Emergence of Plasmid-Mediated Quinolone Resistance (PMQR) Genes in Campylobacter coli in Tunisia and Detection of New Sequence Type ST13450
Abstract
1. Introduction
2. Results
2.1. Antimicrobial Susceptibility of C. coli Isolates
2.2. Screening for PMQR Genes
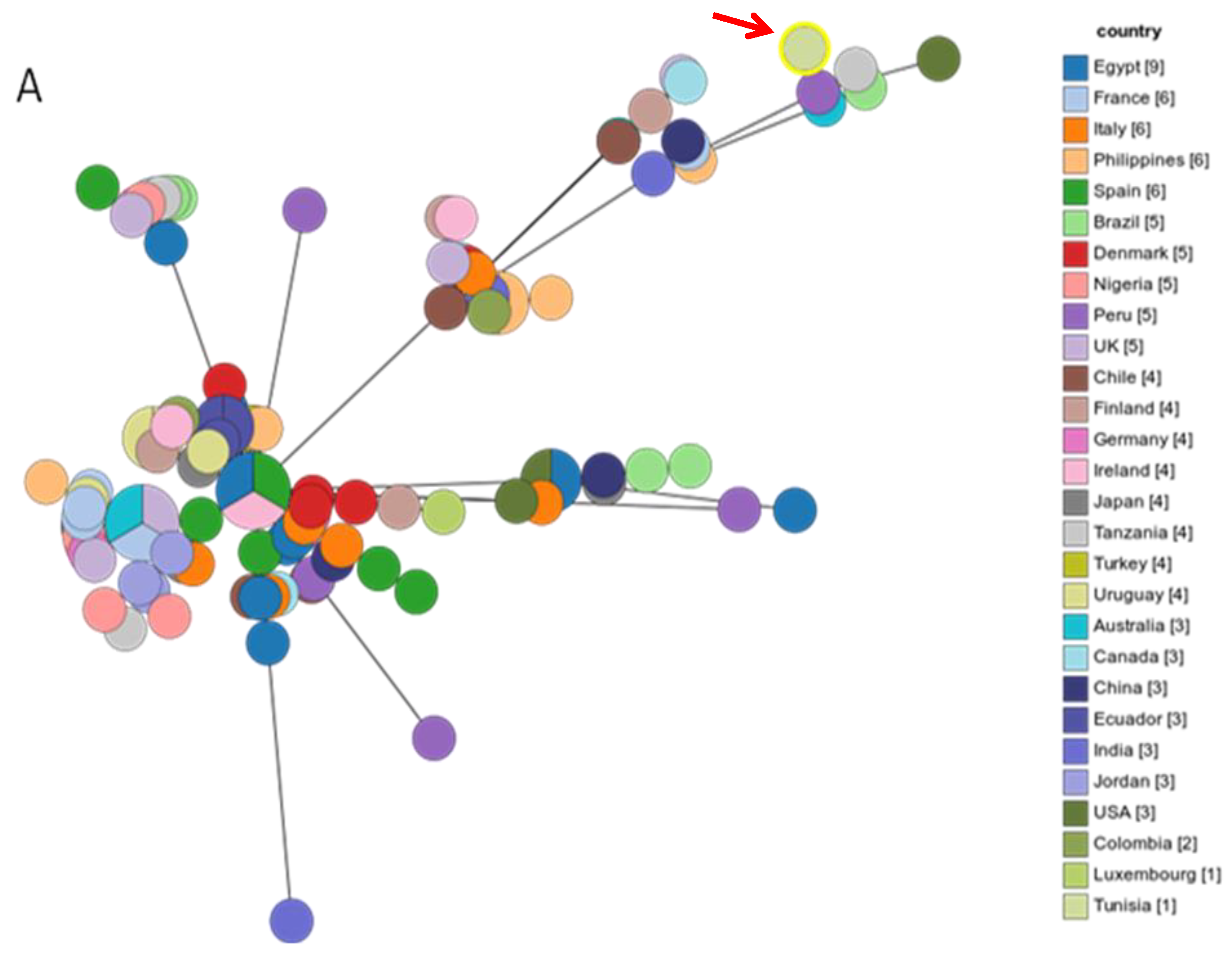
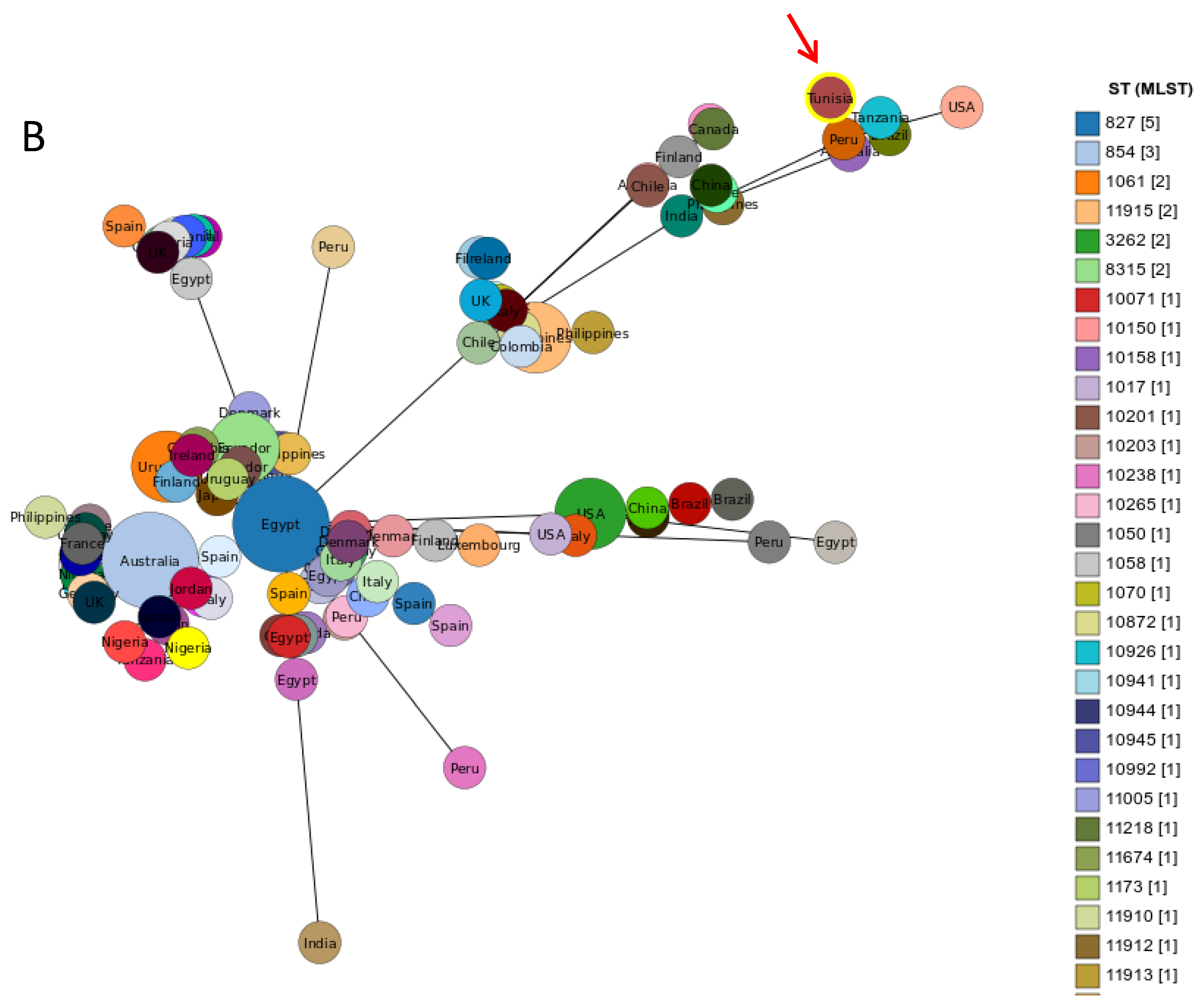
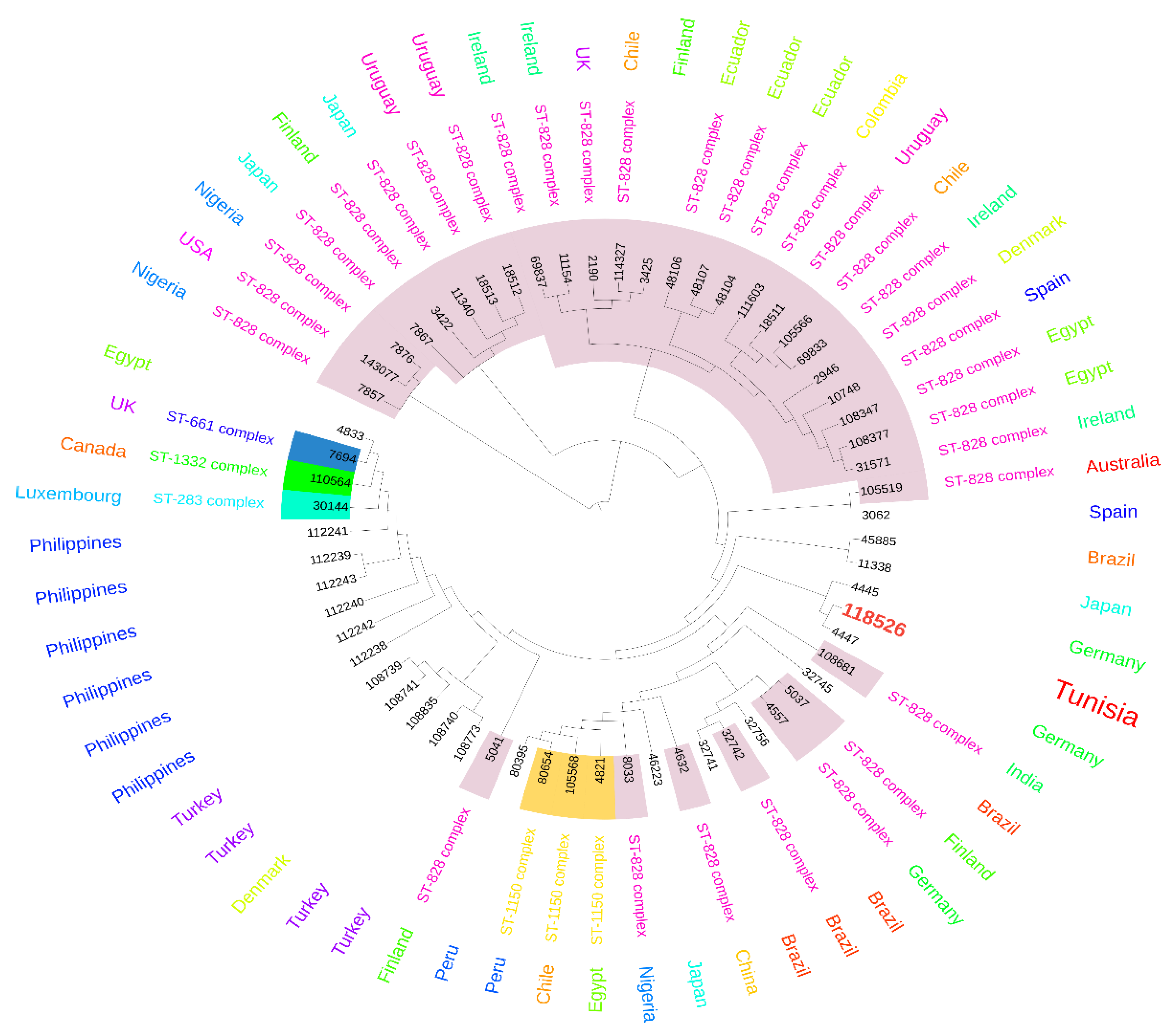
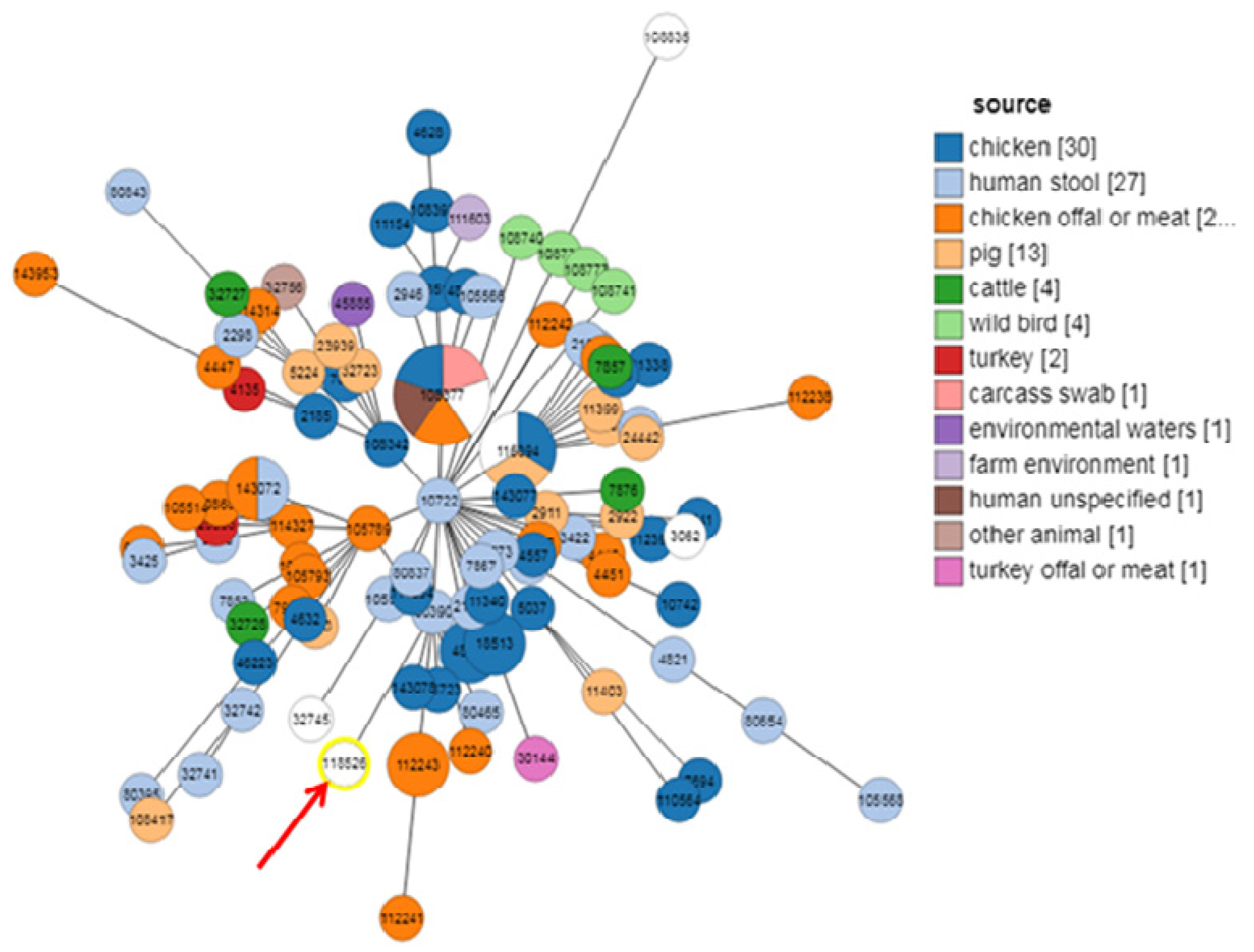
2.3. MLST Typing
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Bacterial Strains
4.3. Antimicrobial Susceptibility Testing
4.4. Screening of PMQR Genes from Campylobacter Isolates
4.5. Phylogenetic Analysis by Multilocus Sequence Typing (MLST)
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nachamkin, I. Campylobacter and Archobacter. In Manual of Clinical Microbiology, 8th ed.; Murtaleray, P.R., Baron, E.J., Jorgensen, J.A., Eds.; American Society for Microbiology (ASM) Press: Washington, DC, USA, 2003; pp. 902–914. [Google Scholar]
- Man, S.M. The clinical importance of emerging Campylobacter species. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Epps, S.V.; Harvey, R.B.; Hume, M.E.; Phillips, T.D.; Anderson, R.C.; Nisbet, D.J. Foodborne Campylobacter: Infections, metabolism, pathogenesis and reservoirs. Int. J. Environ. Res. Public Health 2013, 10, 6292–6304. [Google Scholar] [CrossRef] [PubMed]
- García-Sánchez, L.; Melero, B.; Rovira, J. Campylobacter in the food chain. Adv. Food Nutr. Res. 2018, 86, 215–252. [Google Scholar] [PubMed]
- Igwaran, A.; Okoh, A.I. Human campylobacteriosis: A public health concern of global importance. Heliyon 2019, 5, e02814. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.A.; Gulhan, T. Campylobacter in wild birds: Is it an animal and public health concern? Front. Microbiol. 2022, 12, 812591. [Google Scholar] [CrossRef]
- Coker, O.; Akitoye, I.; Raphael, D.; Thomas, N.; Bolaji, A.; Kehinde, O.; Obi, L.C. Human campylobacteriosis in developing countries. Emerg. Infect. Dis. 2002, 8, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Whiley, H.; Van den Akker, B.; Giglio, S.; Bentham, R. The role of environmental reservoirs in human Campylobacteriosis. Int. J. Environ. Res. Public Health 2013, 10, 5886–5907. [Google Scholar] [CrossRef] [PubMed]
- Iovine, N.M. Resistance mechanisms in Campylobacter jejuni. Virulence 2013, 4, 230–240. [Google Scholar] [CrossRef] [PubMed]
- EFSA (European Food Safety Authority). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 2017, 15, 5077. [Google Scholar] [CrossRef] [PubMed]
- Portes, A.B.; Panzenhagen, P.; Pereira Dos Santos, A.M.; Junior, C.A.C. Antibiotic resistance in Campylobacter: A systematic review of South American isolates. Antibiotics 2023, 12, 548. [Google Scholar] [CrossRef]
- Wieczorek, K.; Osek, J. Antimicrobial resistance mechanisms among Campylobacter. Biomed. Res. Int. 2013, 2013, 340605. [Google Scholar] [CrossRef] [PubMed]
- Luo, N.; Sahin, O.; Lin, J.; Michel, L.O.; Zhang, Q. In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob. Agents Chemother. 2003, 47, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J. Transferable mechanisms of quinolone resistance from 1998 onward. Clin. Microbiol. Rev. 2019, 32, e00007-19. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, M.; Béjaoui, A.; Ben Hamda, C.; Jouini, A.; Ghedira, K.; Zrelli, C.; Hamrouni, S.; Aouadhi, C.; Bessoussa, G.; Ghram, A.; et al. Prevalence and antibiotic resistance patterns of Campylobacter spp. isolated from broiler chickens in the north of Tunisia. Biomed. Res. Int. 2018, 2018, 7943786. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, M.; Béjaoui, A.; Ben Hamda, C.; Ghedira, K.; Ghram, A.; Maaroufi, A. Distribution of virulence and antibiotic resistance genes in Campylobacter jejuni and Campylobacter coli isolated from broiler chickens in Tunisia. J. Microbiol. Immunol. Infect. 2022, 55 Pt 2, 1273–1282. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, M.; Béjaoui, A.; Ben Hamda, C.; Alaya, N.; Hamrouni, S.; Bessoussa, G.; Ghram, A.; Maaroufi, A. Campylobacter spp. in eggs and laying hens in the North-East of Tunisia: High prevalence and multidrug-resistance phenotypes. Vet. Sci. 2022, 9, 108. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, M.; Kamoun, S.; Hkimi, C.; Ghedira, K.; Béjaoui, A.; Maaroufi, A. Relationships between virulence genes and antibiotic resistance phenotypes/genotypes in Campylobacter spp. isolated from layer hens and eggs in the North of Tunisia: Statistical and computational insights. Foods 2022, 11, 3554. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, M.; Béjaoui, A.; Hamrouni, S.; Arfaoui, A.; Maaroufi, A. Persistence of Campylobacter spp. in poultry flocks after disinfection, virulence, and antimicrobial resistance traits of recovered isolates. Antibiotics 2023, 12, 890. [Google Scholar] [CrossRef] [PubMed]
- Béjaoui, A.; Gharbi, M.; Bitri, S.; Nasraoui, D.; Ben Aziza, W.; Ghedira, K.; Rfaik, M.; Marzougui, L.; Ghram, A.; Maaroufi, A. Virulence profiling, multidrug resistance and molecular mechanisms of Campylobacter strains from chicken carcasses in Tunisia. Antibiotics 2022, 11, 830. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef]
- Tang, Y.; Fang, L.; Xu, C.; Zhang, Q. Antibiotic resistance trends and mechanisms in the foodborne pathogen, Campylobacter. Anim. Health Res. Rev. 2017, 18, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Madec, J.Y.; Lupo, A.; Schink, A.K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial resistance in Escherichia coli. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Yanat, B.; Rodríguez-Martínez, J.M.; Touati, A. Plasmid-mediated quinolone resistance in Enterobacteriaceae: A systematic review with a focus on Mediterranean countries. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Sana, F.; Mabrouka, S.; Claudine, Q.; Faouzi, S.A.; Ilhem, B.B.; Véronique, D. Prevalence and characterization of uropathogenic Escherichia coli harboring plasmid-mediated quinolone resistance in a Tunisian university hospital. Diagn. Microbiol. Infect. Dis. 2014, 79, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Hassen, B.; Saloua, B.; Abbassi, M.S.; Ruiz-Ripa, L.; Mama, O.M.; Hassen, A.; Hammami, S.; Torres, C. mcr-1 encoding colistin resistance in CTX-M-1/CTX-M-15- producing Escherichia coli isolates of bovine and caprine origins in Tunisia. First report of CTX-M-15-ST394/D E. coli from goats. Comp. Immunol. Microbiol. Infect. Dis. 2019, 67, 101366. [Google Scholar] [CrossRef]
- Chatzipanagiotou, S.; Ioannidou, V.; Ioannidis, A.; Nicolaou, C.; Papavasileiou, E.; Chaniotaki, S.; Vatopoulos, A.; Tzouvelekis, L.; Legakis, N.J. Absence of the plasmid-mediated quinolone resistance qnrA gene among Campylobacter jejuni clinical isolates from Greece. Int. J. Antimicrob. Agents 2005, 26, 261–262. [Google Scholar] [CrossRef] [PubMed]
- Alfredson, D.A.; Korolik, V. Sequence analysis of a cryptic plasmid pCJ419 from Campylobacter jejuni and construction of an Escherichia coli–Campylobacter shuttle vector. Plasmid 2003, 50, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo-Grass, C.; Strahilevitz, J. High-resolution melt curve analysis for identification of single nucleotide mutations in the quinolone resistance gene aac(6′)-Ib-cr. Antimicrob. Agents Chemother. 2010, 54, 3509–3511. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Martínez, L.; Cano, M.E.; Rodríguez-Martínez, J.M.; Calvo, J.; Pascual, Á. Plasmid-mediated quinolone resistance. Expert Rev. Anti. Infect. Ther. 2008, 6, 685–711. [Google Scholar] [CrossRef]
- Collado, L.; Muñoz, N.; Porte, L.; Ochoa, S.; Varela, C.; Muñoz, I. Genetic diversity and clonal characteristics of ciprofloxacin-resistant Campylobacter jejuni isolated from Chilean patients with gastroenteritis. Infect. Genet. Evol. 2018, 58, 290–293. [Google Scholar] [CrossRef]
- Levican, A.; Ramos-Tapia, I.; Briceño, I.; Guerra, F.; Mena, B.; Varela, C.; Porte, L. Genomic analysis of Chilean strains of Campylobacter jejuni from human faeces. Biomed. Res. Int. 2019, 2019, 1902732. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.N.; Barker, D.O.R.; Duque, S.D.S.; Che, E.V.; Jayamanna, V.; Taboada, E.N.; Falcão, J.P. Campylobacter coli isolated in Brazil typed by core genome Multilocus Sequence Typing shows high genomic diversity in a global context. Infect. Genet. Evol. 2021, 95, 105018. [Google Scholar] [CrossRef] [PubMed]
- Di Donato, G.; Marotta, F.; Nuvoloni, R.; Zilli, K.; Neri, D.; Di Sabatino, D.; Calistri, P.; Di Giannatale, E. Prevalence, population diversity and antimicrobial resistance of Campylobacter coli isolated in Italian swine at slaughterhouse. Microorganisms 2020, 8, 222. [Google Scholar] [CrossRef] [PubMed]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing. 2017. Available online: http://mic.eucast.org (accessed on 1 October 2017).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.J.; Nordmann, P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef] [PubMed]

| Origins of Isolates | qnrA n (%) | qnrB n (%) | qnrC n (%) | qnrD n (%) | qnrS n (%) | qepA n (%) | aac(6′)-Ib n (%) | aac(6′)-Ib-cr n (%) | RE-cmeABC n (%) |
|---|---|---|---|---|---|---|---|---|---|
| Broiler (n = 41) | 0 | 23 (56) | 0 | 0 | 23 (56) | 0 | 31 (75.6) | 10 (24.39) | 25 (60) |
| Layer (n = 53) | 0 | 35 (66) | 0 | 0 | 33 (62.3) | 17 (32) | 11 (26.86) | 4 (9.7) | 19 (35.84) |
| Eggs (n = 4) | 0 | 4 (100) | 0 | 0 | 2 (50) | 1 (25) | 0 (0) | 0 (0) | 3 (75) |
| Environment (n = 41) | 0 | 18 (43.9) | 0 | 0 | 27 (65.8) | 12 (29.26) | 0 (0) | 0 (0) | 9 (21.95) |
| Total (n = 139) | 0 | 80 (57.7) | 0 | 0 | 85 (61.15) | 30 (21.58) | 42 (30.21) | 14 (10) | 56 (40.28) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gharbi, M.; Tiss, R.; Chaouch, M.; Hamrouni, S.; Maaroufi, A. Emergence of Plasmid-Mediated Quinolone Resistance (PMQR) Genes in Campylobacter coli in Tunisia and Detection of New Sequence Type ST13450. Antibiotics 2024, 13, 527. https://doi.org/10.3390/antibiotics13060527
Gharbi M, Tiss R, Chaouch M, Hamrouni S, Maaroufi A. Emergence of Plasmid-Mediated Quinolone Resistance (PMQR) Genes in Campylobacter coli in Tunisia and Detection of New Sequence Type ST13450. Antibiotics. 2024; 13(6):527. https://doi.org/10.3390/antibiotics13060527
Chicago/Turabian StyleGharbi, Manel, Rihab Tiss, Melek Chaouch, Safa Hamrouni, and Abderrazak Maaroufi. 2024. "Emergence of Plasmid-Mediated Quinolone Resistance (PMQR) Genes in Campylobacter coli in Tunisia and Detection of New Sequence Type ST13450" Antibiotics 13, no. 6: 527. https://doi.org/10.3390/antibiotics13060527
APA StyleGharbi, M., Tiss, R., Chaouch, M., Hamrouni, S., & Maaroufi, A. (2024). Emergence of Plasmid-Mediated Quinolone Resistance (PMQR) Genes in Campylobacter coli in Tunisia and Detection of New Sequence Type ST13450. Antibiotics, 13(6), 527. https://doi.org/10.3390/antibiotics13060527






