Antimicrobial Resistance in Livestock: A Serious Threat to Public Health
Abstract
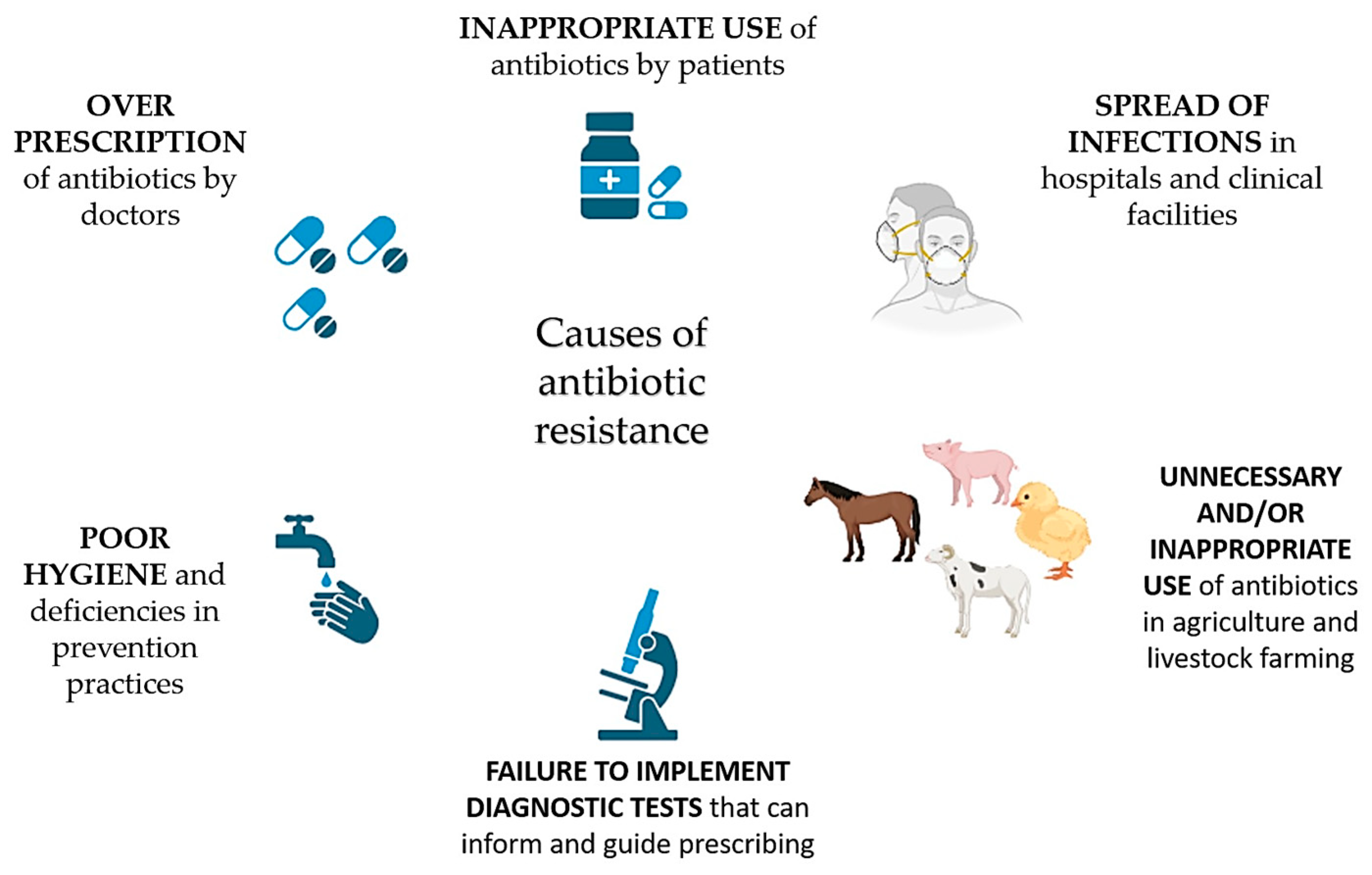
1. Introduction
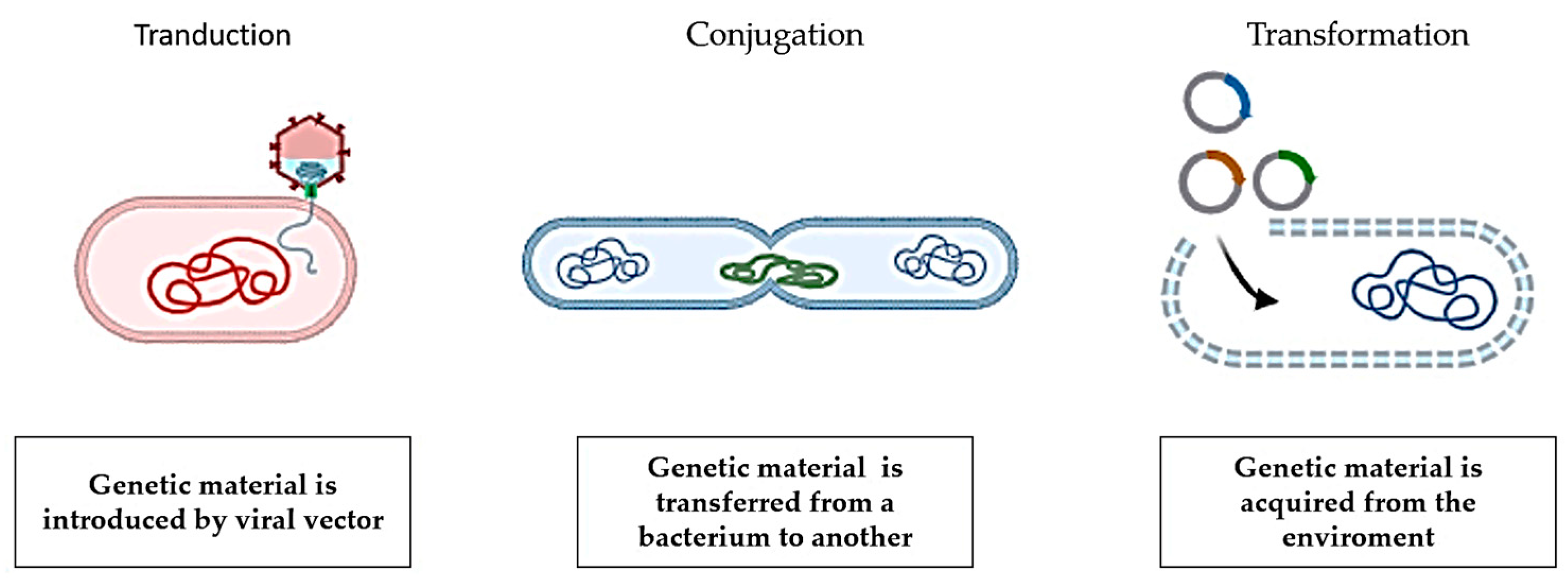
2. Antimicrobial Resistance Transmission
3. The Mechanisms of Antimicrobial Resistance
4. Relationship between Animal and Human Bacteria
5. Antimicrobial Resistance and Animal Breeding
6. Alternatives to Antibiotics
7. Actions to Counteract Antimicrobial Resistance
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Westblade, L.F.; Errington, J.; Dörr, T. Antibiotic tolerance. PLoS Pathog. 2020, 16, e1008892. [Google Scholar] [CrossRef] [PubMed]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Virulence Mech. Bact. Pathog. 2016, 481–511. [Google Scholar] [CrossRef]
- World Health Organization. The World is Running Out of Antibiotics, WHO Report Confirms. September 2017. Available online: https://www.who.int/news/item/20-09-2017-the-world-is-running-out-of-antibiotics-who-report-confirms (accessed on 17 May 2024).
- European Commission. Public Health. EU Action on Antimicrobial Resistance. Available online: https://ec.europa.eu/health/antimicrobial-resistance/eu-action-antimicrobial-resistance_en (accessed on 17 May 2024).
- McEwen, S.A.; Collignon, P.J. Antimicrobial resistance: A one health perspective. Antimicrob. Resist. Bact. Livest. Companion Anim. 2018, 521–547. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Farooqui, H.H.; Selvaraj, S.; Mehta, A.; Heymann, D.L. Community level antibiotic utilization in India and its comparison vis-à-vis European countries: Evidence from pharmaceutical sales data. PLoS ONE 2018, 13, e0204805. [Google Scholar] [CrossRef]
- Littmann, J.; Viens, A.M. The ethical significance of antimicrobial resistance. Public Health Ethics 2015, 8, 209–224. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.J.; Thottathil, S.E.; Newman, T.B. Antibiotics overuse in animal agriculture: A call to action for health care providers. Am. J. Public Health 2015, 105, 2409–2410. [Google Scholar] [CrossRef] [PubMed]
- Mackie, R.I.; Koike, S.; Krapac, I.; Chee-Sanford, J.; Maxwell, S.; Aminov, R.I. Tetracycline residues and tetracycline resistance genes in groundwater impacted by swine production facilities. Anim. Biotechnol. 2006, 17, 157–176. [Google Scholar] [CrossRef]
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef]
- Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; De Schaetzen, M.-A.; Van Huffel, X.; Imberechts, H.; Dierick, K. Antimicrobial resistance in the food chain: A review. Int. J. Environ. Res. Public Health 2013, 10, 2643–2669. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Florez-Cuadrado, D.; Moreno, M.A.; Ugarte-Ruíz, M.; Domínguez, L. Antimicrobial resistance in the food chain in the European Union. Adv. Food Nutr. Res. 2018, 86, 115–136. [Google Scholar]
- Stockwell, V.O.; Duffy, B. Use of antibiotics in plant agriculture. OIE Rev. Sci. Tech. 2012, 31, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.H.; Moore, L.S.P.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J.V. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Chaslus-Dancla, E. Use of antimicrobials in veterinary medicine and mechanisms of resistance. Vet. Res. 2001, 32, 201–225. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, H.; Van Nguyen, K.; Hara, G.L.; Gelband, H.; Laxminarayan, R.; Mouton, J.; Cars, O. Global survey of polymyxin use: A call for international guidelines. J. Glob. Antimicrob. Resist. 2013, 1, 131–134. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014; ISBN 9241564741. [Google Scholar]
- Schwarz, S.; Kehrenberg, C.; Walsh, T.R. Use of antimicrobial agents in veterinary medicine and food animal production. Int. J. Antimicrob. Agents 2001, 17, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Bacterial monopolists: The bundling and dissemination of antimicrobial resistance genes in gram-positive bacteria. Clin. Infect. Dis. 2000, 31, 762–769. [Google Scholar] [CrossRef]
- Aliabadi, F.S.; Lees, P. Antibiotic treatment for animals: Effect on bacterial population and dosage regimen optimisation. Int. J. Antimicrob. Agents 2000, 14, 307–313. [Google Scholar] [CrossRef]
- Magee, J.T. Antibiotic resistance and prescribing in the community. Rev. Res. Med. Microbiol. 2001, 12, 87–96. [Google Scholar] [CrossRef]
- Yoshida, H.; Bogaki, M.; Nakamura, M.; Nakamura, S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 1990, 34, 1271–1272. [Google Scholar] [CrossRef] [PubMed]
- Willmott, C.J.; Maxwell, A. A single point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob. Agents Chemother. 1993, 37, 126–127. [Google Scholar] [CrossRef] [PubMed]
- Strahilevitz, J.; Jacoby, G.A.; Hooper, D.C.; Robicsek, A. Plasmid-mediated quinolone resistance: A multifaceted threat. Clin. Microbiol. Rev. 2009, 22, 664–689. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Wang, R.; Liu, D.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Nesporova, K.; Valcek, A.; Papagiannitsis, C.; Kutilova, I.; Jamborova, I.; Davidova-Gerzova, L.; Bitar, I.; Hrabak, J.; Literak, I.; Dolejska, M. Multi-drug resistant plasmids with ESBL/Ampc and mcr-5.1 in paraguayan poultry farms: The linkage of antibiotic resistance and hatcheries. Microorganisms 2021, 9, 866. [Google Scholar] [CrossRef] [PubMed]
- Baucheron, S.; Tyler, S.; Boyd, D.; Mulvey, M.R.; Chaslus-Dancla, E.; Cloeckaert, A. AcrAB-TolC directs efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium DT104. Antimicrob. Agents Chemother. 2004, 48, 3729–3735. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.A.; Bavro, V.N.; Ricci, V.; Modi, N.; Cacciotto, P.; Kleinekathöfer, U.; Ruggerone, P.; Vargiu, A.V.; Baylay, A.J.; Smith, H.E. AcrB drug-binding pocket substitution confers clinically relevant resistance and altered substrate specificity. Proc. Natl. Acad. Sci. USA 2015, 112, 3511–3516. [Google Scholar] [CrossRef] [PubMed]
- Vidovic, S.; An, R.; Rendahl, A. Molecular and physiological characterization of fluoroquinolone-highly resistant Salmonella enteritidis strains. Front. Microbiol. 2019, 10, 446062. [Google Scholar] [CrossRef] [PubMed]
- Hartog, E.; Ben-Shalom, L.; Shachar, D.; Matthews, K.R.; Yaron, S. Regulation of marA, soxS, rob, acrAB and micF in Salmonella enterica serovar Typhimurium. Microbiol. Immunol. 2008, 52, 565–574. [Google Scholar] [CrossRef]
- Mcmurry, L.M.; Oethinger, M.; Levy, S.B. Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli. FEMS Microbiol. Lett. 1998, 166, 305–309. [Google Scholar] [CrossRef]
- Ruiz, C.; Levy, S.B. Regulation of acrAB expression by cellular metabolites in Escherichia coli. J. Antimicrob. Chemother. 2014, 69, 390–399. [Google Scholar] [CrossRef]
- Rosenberg, E.Y.; Bertenthal, D.; Nilles, M.L.; Bertrand, K.P.; Nikaido, H. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 2003, 48, 1609–1619. [Google Scholar] [CrossRef]
- Olliver, A.; Vallé, M.; Chaslus-Dancla, E.; Cloeckaert, A. Role of an acrR mutation in multidrug resistance of in vitro-selected fluoroquinolone-resistant mutants of Salmonella enterica serovar Typhimurium. FEMS Microbiol. Lett. 2004, 238, 267–272. [Google Scholar]
- Baucheron, S.; Le Hello, S.; Doublet, B.; Giraud, E.; Weill, F.-X.; Cloeckaert, A. ramR mutations affecting fluoroquinolone susceptibility in epidemic multidrug-resistant Salmonella enterica serovar Kentucky ST198. Front. Microbiol. 2013, 4, 213. [Google Scholar] [CrossRef]
- Roberts, M.C. Tetracycline resistance determinants: Mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 1996, 19, 1–24. [Google Scholar] [CrossRef]
- Liu, Y.; Tong, Z.; Shi, J.; Jia, Y.; Yang, K.; Wang, Z. Correlation between exogenous compounds and the horizontal transfer of plasmid-borne antibiotic resistance genes. Microorganisms 2020, 8, 1211. [Google Scholar] [CrossRef]
- Berends, B.R.; van den Bogaard, A.E.J.M.; Snijders, J.M.A. Human health hazards associated with the administration of antimicrobials to slaughter animals: Part II. An assessment of the risks of resistant bacteria in pigs and pork. Vet. Q. 2001, 23, 10–21. [Google Scholar] [CrossRef]
- Hall, R.M.; Collis, C.M. Mobile gene cassettes and integrons: Capture and spread of genes by site-specific recombination. Mol. Microbiol. 1995, 15, 593–600. [Google Scholar] [CrossRef]
- Bennett, P.M. Integrons and gene cassettes: A genetic construction kit for bacteria. J. Antimicrob. Chemother. 1999, 43, 1–4. [Google Scholar] [CrossRef]
- Von Wintersdorff, C.J.H.; Penders, J.; Van Niekerk, J.M.; Mills, N.D.; Majumder, S.; Van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef]
- Poirel, L.; Liard, A.; Rodriguez-Martinez, J.M.; Nordmann, P. Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. J. Antimicrob. Chemother. 2005, 56, 1118–1121. [Google Scholar] [CrossRef]
- Poirel, L.; Potron, A.; Nordmann, P. OXA-48-like carbapenemases: The phantom menace. J. Antimicrob. Chemother. 2012, 67, 1597–1606. [Google Scholar] [CrossRef]
- Cantón, R.; Coque, T.M. The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 2006, 9, 466–475. [Google Scholar] [CrossRef]
- Gillings, M.R. Integrons: Past, Present, and Future. Microbiol. Mol. Biol. Rev. 2014, 78, 257–277. [Google Scholar] [CrossRef]
- Abraham, S.; O’Dea, M.; Trott, D.J.; Abraham, R.J.; Hughes, D.; Pang, S.; McKew, G.; Cheong, E.Y.L.; Merlino, J.; Saputra, S. Isolation and plasmid characterization of carbapenemase (IMP-4) producing Salmonella enterica Typhimurium from cats. Sci. Rep. 2016, 6, 35527. [Google Scholar] [CrossRef]
- Partridge, S.R.; Tsafnat, G.; Coiera, E.; Iredell, J.R. Gene cassettes and cassette arrays in mobile resistance integrons: Review article. FEMS Microbiol. Rev. 2009, 33, 757–784. [Google Scholar] [CrossRef]
- Kumar, S.; Varela, M.F. Molecular mechanisms of bacterial resistance to antimicrobial agents. Chemotherapy 2013, 14, 522–534. [Google Scholar]
- Abraham, E.P.; Chain, E. An enzyme from bacteria able to destroy penicillin. 1940. Rev. Infect. Dis. 1988, 10, 677–678. [Google Scholar]
- Rammelkamp, C.H.; Maxon, T. Resistance of Staphylococcus aureus to the action of penicillin. Proc. Soc. Exp. Biol. Med. 1942, 51, 386–389. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum β-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef] [PubMed]
- Sirot, J.; Chanal, C.; Petit, A.; Sirot, D.; Labia, R.; Gerbaud, G. Klebsiella pneumoniae and other Enterobacteriaceae producing novel plasmid-mediated β-lactamases markedly active against third-generation cephalosporins: Epidemiologic studies. Clin. Infect. Dis. 1988, 10, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Abraham, E.P.; Newton, G.G.F. The structure of cephalosporin C. Biochem. J. 1961, 79, 377. [Google Scholar] [CrossRef] [PubMed]
- González-Candelas, F.; Comas, I.; Martínez, J.L.; Galán, J.C.; Baquero, F. The evolution of antibiotic resistance. In Genetics and Evolution of Infectious Disease; Elsevier: Amsterdam, The Netherlands, 2011; pp. 305–337. [Google Scholar]
- Morar, M.; Pengelly, K.; Koteva, K.; Wright, G.D. Mechanism and diversity of the erythromycin esterase family of enzymes. Biochemistry 2012, 51, 1740–1751. [Google Scholar] [CrossRef] [PubMed]
- Golkar, T.; Zieliński, M.; Berghuis, A.M. Look and outlook on enzyme-mediated macrolide resistance. Front. Microbiol. 2018, 9, 404144. [Google Scholar] [CrossRef] [PubMed]
- Castañeda-García, A.; Blázquez, J.; Rodríguez-Rojas, A. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics 2013, 2, 217–236. [Google Scholar] [CrossRef]
- Silver, L.L. Fosfomycin: Mechanism and resistance. Cold Spring Harb. Perspect. Med. 2017, 7, a025262. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Bacterial resistance to antibiotics: Enzymatic degradation and modification. Adv. Drug Deliv. Rev. 2005, 57, 1451–1470. [Google Scholar] [CrossRef] [PubMed]
- Garneau-Tsodikova, S.; Labby, K.J. Mechanisms of resistance to aminoglycoside antibiotics: Overview and perspectives. Medchemcomm 2016, 7, 11–27. [Google Scholar] [CrossRef]
- Baysarowich, J.; Koteva, K.; Hughes, D.W.; Ejim, L.; Griffiths, E.; Zhang, K.; Junop, M.; Wright, G.D. Rifamycin antibiotic resistance by ADP-ribosylation: Structure and diversity of Arr. Proc. Natl. Acad. Sci. USA 2008, 105, 4886–4891. [Google Scholar] [CrossRef]
- Shaw, W. V Chloramphenicol acetyltransferase: Enzymology and molecular biology. Crit. Rev. Biochem. 1983, 14, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Weisblum, B. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 1995, 39, 577–585. [Google Scholar] [CrossRef]
- Vannuffel, P.; Di Giambattista, M.; Morgan, E.A.; Cocito, C. Identification of a single base change in ribosomal RNA leading to erythromycin resistance. J. Biol. Chem. 1992, 267, 8377–8382. [Google Scholar] [CrossRef]
- Ross, J.I.; Eady, E.A.; Cove, J.H.; Jones, C.E.; Ratyal, A.H.; Miller, Y.W.; Vyakrnam, S.; Cunliffe, W.J. Clinical resistance to erythromycin and clindamycin in cutaneous propionibacteria isolated from acne patients is associated with mutations in 23S rRNA. Antimicrob. Agents Chemother. 1997, 41, 1162–1165. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.E.; Taylor, D.E. Site-specific mutations in the 23S rRNA gene of Helicobacter pylori confer two types of resistance to macrolide-lincosamide-streptogramin B antibiotics. Antimicrob. Agents Chemother. 1998, 42, 1952–1958. [Google Scholar] [CrossRef] [PubMed]
- Canu, A.; Malbruny, B.; Coquemont, M.; Davies, T.A.; Appelbaum, P.C.; Leclercq, R. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2002, 46, 125–131. [Google Scholar] [CrossRef]
- Recht, M.I.; Douthwaite, S.; Dahlquist, K.D.; Puglisi, J.D. Effect of mutations in the A site of 16 S rRNA on aminoglycoside antibiotic-ribosome interaction. J. Mol. Biol. 1999, 286, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Recht, M.I.; Puglisi, J.D. Aminoglycoside resistance with homogeneous and heterogeneous populations of antibiotic-resistant ribosomes. Antimicrob. Agents Chemother. 2001, 45, 2414–2419. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Katsukawa, C.; Tamaru, A.K.I.; Abe, C.; Makino, M.; Mizuguchi, Y.; Taniguchi, H. Detection of kanamycin-resistant Mycobacterium tuberculosis by identifying mutations in the 16S rRNA gene. J. Clin. Microbiol. 1998, 36, 1220–1225. [Google Scholar] [CrossRef]
- Shi, Y. Common folds and transport mechanisms of secondary active transporters. Annu. Rev. Biophys. 2013, 42, 51–72. [Google Scholar] [CrossRef]
- West, I.C. Energy coupling in secondary active transport. Biochim. Biophys. Acta (BBA)-Rev. Biomembr. 1980, 604, 91–126. [Google Scholar]
- Davidson, A.L.; Maloney, P.C. ABC transporters: How small machines do a big job. Trends Microbiol. 2007, 15, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Stieger, B.; Higgins, C.F. Twenty years of ATP-binding cassette (ABC) transporters. Pflügers Arch. J. Physiol. 2007, 453, 543. [Google Scholar] [CrossRef] [PubMed]
- Holland, I.B. Rise and rise of the ABC transporter families. Res. Microbiol. 2019, 170, 304–320. [Google Scholar] [CrossRef] [PubMed]
- Krämer, R. Functional principles of solute transport systems: Concepts and perspectives. Biochim. Biophys. Acta (BBA)-Bioenerg. 1994, 1185, 1–34. [Google Scholar] [CrossRef]
- Poolman, B.; Konings, W.N. Secondary solute transport in bacteria. Biochim. Biophys. Acta (BBA)-Bioenerg. 1993, 1183, 5–39. [Google Scholar] [CrossRef]
- Paulsen, I.T.; Brown, M.H.; Skurray, R.A. Proton-dependent multidrug efflux systems. Microbiol. Rev. 1996, 60, 575–608. [Google Scholar] [CrossRef] [PubMed]
- Jack, D.L.; Yang, N.M.; Saier, M.H. The drug/metabolite transporter superfamily. Eur. J. Biochem. 2001, 268, 3620–3639. [Google Scholar] [CrossRef]
- Hvorup, R.N.; Winnen, B.; Chang, A.B.; Jiang, Y.; Zhou, X.F.; Saier, M.H. The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur. J. Biochem. 2003, 270, 799–813. [Google Scholar] [CrossRef]
- Hassan, K.A.; Liu, Q.; Elbourne, L.D.H.; Ahmad, I.; Sharples, D.; Naidu, V.; Chan, C.L.; Li, L.; Harborne, S.P.D.; Pokhrel, A.; et al. Pacing across the membrane: The novel PACE family of efflux pumps is widespread in Gram-negative pathogens. Res. Microbiol. 2018, 169, 450–454. [Google Scholar] [CrossRef]
- Tseng, T.T.; Gratwick, K.S.; Kollman, J.; Park, D.; Nies, D.H.; Goffeau, A.; Saier, M.H. The RND permease superfamily: An ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1999, 1, 107–125. [Google Scholar]
- Kuroda, T.; Tsuchiya, T. Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta—Proteins Proteom. 2009, 1794, 763–768. [Google Scholar] [CrossRef]
- McDaniel, C.J.; Cardwell, D.M.; Moeller, R.B.; Gray, G.C. Humans and cattle: A review of bovine zoonoses. Vector-Borne Zoonotic Dis. 2014, 14, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Meijs, A.P.; Gijsbers, E.F.; Hengeveld, P.D.; Dierikx, C.M.; de Greeff, S.C.; van Duijkeren, E. ESBL/pAmpC-producing Escherichia coli and Klebsiella pneumoniae carriage among veterinary healthcare workers in the Netherlands. Antimicrob. Resist. Infect. Control 2021, 10, 147. [Google Scholar] [CrossRef] [PubMed]
- Nadimpalli, M.L.; Stewart, J.R.; Pierce, E.; Pisanic, N.; Love, D.C.; Hall, D.; Larsen, J.; Carroll, K.C.; Tekle, T.; Perl, T.M.; et al. Face mask use and persistence of livestock-associated staphylococcus aureus nasal carriage among industrial hog operation workers and household contacts, USA. Environ. Health Perspect. 2018, 126, 127005. [Google Scholar] [CrossRef]
- Nadimpalli, M.; Stewart, J.R.; Pierce, E.; Pisanic, N.; Love, D.C.; Hall, D.; Larsen, J.; Carroll, K.C.; Tekle, T.; Perl, T.M.; et al. Livestock-associated, antibiotic-resistant Staphylococcus aureus nasal carriage and recent skin and soft tissue infection among industrial HOG operation workers. PLoS ONE 2016, 11, e0165713. [Google Scholar] [CrossRef]
- Jackson, J.; Villarroel, A. A Survey of The Risk of Zoonoses for Veterinarians. Zoonoses Public Health 2012, 59, 193–201. [Google Scholar] [CrossRef]
- Davis, G.S.; Waits, K.; Nordstrom, L.; Weaver, B.; Aziz, M.; Gauld, L.; Grande, H.; Bigler, R.; Horwinski, J.; Porter, S. Intermingled Klebsiella pneumoniae populations between retail meats and human urinary tract infections. Clin. Infect. Dis. 2015, 61, 892–899. [Google Scholar] [CrossRef] [PubMed]
- Price, L.B.; Stegger, M.; Hasman, H.; Aziz, M.; Larsen, J.; Andersen, P.S.; Pearson, T.; Waters, A.E.; Foster, J.T.; Schupp, J. Staphylococcus aureus CC398: Host adaptation and emergence of methicillin resistance in livestock. MBio 2012, 3, 10–1128. [Google Scholar] [CrossRef]
- Ward, M.J.; Gibbons, C.L.; McAdam, P.R.; van Bunnik, B.A.D.; Girvan, E.K.; Edwards, G.F.; Fitzgerald, J.R.; Woolhouse, M.E.J. Time-scaled evolutionary analysis of the transmission and antibiotic resistance dynamics of Staphylococcus aureus clonal complex 398. Appl. Environ. Microbiol. 2014, 80, 7275–7282. [Google Scholar] [CrossRef]
- Levy, S.B.; Fitzgerald, G.B.; Macone, A.B. Spread of antibiotic-resistant plasmids from chicken to chicken and from chicken to man. Nature 1976, 260, 40–42. [Google Scholar] [CrossRef] [PubMed]
- Viñes, J.; Cuscó, A.; Napp, S.; Alvarez, J.; Saez-Llorente, J.L.; Rosàs-Rodoreda, M.; Francino, O.; Migura-Garcia, L. Transmission of similar mcr-1 carrying plasmids among different Escherichia coli lineages isolated from livestock and the farmer. Antibiotics 2021, 10, 313. [Google Scholar] [CrossRef] [PubMed]
- Vidovic, S.; Korber, D.R. Prevalence of Escherichia coli O157 in Saskatchewan cattle: Characterization of isolates by using random amplified polymorphic DNA PCR, antibiotic resistance profiles, and pathogenicity determinants. Appl. Environ. Microbiol. 2006, 72, 4347–4355. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Cao, M.; Wang, W.; Zhang, L.; Ma, T.; Liu, G.; Zhang, Y.; Shang, Z.; Chen, X.; Shi, Y. Exploring the prevalence and distribution patterns of antibiotic resistance genes in bovine gut microbiota using a metagenomic approach. Microb. Drug Resist. 2021, 27, 980–990. [Google Scholar] [CrossRef]
- Fournier, C.; Nordmann, P.; Pittet, O.; Poirel, L. Does an antibiotic stewardship applied in a pig farm lead to low ESBL prevalence? Antibiotics 2021, 10, 574. [Google Scholar] [CrossRef] [PubMed]
- Thanner, S.; Drissner, D.; Walsh, F. Antimicrobial resistance in agriculture. MBio 2016, 7, e02227-15. [Google Scholar] [CrossRef] [PubMed]
- Swann, M.M.; Blaxter, K.L.; Field, H.I.; Howie, J.W.; Lucas, I.A.M.; Millar, E.L.M.; Murdoch, J.C.; Parsons, J.H.; White, E.G. Report to the Joint Committee on the Use of Antibiotics in Animal Husbandry and Veterinary Medicine; Her Majesty’s Stationary Office: London, UK, 1969; Volume 4190. [Google Scholar]
- Castanon, J.I.R. History of the use of antibiotic as growth promoters in European poultry feeds. Poult. Sci. 2007, 86, 2466–2471. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.L.; Caffrey, N.P.; Nóbrega, D.B.; Cork, S.C.; Ronksley, P.E.; Barkema, H.W.; Polachek, A.J.; Ganshorn, H.; Sharma, N.; Kellner, J.D.; et al. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: A systematic review and meta-analysis. Lancet Planet. Health 2017, 1, e316–e327. [Google Scholar] [CrossRef]
- McEwen, S.A.; Angulo, F.J.; Collignon, P.J.; Conly, J.M. Unintended consequences associated with national-level restrictions on antimicrobial use in food-producing animals. Lancet Planet. Health 2018, 2, e279–e282. [Google Scholar] [CrossRef]
- The World Health Organization. Impacts of Antimicrobial Growth Promoter Termination in Denmark. The WHO International Review Panel’s Evaluation of the Termination of the Use of Antimicrobial Growth Promoters in Denmark: Foulum, Denmark 6–9 November 2002. Available online: https://apps.who.int/iris/handle/10665/68357 (accessed on 20 May 2024).
- Wierup, M. The Swedish experience of the 1986 year ban of antimicrobial growth promoters, with special reference to animal health, disease prevention, productivity, and usage of antimicrobials. Microb. Drug Resist. 2001, 7, 183–190. [Google Scholar] [CrossRef]
- Langlois, B.E.; Dawson, K.A.; Leak, I.; Aaron, D.K. Effect of age and housing location on antibiotic resistance of fecal coliforms from pigs in a non-antibiotic-exposed herd. Appl. Environ. Microbiol. 1988, 54, 1341–1344. [Google Scholar] [CrossRef] [PubMed]
- Mevius, D.J.; Hartman, E.G. In vitro-activiteit van 12 veterinair gebruikte antibacteriële middelen tegen Mannheimia haemolytica en Pasteurella multocida geïsoleerd uit kalveren in Nederland. Tijdschr. Diergeneeskd. 2000, 125, 147–152. [Google Scholar] [PubMed]
- Berge, A.C.B.; Atwill, E.R.; Sischo, W.M. Assessing dynamics of antibiotic resistance in faecal Escherichia coli and in young calves using cluster analysis techniques. In Proceedings of the Society for Veterinary Epidemiology and Preventive Medicine, Proceedings, Noordwijkerhout, The Netherlands, 28–30 March 2001. [Google Scholar]
- Walson, J.L.; Marshall, B.; Pokhrel, B.M.; Kafle, K.K.; Levy, S.B. Carriage of antibiotic-resistant fecal bacteria in Nepal reflects proximity to Kathmandu. J. Infect. Dis. 2001, 184, 1163–1169. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Levine, U.Y.; Looft, T.; Bandrick, M.; Casey, T.A. Treatment, promotion, commotion: Antibiotic alternatives in food-producing animals. Trends Microbiol. 2013, 21, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Hagens, S.; Loessner, M.J. Application of bacteriophages for detection and control of foodborne pathogens. Appl. Microbiol. Biotechnol. 2007, 76, 513–519. [Google Scholar] [CrossRef] [PubMed]
- PhageGuard. Available online: https://phageguard.com/ (accessed on 20 May 2024).
- Soni, K.A.; Nannapaneni, R. Removal of listeria monocytogenes biofilms with bacteriophage P100. J. Food Prot. 2010, 73, 1519. [Google Scholar] [CrossRef]
- Iacumin, L.; Manzano, M.; Comi, G. Phage inactivation of listeria monocytogenes on san daniele dry-cured ham and elimination of biofilms from equipment and working environments. Microorganisms 2016, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Sadekuzzaman, M.; Yang, S.; Mizan, M.F.R.; Kim, H.S.; Ha, S. Do Effectiveness of a phage cocktail as a biocontrol agent against L. monocytogenes biofilms. Food Control 2017, 78, 256–263. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C.; Ross, P.R. Bacteriocins: Developing innate immunity for food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef]
- Breukink, E.; de Kruijff, B. Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 2006, 5, 321–323. [Google Scholar] [CrossRef]
- Machaidze, G.; Seelig, J. Specific Binding of Cinnamycin (Ro 09-0198) to Phosphatidylethanolamine. Comparison between Micellar and Membrane Environments. Biochemistry 2003, 42, 12570–12576. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.C.G.; Silva, S.P.M.; Ribeiro, S.C. Application of bacteriocins and protective cultures in dairy food preservation. Front. Microbiol. 2018, 9, 345996. [Google Scholar] [CrossRef] [PubMed]
- Gómez, N.C.; Ramiro, J.M.P.; Quecan, B.X.V.; de Melo Franco, B.D.G. Use of potential probiotic lactic acid bacteria (LAB) biofilms for the control of Listeria monocytogenes, Salmonella Typhimurium, and Escherichia coli O157: H7 biofilms formation. Front. Microbiol. 2016, 7, 863. [Google Scholar] [CrossRef] [PubMed]
- Minei, C.C.; Gomes, B.C.; Ratti, R.P.; D’Angelis, C.E.M.; De Martinis, E.C.P. Influence of peroxyacetic acid and nisin and coculture with Enterococcus faecium on Listeria monocytogenes biofilm formation. J. Food Prot. 2008, 71, 634–638. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of lactic acid bacteria: Extending the family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef] [PubMed]
- Bak, H.; Rathkjen, P.H. Reduced use of antimicrobials after vaccination of pigs against porcine proliferative enteropathy in a Danish SPF herd. Acta Vet. Scand. 2009, 51, 1. [Google Scholar] [CrossRef] [PubMed]
- Lyall, J.; Irvine, R.M.; Sherman, A.; McKinley, T.J.; Núñez, A.; Purdie, A.; Outtrim, L.; Brown, I.H.; Rolleston-Smith, G.; Sang, H. Suppression of avian influenza transmission in genetically modified chickens. Science 2011, 331, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Harrison, E.M.; Ba, X.; Coll, F.; Blane, B.; Restif, O.; Carvell, H.; Köser, C.U.; Jamrozy, D.; Reuter, S.; Lovering, A.; et al. Genomic identification of cryptic susceptibility to penicillins and β-lactamase inhibitors in methicillin-resistant Staphylococcus aureus. Nat. Microbiol. 2019, 4, 1680–1691. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 2019, 365, eaaw1944. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.M.; Wertheim, H.F.L.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H. Antibiotic resistance—The need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Antibiotic Resistance: Synthesis of Recommendations by Expert Policy Groups; World Health Organization: Geneva, Switzerland, 2001. [Google Scholar]
| Modification of cell permeability | reduction of entry channels; efflux pumps |
| Production of inactivating enzymes | β-lactamase; acetyltransferase; phosphotransferase; adenyltransferase |
| Modification of the attachment site | penicillin-binding proteins (PBP); RNA polymerase |
| Activation of alternative metabolic pathway | modified enzymes |
| Bacteria | ARGs |
|---|---|
| Gram positive: Acinetobacter, Aeromonas, Clostridium, E. cloacae, E. coli, K. pneumoniae, P. aeruginosa, Salmonella, Sphingomonas, Vibrio, etc. | β-Lactams: blaCTX−M−1, blaCTX−M−8, blaCTX−M−14, mecA, ampC, etc. Aminoglycosides: aac, aad, etc. |
| Gram negative: E. faecium, E. faecalis, E. hirae, E. durans, E. caaeliflavus, E. avium, S. agalactiae, S. aureus, S. intermedius, S. hyicus, etc. | Tetracyclines: tetA, tetB, tetC, tetG, tetO, etc. Sulfonamides: sulI, sulII, sul3, etc. MLSB: ermA, ermB, etc. Vancomycin: van Colistin: mcr-1 and mcr-5.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bava, R.; Castagna, F.; Lupia, C.; Poerio, G.; Liguori, G.; Lombardi, R.; Naturale, M.D.; Mercuri, C.; Bulotta, R.M.; Britti, D.; et al. Antimicrobial Resistance in Livestock: A Serious Threat to Public Health. Antibiotics 2024, 13, 551. https://doi.org/10.3390/antibiotics13060551
Bava R, Castagna F, Lupia C, Poerio G, Liguori G, Lombardi R, Naturale MD, Mercuri C, Bulotta RM, Britti D, et al. Antimicrobial Resistance in Livestock: A Serious Threat to Public Health. Antibiotics. 2024; 13(6):551. https://doi.org/10.3390/antibiotics13060551
Chicago/Turabian StyleBava, Roberto, Fabio Castagna, Carmine Lupia, Giusi Poerio, Giovanna Liguori, Renato Lombardi, Maria Diana Naturale, Caterina Mercuri, Rosa Maria Bulotta, Domenico Britti, and et al. 2024. "Antimicrobial Resistance in Livestock: A Serious Threat to Public Health" Antibiotics 13, no. 6: 551. https://doi.org/10.3390/antibiotics13060551
APA StyleBava, R., Castagna, F., Lupia, C., Poerio, G., Liguori, G., Lombardi, R., Naturale, M. D., Mercuri, C., Bulotta, R. M., Britti, D., & Palma, E. (2024). Antimicrobial Resistance in Livestock: A Serious Threat to Public Health. Antibiotics, 13(6), 551. https://doi.org/10.3390/antibiotics13060551










