.; Rainey, P.B.; van Nimwegen, E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 2014, 31, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Kafil, H.S.; Mobarez, A.M.; Moghadam, M.F. Adhesion and virulence factor properties of Enterococci isolated from clinical samples in Iran. Indian J. Pathol. Microbiol. 2013, 56, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Reid, C.J.; Wyrsch, E.R.; Roy Chowdhury, P.; Zingali, T.; Liu, M.; Darling, A.E.; Chapman, T.A.; Djordjevic, S.P. Porcine commensal Escherichia coli: A reservoir for class 1 integrons associated with IS26. Microb. Genom. 2017, 3, e000143. [Google Scholar] [CrossRef]
- Reid, C.J.; DeMaere, M.Z.; Djordjevic, S.P. Australian porcine clonal complex 10 (CC10) Escherichia coli belong to multiple sublineages of a highly diverse global CC10 phylogeny. Microb. Genom. 2019, 5, e000225. [Google Scholar] [CrossRef] [PubMed]
- Perovic, O.; Singh-Moodley, A.; Lowe, M. In vitro activity of ceftolozane-tazobactam against Escherichia coli, Klebsiella pneumoniae and Pseudomonas aeruginosa obtained from blood cultures from sentinel public hospitals in South Africa. Antibiotics 2023, 12, 453. [Google Scholar] [CrossRef] [PubMed]
- Paveenkittiporn, W.; Kamjumphol, W.; Ungcharoen, R.; Kerdsin, A. Whole-genome sequencing of clinically isolated carbapenem-resistant Enterobacterales harboring mcr genes in Thailand, 2016–2019. Front Microbiol 2020, 11, 586368. [Google Scholar] [CrossRef]
- Rathnayake, I.U.; Hargreaves, M.; Huygens, F. Genotyping of Enterococcus faecalis and Enterococcus faecium isolates by use of a set of eight single nucleotide polymorphisms. J. Clin. Microbiol. 2011, 49, 367–372. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Freitas, A.R.; Novais, C.; Ruiz-Garbajosa, P.; Coque, T.M.; Peixe, L. Clonal expansion within clonal complex 2 and spread of vancomycin-resistant plasmids among different genetic lineages of Enterococcus faecalis from Portugal. J. Antimicrob. Chemother. 2009, 63, 1104–1111. [Google Scholar] [CrossRef]
- Kawalec, M.; Pietras, Z.; Danilowicz, E.; Jakubczak, A.; Gniadkowski, M.; Hryniewicz, W.; Willems, R.J. Clonal structure of Enterococcus faecalis isolated from Polish hospitals: Characterization of epidemic clones. J. Clin. Microbiol. 2007, 45, 147–153. [Google Scholar] [CrossRef]
- Sharon, B.M.; Arute, A.P.; Nguyen, A.; Tiwari, S.; Reddy Bonthu, S.S.; Hulyalkar, N.V.; Neugent, M.L.; Palacios Araya, D.; Dillon, N.A.; Zimmern, P.E.; et al. Genetic and functional enrichments associated with Enterococcus faecalis isolated from the urinary tract. mBio 2023, 14, e0251523. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, S.E.; Abia, A.L.K.; Amoako, D.G.; Perrett, K.; Bester, L.A.; Essack, S.Y. From farm-to-fork: E. coli from an intensive pig production system in South Africa shows high resistance to critically important antibiotics for human and animal use. Antibiotics 2021, 10, 178. [Google Scholar] [CrossRef]
- Barroga, T.R.M.; Morales, R.G.; Benigno, C.C.; Castro, S.J.M.; Caniban, M.M.; Cabullo, M.F.B.; Agunos, A.; de Balogh, K.; Dorado-Garcia, A. Antimicrobials used in backyard and commercial poultry and swine farms in the Philippines: A qualitative pilot study. Front. Vet. Sci. 2020, 7, 329. [Google Scholar] [CrossRef]



| Sample ID | E. coli (CFUs) | Salmonella spp. (CFUs) | Enterococci spp. (CFUs) | Campylobacter spp. (CFUs) |
|---|---|---|---|---|
| PC1-S1 | Absent | Absent | Absent | Absent |
| PC2-S2 | Absent | Absent | Absent | Absent |
| PC3-S3 | Absent | Absent | 16 * | Absent |
| PC4-S4 | Absent | Absent | 1 * | Absent |
| PC5-S5 | Absent | Absent | Absent | Absent |
| PC6-B1 | Absent | Absent | Absent | Absent |
| PC7-B2 | Absent | Absent | Absent | Absent |
| PC8-B3 | Absent | Absent | Absent | Absent |
| PC9-B4 | 20 * | Absent | Absent | Absent |
| PC10-B5 | Absent | Absent | 3 * | Absent |
| Antibiotic Class | Antibiotic | E. coli PC9-B4 (µg/mL) | MIC Interpretation # | E. faecalis PC3-S3 (µg/mL) | E. faecalis PC4-S4 (µg/mL) | E. faecalis PC10-B5 (µg/mL) | MIC Interpretation # |
|---|---|---|---|---|---|---|---|
| Aminoglycoside | Amikacin | ≤8 | S | 32 | 32 | 32 | NI * |
| Gentamicin | ≤2 | S | 4 | 4 | 4 | NI * | |
| Gentamicin synergy | NT | - | ≤500 | ≤500 | ≤500 | NI | |
| Streptomycin synergy | NT | - | ≤1000 | ≤1000 | ≤1000 | NI | |
| Tobramycin | ≤2 | S | ≤2/38 | ≤2/38 | ≤2/38 | NI * | |
| Beta-lactam (penicillins) | Ampicillin | ≤8 | S | 4 | 4 | 4 | S |
| Ampicillin/sulbactam | ≤8/4 | S | NT | NT | NT | - | |
| Amoxicillin/clavulanic acid | ≤8/4 | S | ≤4/2 | ≤4/2 | ≤4/2 | S | |
| Oxacillin | NT | - | >2 | >2 | >2 | NI | |
| Penicillin | NT | - | 8 | 8 | 8 | NI | |
| Piperacillin | ≤8 | S | NT | NT | NT | - | |
| Piperacillin/tazobactam | ≤8 | S | NT | NT | NT | - | |
| Beta-lactam (cephalosporins) | Cefepime | ≤1 | S | NT | NT | NT | - |
| Cefotaxime | ≤1 | S | NT | NT | NT | - | |
| Cefotaxime/clavulanic acid | ≤0.5 | NI | NT | NT | NT | - | |
| Cefoxitin | ≤8 | S | NT | NT | NT | - | |
| Cefuroxime | ≤4 | S | NT | NT | NT | - | |
| Ceftazidime | ≤1 | S | NT | NT | NT | - | |
| Ceftazidime/clavulanic acid | ≤0.25 | NI | NT | NT | NT | - | |
| Cephalothin | ≤8 | NI | NT | NT | NT | - | |
| Beta-lactam (carbapenems) | Doripenem | ≤1 | S | NT | NT | NT | - |
| Ertapenem | ≤0.5 | S | NT | NT | NT | - | |
| Imipenem | ≤1 | S | ≤4 | ≤4 | ≤4 | S | |
| Meropenem | ≤1 | S | NT | NT | NT | - | |
| Beta-lactam (monobactams) | Aztreonam | ≤1 | S | NT | NT | NT | - |
| Amphenicol | Chloramphenicol | ≤8 | S | ≤8 | ≤8 | ≤8 | NI |
| Cyclic lipopeptide | Daptomycin | NT | - | ≤1 | ≤1 | ≤1 | NI |
| Fluoroquinolone | Ciprofloxacin | ≤0.5 | S | ≤1 | ≤1 | ≤1 | S |
| Levofloxacin | ≤1 | S | ≤1 | ≤1 | ≤1 | S | |
| Moxifloxacin | NT | - | ≤256 | ≤256 | ≤256 | R | |
| Norfloxacin | ≤0.5 | S | ≤4 | ≤4 | ≤4 | NI | |
| Fusidane | Fusidic acid | NT | - | ≤2 | ≤2 | ≤2 | NI |
| Lincosamides | Clindamycin | NT | - | >2 | >2 | >2 | NI |
| Pristinamycin | NT | - | 2 | 2 | 2 | NI | |
| Macrolide | Erythromycin | NT | - | 1 | 1 | 1 | NI |
| Protein synthesis inhibitor | Mupirocin | >16 | NI | NT | NT | NT | - |
| Nitrofuran | Nitrofurantoin | ≤32 | S | ≤32 | ≤32 | ≤32 | S |
| Phosphonic acid | Fosfomycin | ≤32 | S | ≤32 | ≤32 | ≤32 | NI |
| Polymyxin | Colistin | ≤2 | S | NT | NT | NT | - |
| Rifamycin | Rifampin | NT | - | ≤0.5 | ≤0.5 | ≤0.5 | NI |
| Tetracycline | Minocycline | >8 | NI | ≤1 | ≤1 | ≤1 | NI |
| Tetracycline | >8 | NI | 8 | 8 | 8 | NI | |
| Tigecycline | ≤1 | R | NT | NT | NT | - | |
| Glycopeptide and lipoglycopeptide | Teicoplanin | NT | - | ≤1 | ≤1 | ≤1 | S |
| Vancomycin | NT | - | 2 | 2 | 2 | S | |
| Oxazolidinone | Linezolid | NT | - | 2 | 2 | 2 | S |
| Sulfonamide | Trimethoprim/sulfamethoxazole | >4/76 | R | NT | NT | NT | - |
| Sample ID | PC9-B4 * | PC3-S3 * | PC4-S4 * | PC10-B5 * | ||
|---|---|---|---|---|---|---|
| Organism | E. coli | E. faecalis | E. faecalis | E. faecalis | ||
| CH type | 11–54 | - | - | - | ||
| O type | O69 | - | - | - | ||
| H type | H32 | - | - | - | ||
| MLST | 10 ^ | 30 # | 30 # | 30 # | ||
| ARGs | Aminoglycoside | aadA1 | Y | - | - | - |
| Fluoroquinolone | gyrA | Y | - | - | - | |
| Lincosamide | isaA | - | Y | Y | Y | |
| Sulphonamide | sul2 | Y | - | - | - | |
| Tetracycline | tetB | Y | - | - | - | |
| tetM | - | Y | Y | Y | ||
| Trimethoprim | dfrA1 | Y | - | - | - | |
| VF genes | Adhesin | ace | - | Y | Y | Y |
| efaAfs | - | Y | Y | Y | ||
| Colicin | cba | Y | - | - | - | |
| cea | Y | - | - | - | ||
| cia | Y | - | - | - | ||
| cma | Y | - | - | - | ||
| Cytolysin toxin | cylA | - | Y | Y | Y | |
| cylL | - | Y | Y | Y | ||
| cylM | - | Y | Y | Y | ||
| Endocarditis and biofilm-associated pili genes | ebpA | - | Y | Y | Y | |
| ebpB | - | Y | Y | Y | ||
| Enterococcus faecalis leucine-rich protein A | elrA | - | Y | Y | Y | |
| Glutamate decarboxylase | gad | Y | - | - | - | |
| gelE | - | Y | Y | Y | ||
| Heat stable toxin | astA | Y | - | - | - | |
| Hyaluronidase | hylA | - | Y | Y | Y | |
| Increased serum survival | iss | Y | - | - | - | |
| Outer membrane protease | ompT | Y | - | - | - | |
| Plasmid-encoded catalase peroxidase | katP | Y | - | - | - | |
| Sex pheromone | cad | - | Y | Y | Y | |
| camE | - | Y | Y | Y | ||
| cCF10 | - | Y | Y | Y | ||
| cOB1 | - | Y | Y | Y | ||
| Tellurium ion resistance | terC | Y | - | - | - | |
| Thiol peroxidase | tpx | - | Y | Y | Y | |
| Outer membrane protein complement resistance | traT | Y | - | - | - | |
| Sortase | SrtA | - | Y | Y | Y | |
| Plasmids | IncB/O/K/Z | Y | - | - | - | |
| IncFII(pCoo) | Y | - | - | - | ||
| repUS43 | - | Y | Y | Y | ||
| repUS11 | - | Y | Y | Y | ||
| rep9a | - | Y | Y | Y | ||
| Antibiotic Class | Antibiotic | Sample ID | Acceptable Maximum Residue Level (µg/kg) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PC1-S1 * (µg/kg) | PC2-S2 * (µg/kg) | PC3-S3 * (µg/kg) | PC4-S4 * (µg/kg) | PC5-S5 * (µg/kg) | PC6-B1 * (µg/kg) | PC7-B2 * (µg/kg) | PC8-B3 * (µg/kg) | PC9-B4 * (µg/kg) | PC10-B5 * (µg/kg) | |||
| Fluoroquinolones | Ciprofloxacin | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | 100 # |
| Enrofloxacin | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | 100 # | |
| Norfloxacin | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | - | |
| Lincosamides | Lincomycin | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | 200 ^ |
| Macrolides | Tylosin | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | 100 ^ |
| Sulfonamides | Sulfadiazine | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | - |
| Sulfadimidine | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | 100 ^ | |
| Sulfamethoxazole | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | - | |
| Tetracyclines | Chlortetracycline | <50 | <50 | <50 | <50 | 71.5 | <50 | <50 | <50 | <50 | <50 | 200 ^ |
| Doxycycline | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | - | |
| Oxytetracycline | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | 200 ^ | |
| Tetracycline | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | 200 ^/600 # | |
| Pleuromutilin | Tiamulin | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | - |
| Diaminopyrimidines | Trimethoprim | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | - |
| Quindoxin | Olaquindox metabolite | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | - |
| Sample ID | Raw Paired-End Reads (n = NGS Reads) | Paired-End Reads after Host Removal (n = NGS Reads) | Paired-End Reads Mapped to Bacteria (n = NGS Reads) | Predicted ARG (n = Annotated ORF) | Predicted Secreted VF Genes (n = Annotated ORF) | Predicted Secreted Toxin Genes (n = Annotated ORF) |
|---|---|---|---|---|---|---|
| PC1-S1 | 6,559,325 | 761,775 | 249,495 | 47 | 86 | 24 |
| PC2-S2 | 7,276,334 | 372,205 | 4242 | 6 | 2 | 5 |
| PC3-S3 | 9,008,555 | 407,827 | 59,159 | 5 | 11 | 2 |
| PC4-S4 | 8,346,930 | 353,759 | 25,353 | 1 | 0 | 0 |
| PC5-S5 | 7,706,014 | 423,019 | 48,284 | 8 | 10 | 3 |
| PC6-B1 | 6,948,972 | 260,638 | 2460 | 0 | 1 | 0 |
| PC7-B2 | 6,318,244 | 388,517 | 47,285 | 7 | 12 | 3 |
| PC8-B3 | 6,811,831 | 400,456 | 71,941 | 11 | 15 | 3 |
| PC9-B4 | 8,083,308 | 336,636 | 2049 | 2 | 0 | 0 |
| PC10-B5 | 6,727,542 | 295,466 | 2043 | 2 | 1 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lowe, M.; Strasheim, W.; Chan, W.Y.; Perovic, O. Bacterial and Genetic Features of Raw Retail Pork Meat: Integrative Analysis of Antibiotic Susceptibility, Whole-Genome Sequencing, and Metagenomics. Antibiotics 2024, 13, 700. https://doi.org/10.3390/antibiotics13080700
Lowe M, Strasheim W, Chan WY, Perovic O. Bacterial and Genetic Features of Raw Retail Pork Meat: Integrative Analysis of Antibiotic Susceptibility, Whole-Genome Sequencing, and Metagenomics. Antibiotics. 2024; 13(8):700. https://doi.org/10.3390/antibiotics13080700
Chicago/Turabian StyleLowe, Michelle, Wilhelmina Strasheim, Wai Yin Chan, and Olga Perovic. 2024. "Bacterial and Genetic Features of Raw Retail Pork Meat: Integrative Analysis of Antibiotic Susceptibility, Whole-Genome Sequencing, and Metagenomics" Antibiotics 13, no. 8: 700. https://doi.org/10.3390/antibiotics13080700
APA StyleLowe, M., Strasheim, W., Chan, W. Y., & Perovic, O. (2024). Bacterial and Genetic Features of Raw Retail Pork Meat: Integrative Analysis of Antibiotic Susceptibility, Whole-Genome Sequencing, and Metagenomics. Antibiotics, 13(8), 700. https://doi.org/10.3390/antibiotics13080700






