Genomic Study of High-Risk Clones of Enterobacter hormaechei Collected from Tertiary Hospitals in the United Arab Emirates
Abstract
1. Introduction
2. Results
2.1. Genome-Based Species and ST Identification of Enterobacter spp.
2.2. Antimicrobial Susceptibility Profile of Enterobacter hormaechei
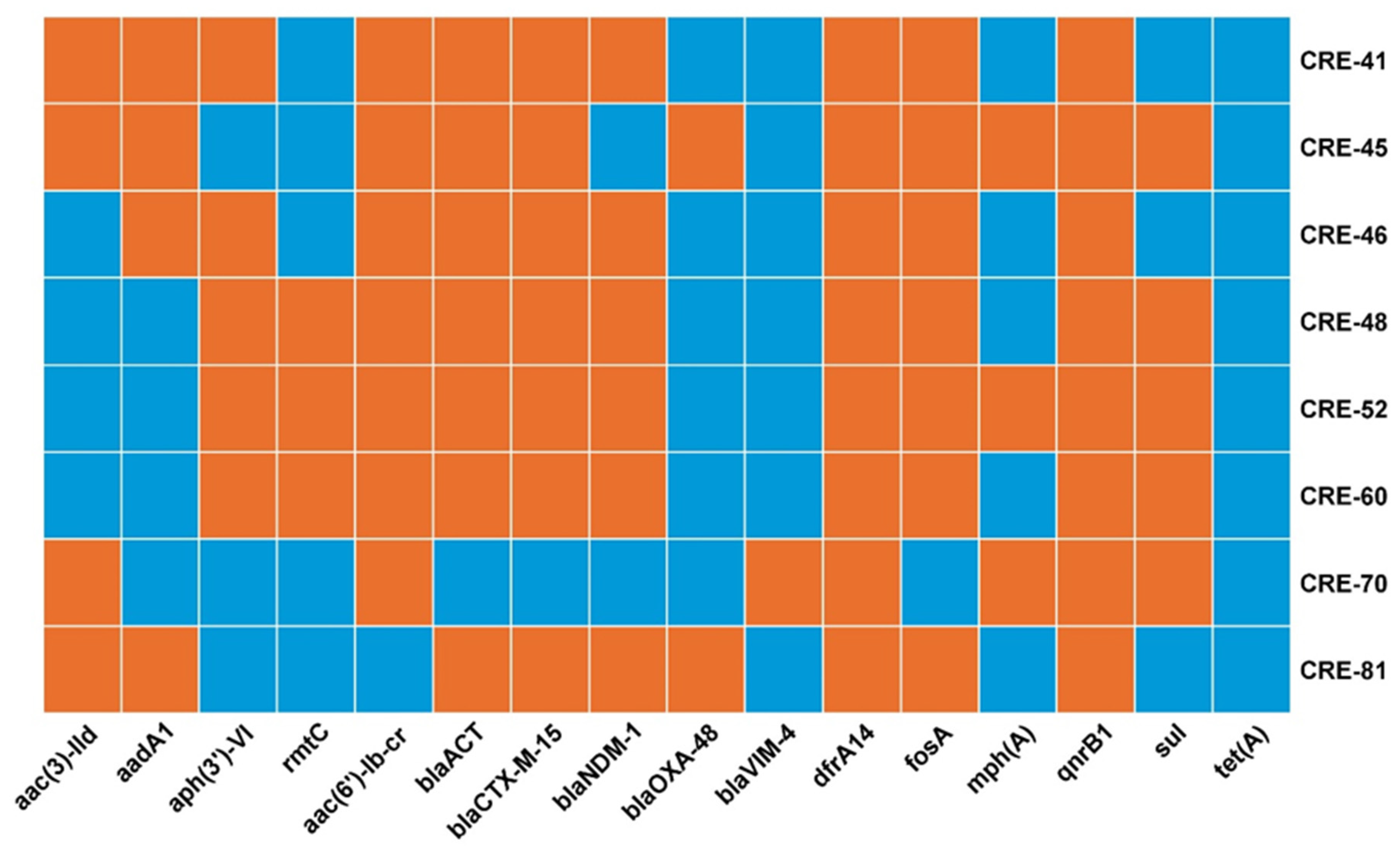
2.3. Antimicrobial Resistance Genes
2.4. Virulence Genes
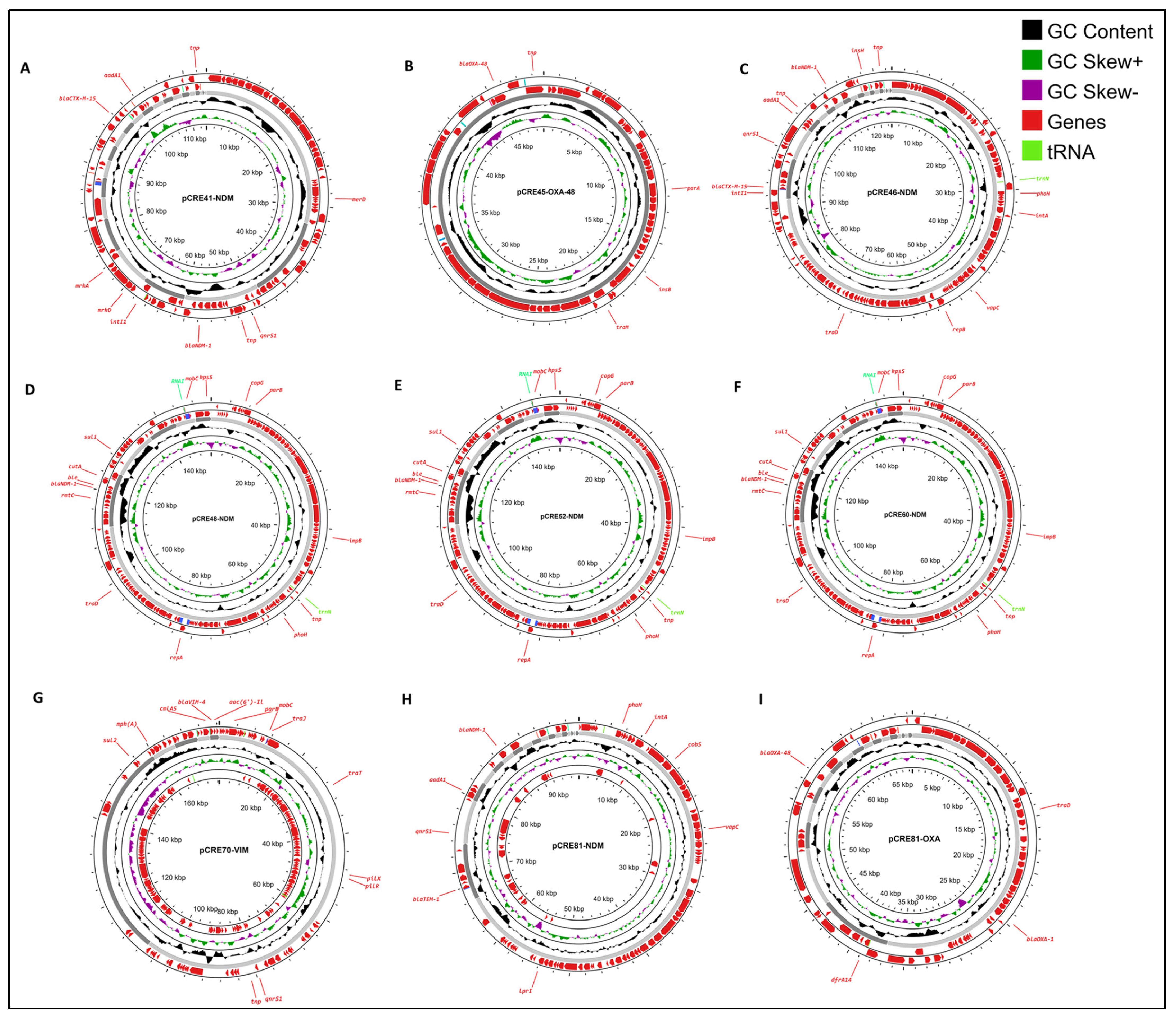
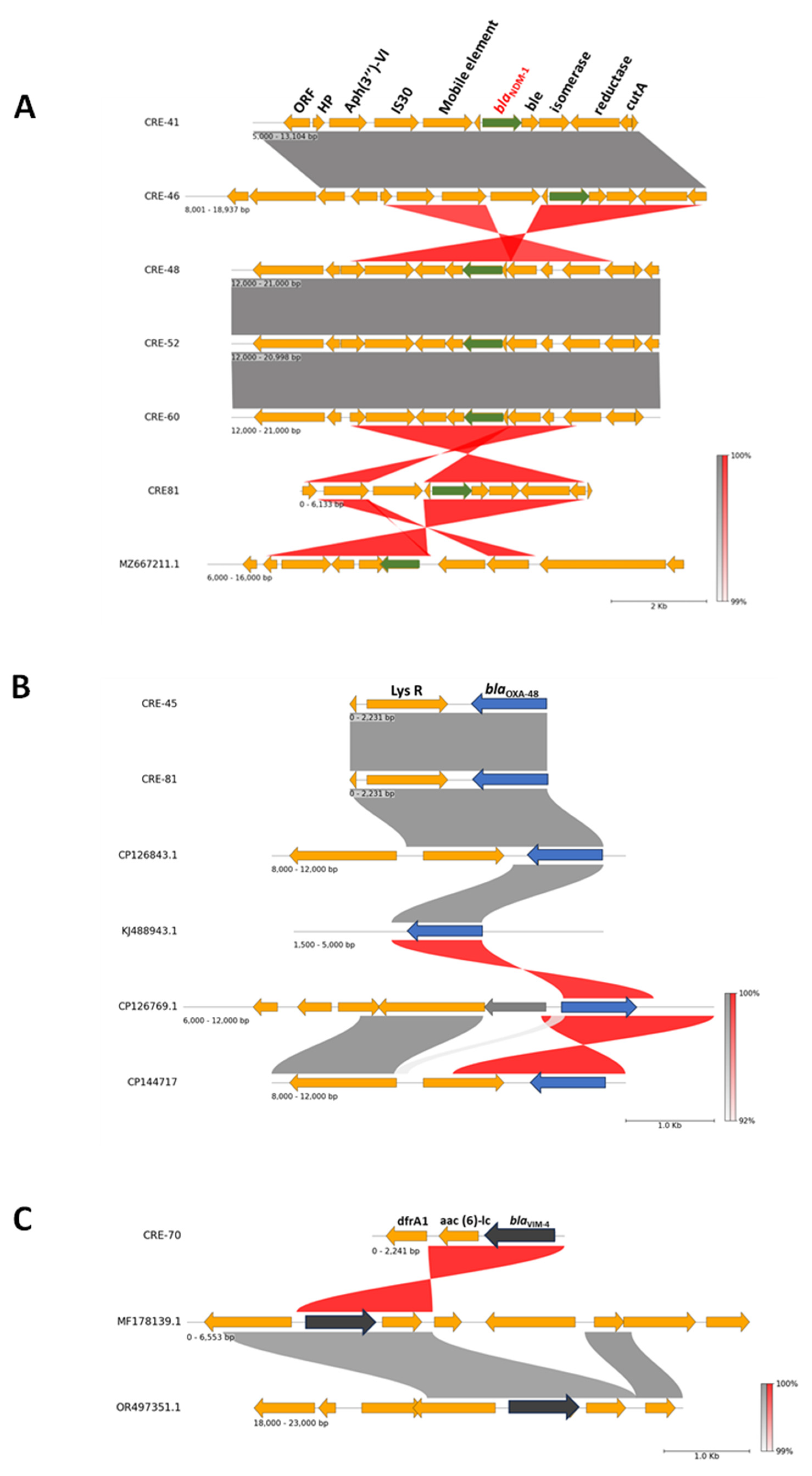
2.5. Plasmids and Genetic Environment of Carbapenemase Genes
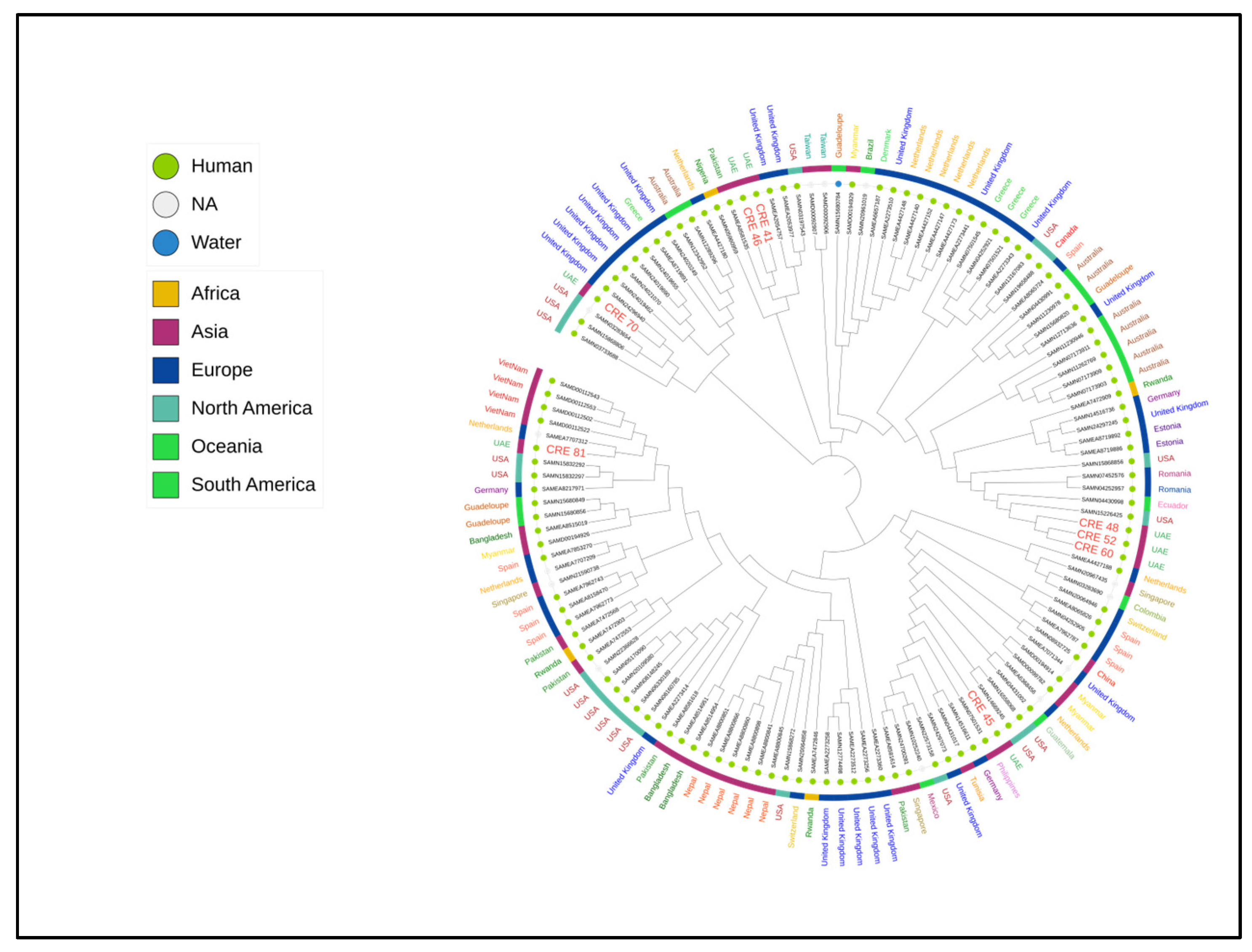
2.6. Phylogenetic Tree of Enterobacter hormaechei
3. Discussion
4. Materials and Methods
4.1. Strain Collection
4.2. Antibiotic Susceptibility Testing
4.3. Whole-Genome Sequencing
4.4. Resistance Gene Content, Virulence Genes, and Plasmid Analysis
4.5. Plasmid Analysis and Genetic Environment
4.6. Phylogenetic Tree
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- St John, A.; Perault, A.I.; Giacometti, S.I.; Sommerfield, A.G.; DuMont, A.L.; Lacey, K.A.; Zheng, X.; Sproch, J.; Petzold, C.; Dancel-Manning, K.; et al. Capsular Polysaccharide Is Essential for the Virulence of the Antimicrobial-Resistant Pathogen Enterobacter hormaechei. mBio 2023, 14, e0259022. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Zhu, M.; Li, Y.; Huang, D.; Wang, L.; Yan, C.; Zhang, L.; Dong, F.; Lu, J.; Lin, X.; et al. Whole-Genome Sequencing-Based Species Classification, Multilocus Sequence Typing, and Antimicrobial Resistance Mechanism Analysis of the Enterobacter cloacae Complex in Southern China. Microbiol. Spectr. 2022, 10, e0216022. [Google Scholar] [CrossRef] [PubMed]
- Wendel, A.F.; Peter, D.; Mattner, F.; Weiss, M.; Hoppenz, M.; Wolf, S.; Bader, B.; Peter, S.; Liese, J. Surveillance of Enterobacter cloacae Complex Colonization and Comparative Analysis of Different Typing Methods on a Neonatal Intensive Care Unit in Germany. Antimicrob. Resist. Infect. Control 2022, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, F.; Fang, Y.; Zhong, Q.; Xiao, Y.; Zheng, Y.; Zhu, J.; Zhao, C.; Cao, X.; Xiong, J.; et al. Clinical Characteristics, Prognosis and Treatment of Bloodstream Infections with Enterobacter Cloacae Complex in a Chinese Tertiary Hospital: A Retrospective Study. Infect. Drug Resist. 2024, 17, 1811–1825. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Huang, C.; Ye, X.; Jiang, H.; Zhang, R.; Hu, X.; Xu, D. Carbapenem-Resistant Enterobacter cloacae Causing Nosocomial Infections in Southwestern China: Molecular Epidemiology, Risk Factors, and Predictors of Mortality. Infect. Drug Resist. 2020, 13, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Zong, Z.; Feng, Y.; McNally, A. Carbapenem and Colistin Resistance in Enterobacter: Determinants and Clones. Trends Microbiol. 2021, 29, 473–476. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Wang, P.; Xu, T.; Zhou, Y.; Chen, R.; Zhu, H.; Zhou, K. Development of a One-Step Multiplex PCR Assay for Differential Detection of Four Species (Enterobacter cloacae, Enterobacter hormaechei, Enterobacter roggenkampii, and Enterobacter kobei) Belonging to Enterobacter cloacae Complex with Clinical Significance. Front. Cell. Infect. Microbiol. 2021, 11, 677089. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Q.; Huang, J.; Zhang, X.; Zhou, L.; Quan, J.; Wang, Z.; Zhou, H.; Li, R.; Tu, Y. Clonal Outbreak of NDM-1-Producing Enterobacter hormaechei Belonging to High-Risk International Clone ST78 with the Coexistence of tmexCD2-toprJ2 and mcr-9 in China. Int. J. Antimicrob. Agents 2023, 61, 106790. [Google Scholar] [CrossRef]
- Hawken, S.E.; Washer, L.L.; Williams, C.L.; Newton, D.W.; Snitkin, E.S. Genomic Investigation of a Putative Endoscope-Associated Carbapenem-Resistant Enterobacter cloacae Outbreak Reveals a Wide Diversity of Circulating Strains and Resistance Mutations. Clin. Infect. Dis. 2018, 66, 460–463. [Google Scholar] [CrossRef]
- Gomez-Simmonds, A.; Annavajhala, M.K.; Wang, Z.; Macesic, N.; Hu, Y.; Giddins, M.J.; O’Malley, A.; Toussaint, N.C.; Whittier, S.; Torres, V.J.; et al. Genomic and Geographic Context for the Evolution of High-Risk Carbapenem-Resistant Enterobacter cloacae Complex Clones ST171 and ST78. mBio 2018, 9, e00542-18. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; Matsumura, Y.; Adams, M.D.; Bradford, P.; Motyl, M.; Chen, L.; Kreiswirth, B.N.; Pitout, J.D.D. Genomic Epidemiology of Global Carbapenemase-Producing Enterobacter spp., 2008–2014. Emerg. Infect. Dis. 2018, 24, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Morhart, P.; Gerlach, R.G.; Kunz, C.; Held, J.; Valenza, G.; Wölfle, J.; Reutter, H.; Hanslik, G.J.; Fahlbusch, F.B. Application of Next-Generation Sequencing to Enterobacter hormaechei Subspecies Analysis during a Neonatal Intensive Care Unit Outbreak. Children 2023, 10, 1696. [Google Scholar] [CrossRef] [PubMed]
- Davin-Regli, A.; Pagès, J.M. Enterobacter aerogenes and Enterobacter cloacae; Versatile Bacterial Pathogens Confronting Antibiotic Treatment. Front. Microbiol. 2015, 6, 392. [Google Scholar] [CrossRef] [PubMed]
- Beyrouthy, R.; Barets, M.; Marion, E.; Dananché, C.; Dauwalder, O.; Robin, F.; Gauthier, L.; Jousset, A.; Dortet, L.; Guérin, F.; et al. Novel Enterobacter Lineage as Leading Cause of Nosocomial Outbreak Involving Carbapenemase-Producing Strains. Emerg. Infect. Dis. 2018, 24, 1505–1515. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, H.; Schmoldt, S.; Trülzsch, K.; Stumpf, A.; Bengsch, S.; Blankenstein, T.; Heesemann, J.; Roggenkamp, A. Nosocomial Urosepsis Caused by Enterobacter kobei with Aberrant Phenotype. Diagn. Microbiol. Infect. Dis. 2005, 53, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Wendel, A.F.; Meyer, S.; Deenen, R.; Köhrer, K.; Kolbe-Busch, S.; Pfeffer, K.; Willmann, M.; Kaasch, A.J.; MacKenzie, C.R. Long-Term, Low-Frequency Cluster of a German-Imipenemase-1-Producing Enterobacter hormaechei ssp. steigerwaltii ST89 in a Tertiary Care Hospital in Germany. Microb. Drug Resist. 2018, 24, 1305–1315. [Google Scholar] [CrossRef]
- Gou, J.J.; Liu, N.; Guo, L.H.; Xu, H.; Lv, T.; Yu, X.; Chen, Y.B.; Guo, X.B.; Rao, Y.T.; Zheng, B.W. Carbapenem-Resistant Enterobacter hormaechei ST1103 with IMP-26 Carbapenemase and ESBL Gene blaSHV-178. Infect. Drug Resist. 2020, 13, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Wang, J.; Zhang, P.; Wang, N.; Yuan, Q.; Shi, W.; Zhang, X.; Li, X.; Qu, T. Emergence of carbapenem-resistant Enterobacter hormaechei ST93 plasmids co-harbouring blaNDM-1, blaKPC-2, and mcr-9 in bloodstream infection. J. Glob. Antimicrob. Resist. 2023, 34, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Liu, Y.; Wang, R.; Wang, Q.; Jin, L.; Wang, H. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. EBioMedicine 2020, 51, 102599. [Google Scholar] [CrossRef]
- Bes, T.; Nagano, D.; Martins, R.; Marchi, A.P.; Perdigão-Neto, L.; Higashino, H.; Prado, G.; Guimaraes, T.; Levin, A.S.; Costa, S. Bloodstream Infections caused by Klebsiella pneumoniae and Serratia marcescens isolates co-harboring NDM-1 and KPC-2. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 57. [Google Scholar] [CrossRef] [PubMed]
- Rozales, F.P.; Lovison, O.A.; Magagnin, C.M.; Martins, A.S.; Crispim, M.N.; Zavascki, A.P.; Barth, A.L. Characteristics of Enterobacteriaceae Isolates Coharboring Distinct Carbapenemase Genes. Infect. Control. Hosp. Epidemiol. 2017, 38, 1123–1126. [Google Scholar] [CrossRef] [PubMed]
- Ham, D.C.; Mahon, G.; Bhaurla, S.K.; Horwich-Scholefield, S.; Klein, L.; Dotson, N.; Rasheed, J.K.; McAllister, G.; Stanton, R.A.; Karlsson, M.; et al. Gram-Negative Bacteria Harboring Multiple Carbapenemase Genes, United States, 2012–2019. Emerg. Infect. Dis. 2021, 27, 2475–2479. [Google Scholar] [CrossRef]
- Lumbreras-Iglesias, P.; de Toro, M.; Vázquez, X.; García-Carús, E.; Rodicio, M.R.; Fernández, J. High-risk international clones ST66, ST171 and ST78 of Enterobacter cloacae complex causing blood stream infections in Spain and carrying blaOXA-48 with or without mcr-9. J. Infect. Public Health 2023, 16, 272–279. [Google Scholar] [CrossRef]
- Chavda, K.D.; Chen, L.; Fouts, D.E.; Sutton, G.; Brinkac, L.; Jenkins, S.G.; Bonomo, R.A.; Adams, M.D.; Kreiswirth, B.N. Comprehensive Genome Analysis of Carbapenemase-Producing Enterobacter spp.: New Insights into Phylogeny, Population Structure, and Resistance Mechanisms. mBio 2016, 7, e02093-16. [Google Scholar] [CrossRef]
- Bonnin, R.A.; Girlich, D.; Jousset, A.B.; Emeraud, C.; Creton, E.; Gauthier, L.; Jové, T.; Dortet, L.; Naas, T. Genomic analysis of VIM-2-producing Enterobacter hormaechei subsp. steigerwaltii. Int. J. Antimicrob. Agents 2021, 57, 106285. [Google Scholar] [CrossRef] [PubMed]
- Jamal, W.; Rotimi, V.O.; Albert, M.J.; Khodakhast, F.; Nordmann, P.; Poirel, L. High prevalence of VIM-4 and NDM-1 metallo-β-lactamase among carbapenem-resistant Enterobacteriaceae. J. Med. Microbiol. 2013, 62 Pt 8, 1239–1244. [Google Scholar] [CrossRef] [PubMed]
- Sonnevend, Á.; Ghazawi, A.; Yahfoufi, N.; Al-Baloushi, A.; Hashmey, R.; Mathew, M.; Tariq, W.Z.; Pál, T. VIM-4 carbapenemase-producing Enterobacter cloacae in the United Arab Emirates. Clin. Microbiol. Infect. 2012, 18, E494–E496. [Google Scholar] [CrossRef]
- Soliman, A.M.; Maruyama, F.; Zarad, H.O.; Ota, A.; Nariya, H.; Shimamoto, T.; Shimamoto, T. Emergence of a Multidrug-Resistant Enterobacter hormaechei Clinical Isolate from Egypt Co-Harboring mcr-9 and blaVIM-4. Microorganisms 2020, 8, 595. [Google Scholar] [CrossRef]
- Yeh, T.K.; Lin, H.J.; Liu, P.Y.; Wang, J.H.; Hsueh, P.R. Antibiotic resistance in Enterobacter hormaechei. Int. J. Antimicrob. Agents 2022, 60, 106650. [Google Scholar] [CrossRef]
- Hong, Y.K.; Lee, J.Y.; Ko, K.S. Colistin resistance in Enterobacter spp. isolates in Korea. J. Microbiol. 2018, 56, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Khuntayaporn, P.; Thirapanmethee, K.; Chomnawang, M.T. An Update of Mobile Colistin Resistance in Non-Fermentative Gram-Negative Bacilli. Front. Cell. Infect. Microbiol. 2022, 12, 882236. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Cui, Y.; Quan, J.; Zhao, D.; Han, X.; Shi, Q.; Wang, Q.; Jiang, Y.; Du, X.; Li, X.; et al. High prevalence of colistin resistance and mcr-9/10 genes in Enterobacter spp. in a tertiary hospital over a decade. Int. J. Antimicrob. Agents 2022, 59, 106573. [Google Scholar] [CrossRef] [PubMed]
- Mondal, A.H.; Khare, K.; Saxena, P.; Debnath, P.; Mukhopadhyay, K.; Yadav, D. A Review on Colistin Resistance: An Antibiotic of Last Resort. Microorganisms 2024, 12, 772. [Google Scholar] [CrossRef] [PubMed]
- Izdebski, R.; Biedrzycka, M.; Urbanowicz, P.; Papierowska-Kozdój, W.; Dominiak, M.; Żabicka, D.; Gniadkowski, M. Multiple secondary outbreaks of NDM-producing Enterobacter hormaechei in the context of endemic NDM-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 2022, 77, 1561–1569. [Google Scholar] [CrossRef] [PubMed]
- Raun-Petersen, C.; Toft, A.; Nordestgaard, M.M.; Holm, A.; Overballe-Petersen, S.; Hammerum, A.M.; Hasman, H.; Justesen, U.S. Investigation of an Enterobacter hormaechei OXA-436 carbapenemase outbreak: When everything goes down the drain. Infect. Prev. Pract. 2022, 4, 100228. [Google Scholar] [CrossRef] [PubMed]
- Gartzonika, K.; Politi, L.; Mavroidi, A.; Tsantes, A.G.; Spanakis, N.; Priavali, E.; Vrioni, G.; Tsakris, A. High prevalence of clonally related ST182 NDM-1-producing Enterobacter cloacae complex clinical isolates in Greece. Int. J. Antimicrob. Agents 2023, 62, 106837. [Google Scholar] [CrossRef]
- Azam, M.W.; Zarrilli, R.; Khan, A.U. Updates on the Virulence Factors Produced by Multidrug-Resistant Enterobacterales and Strategies to Control Their Infections. Microorganisms 2023, 11, 1901. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; CLSI guideline M100; CLSI: Malvern, PA, USA, 2022. [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 14.0. 2024. Available online: http://www.eucast.org (accessed on 20 February 2024).
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2020, 72, 2764–2768. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Nash, J.H.E. MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef]
- Gardner, S.N.; Slezak, T.; Hall, B.G. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 2015, 31, 2877–2878. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
| Isolate | Species | Hospital | Sample Type | Sequence Type (ST) |
|---|---|---|---|---|
| CRE-41 | Enterobacter hormaechei subsp. Steigerwaltii | A | Wound swab | 124 |
| CRE-45 | Enterobacter hormaechei | B | Sputum | 182 |
| CRE-46 | Enterobacter hormaechei subsp. steigerwaltii | A | Tissue | 124 |
| CRE-48 | Enterobacter hormaechei | A | Sputum | 90 |
| CRE-52 | Enterobacter hormaechei | A | Sputum | 90 |
| CRE-60 | Enterobacter hormaechei | A | Sputum | 90 |
| CRE-70 | Enterobacter hormaechei subsp. Hormaechei | B | Sputum | 269 |
| CRE-81 | Enterobacter hormaechei subsp. Xiangfangensis | B | Urine | 171 |
| Isolate | PTZ | CTX | CAZ | C/A | C/T | CPM | IMI | MEM | AK | GM | CIP | TIG | COL | TS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CRE-41 | ≥128 | ≥64 | ≥64 | ≥16 | ≥32 | ≥32 | 8 | ≥16 | 16 | ≥16 | ≥4 | ≤0.5 | 8 | ≤20 |
| CRE-45 | ≥128 | ≥64 | ≥64 | 1 | ≥32 | ≥32 | 4 | 2 | 4 | ≥16 | ≥4 | ≥8 | ≤0.5 | ≥320 |
| CRE-46 | ≥128 | ≥64 | ≥64 | ≥16 | ≥32 | ≥32 | 8 | ≥16 | 16 | ≥16 | ≥4 | ≤0.5 | 8 | 40 |
| CRE-48 | ≥128 | ≥64 | ≥64 | ≥16 | ≥32 | ≥32 | 8 | ≥16 | 32 | ≥16 | ≥4 | ≤0.5 | 8 | ≥320 |
| CRE-52 | ≥128 | ≥64 | ≥64 | ≥16 | ≥32 | ≥32 | 8 | ≥16 | 32 | ≥16 | ≥4 | ≤0.5 | 8 | ≥320 |
| CRE-60 | ≥128 | ≥64 | ≥64 | ≥16 | ≥32 | ≥32 | ≥16 | ≥16 | 32 | ≥16 | ≥4 | ≤0.5 | 8 | ≥320 |
| CRE-70 | ≥128 | 32 | ≥64 | ≥16 | ≥32 | 2 | 8 | ≥16 | 16 | ≥16 | 1 | ≤0.5 | ≤0.5 | ≥320 |
| CRE-81 | ≥128 | ≥64 | ≥64 | ≥16 | ≥32 | ≥32 | ≥16 | ≥16 | 32 | ≥16 | ≥4 | ≤0.5 | ≤0.5 | ≤20 |
| VF Class | Related Genes | CRE45 | CRE41 | CRE46 | CRE48 | CRE52 | CRE60 | CRE70 | CRE81 |
|---|---|---|---|---|---|---|---|---|---|
| ST 182 | ST 124 | ST 124 | ST 90 | ST 90 | ST 90 | ST 269 | ST 171 | ||
| Adherence | |||||||||
| csgC | ||||||||
| papC | ||||||||
| fimA, fimC, fimD | ||||||||
| mrkA, mrkB, mrkD | ||||||||
| Autotransporter | ehaB | ||||||||
| Invasion | |||||||||
| ibeB | ||||||||
| cheB, cheW, cheR, cheY, motA | ||||||||
| Iron uptake | |||||||||
| iucA, iucB, iucC, iucD, iutA | ||||||||
| iroB, iroC, iroD, iroE, iroN | ||||||||
| irp1, irp2 | ||||||||
| - | ||||||||
| Secretion system (T6SS) | - | ||||||||
| Toxin (Cytotoxin, Shiga-toxin) | - | ||||||||
| Hemolysin | hlyA, hlyB, hlyC, hlyD | ||||||||
| Biofilm formation | adeG, pgaC | ||||||||
| Virulence factor MviM | MviM | ||||||||
| Virulence factor VirK | virK | ||||||||
| Immune evasion | gale, gtrA | ||||||||
| Serum resistance | - | ||||||||
| Virulence protein MsgA | msgA | ||||||||
| Isolate | Plasmid Size (bp) | Resistance Genes | Rep Type(s) | Relaxase Type(s) | Predicted Mobility |
|---|---|---|---|---|---|
| CRE41 | 115,921 | blaNDM1, blaCTX-M-15, qnrS1, aadA1 | IncFII | MOBF | conjugative |
| CRE45 | 45,454 | blaOXA-48 | IncL/M | - | non-mobilizable |
| CRE46 | 116,411 | blaNDM1, blaCTX-M-15, qnrS1, aadA1 | IncFII | MOBF | conjugative |
| CRE48 | 149,926 | blaNDM1, rmtC, sul1, dfrA14 | IncFII | MOBF | conjugative |
| CRE52 | 150,660 | blaNDM1, rmtC, sul1, dfrA14 | IncFII | MOBF | conjugative |
| CRE60 | 151,476 | blaNDM1, rmtC, sul1, dfrA14 | IncFII | MOBF | conjugative |
| CRE70 | 172,757 | blaVIM-4, mphA, sul2, cmlA5, aac(6′)-II | IncC | MOBH | conjugative |
| CRE81 | 95,149 | blaNDM-1, blaTEM-1, qnrS1, aadA1 | IncFII | MOBP | conjugative |
| 69,442 | blaOXA-48, blaOXA-1, dfrA14 | IncL/M | MOBP | conjugative |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghazawi, A.; Anes, F.; Mouftah, S.; Elbediwi, M.; Baig, A.; Alketbi, M.; Almazrouei, F.; Alhashmi, M.; Alzarooni, N.; Manzoor, A.; et al. Genomic Study of High-Risk Clones of Enterobacter hormaechei Collected from Tertiary Hospitals in the United Arab Emirates. Antibiotics 2024, 13, 592. https://doi.org/10.3390/antibiotics13070592
Ghazawi A, Anes F, Mouftah S, Elbediwi M, Baig A, Alketbi M, Almazrouei F, Alhashmi M, Alzarooni N, Manzoor A, et al. Genomic Study of High-Risk Clones of Enterobacter hormaechei Collected from Tertiary Hospitals in the United Arab Emirates. Antibiotics. 2024; 13(7):592. https://doi.org/10.3390/antibiotics13070592
Chicago/Turabian StyleGhazawi, Akela, Febin Anes, Shaimaa Mouftah, Mohammed Elbediwi, Awase Baig, Muna Alketbi, Fatema Almazrouei, Mariam Alhashmi, Norah Alzarooni, Ashrat Manzoor, and et al. 2024. "Genomic Study of High-Risk Clones of Enterobacter hormaechei Collected from Tertiary Hospitals in the United Arab Emirates" Antibiotics 13, no. 7: 592. https://doi.org/10.3390/antibiotics13070592
APA StyleGhazawi, A., Anes, F., Mouftah, S., Elbediwi, M., Baig, A., Alketbi, M., Almazrouei, F., Alhashmi, M., Alzarooni, N., Manzoor, A., Habib, I., Strepis, N., Nabi, A., & Khan, M. (2024). Genomic Study of High-Risk Clones of Enterobacter hormaechei Collected from Tertiary Hospitals in the United Arab Emirates. Antibiotics, 13(7), 592. https://doi.org/10.3390/antibiotics13070592







