A Novel Bacteriophage Infecting Multi-Drug- and Extended-Drug-Resistant Pseudomonas aeruginosa Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Bacteriophage Isolation
2.3. Bacteriophage Propagation and Titration
2.4. Phage Sequencing and Genomic Annotation
2.5. Phylogenetic Tree of the Novel Phage
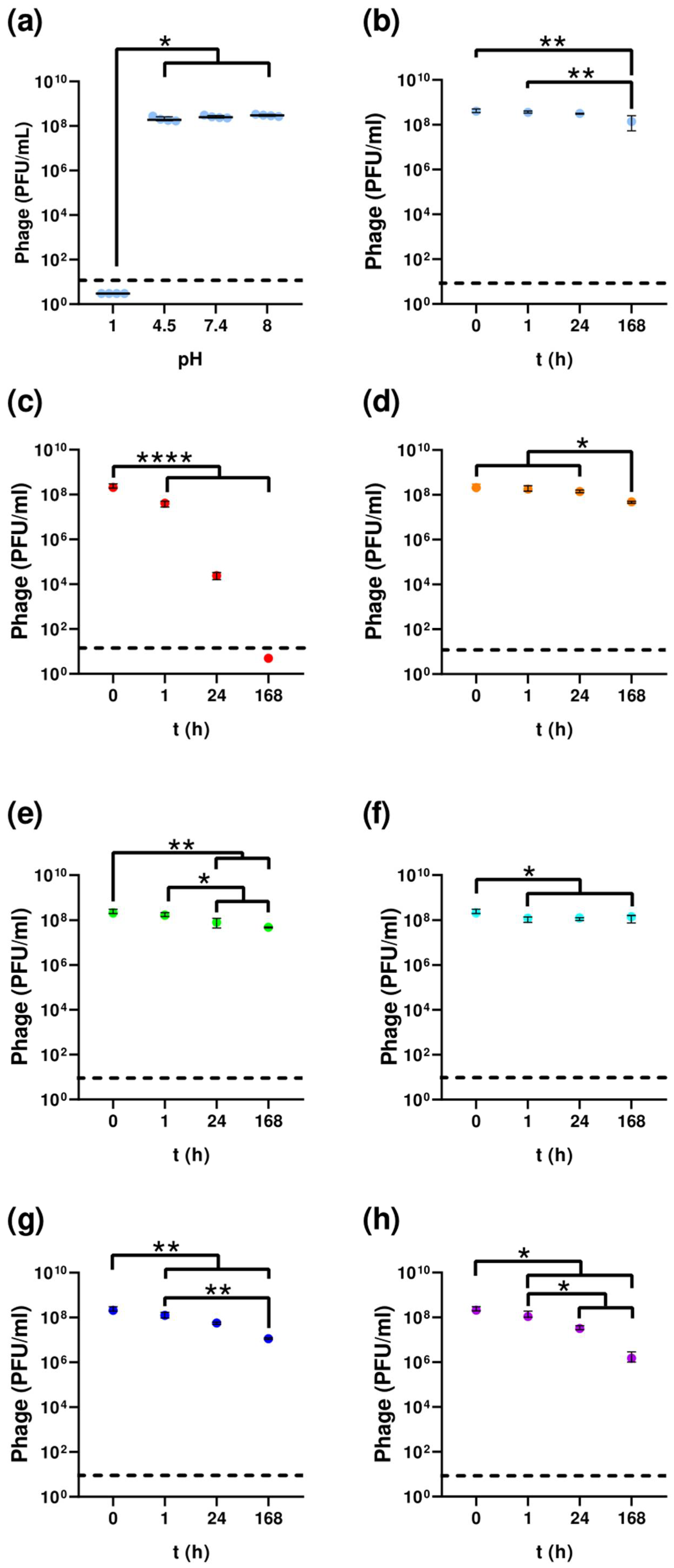
2.6. Temperature and pH Stability
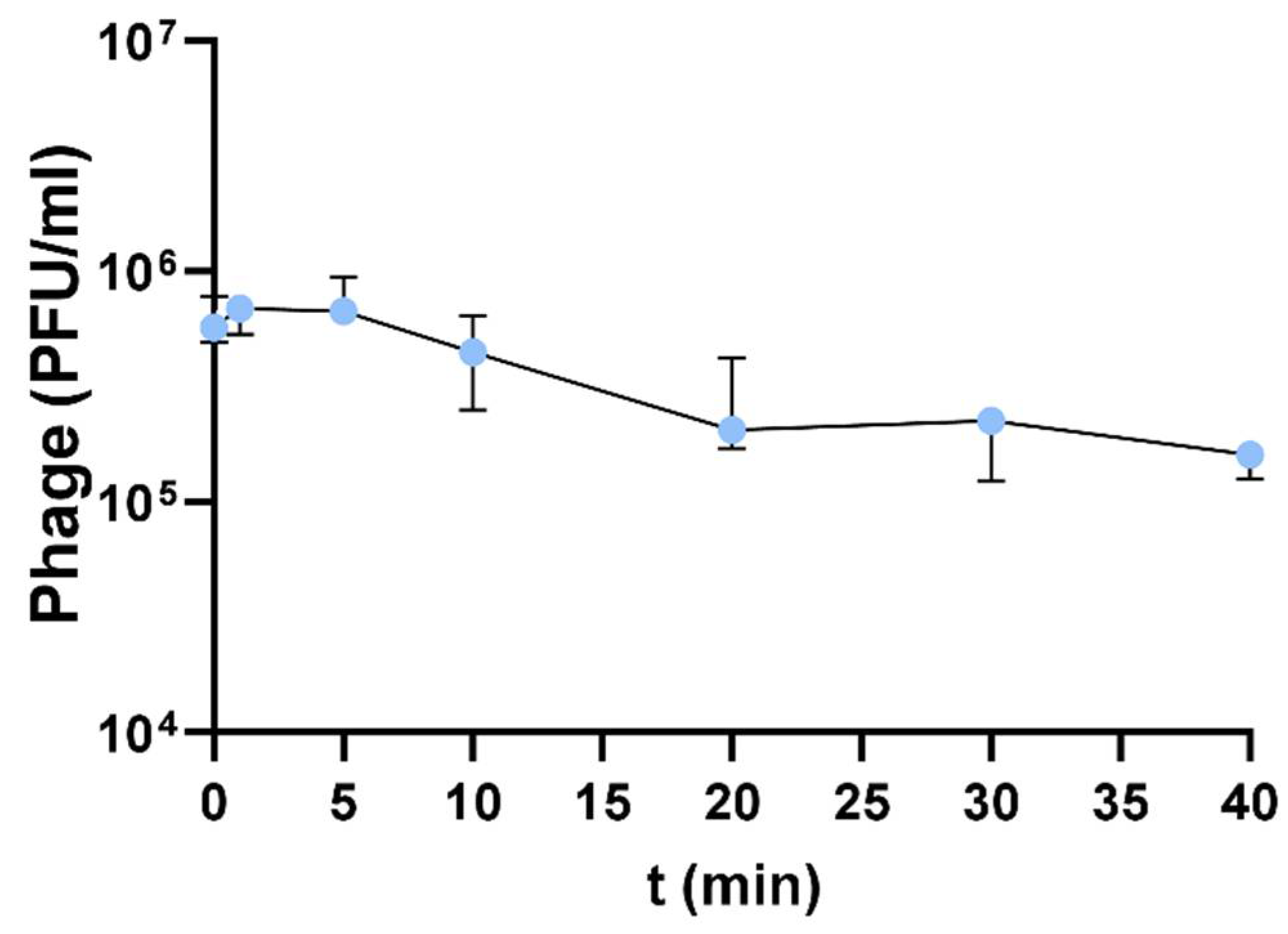
2.7. Adsorption Assays
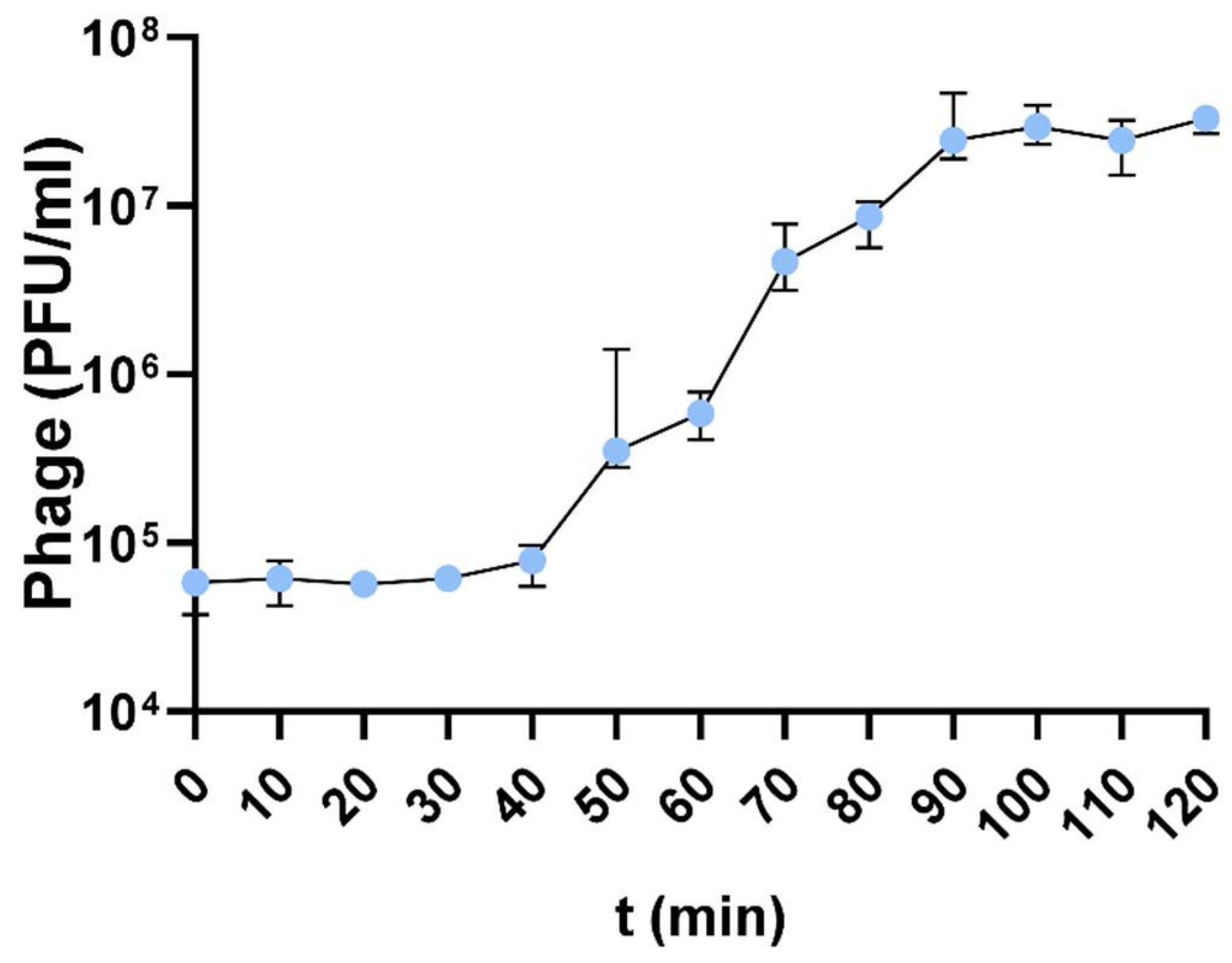
2.8. One-Step Growth Curve
2.9. Host Range Analysis
2.10. Inhibition Assays
2.11. Biofilm Eradication Assays
2.12. Inhibition of Biofilm Formation
2.13. Statistical Analysis
3. Results
3.1. Comparative Genomics
3.2. Temperature and pH Stability
3.3. Adsorption Assays
3.4. One-Step Growth Curve
3.5. Host Range Analysis
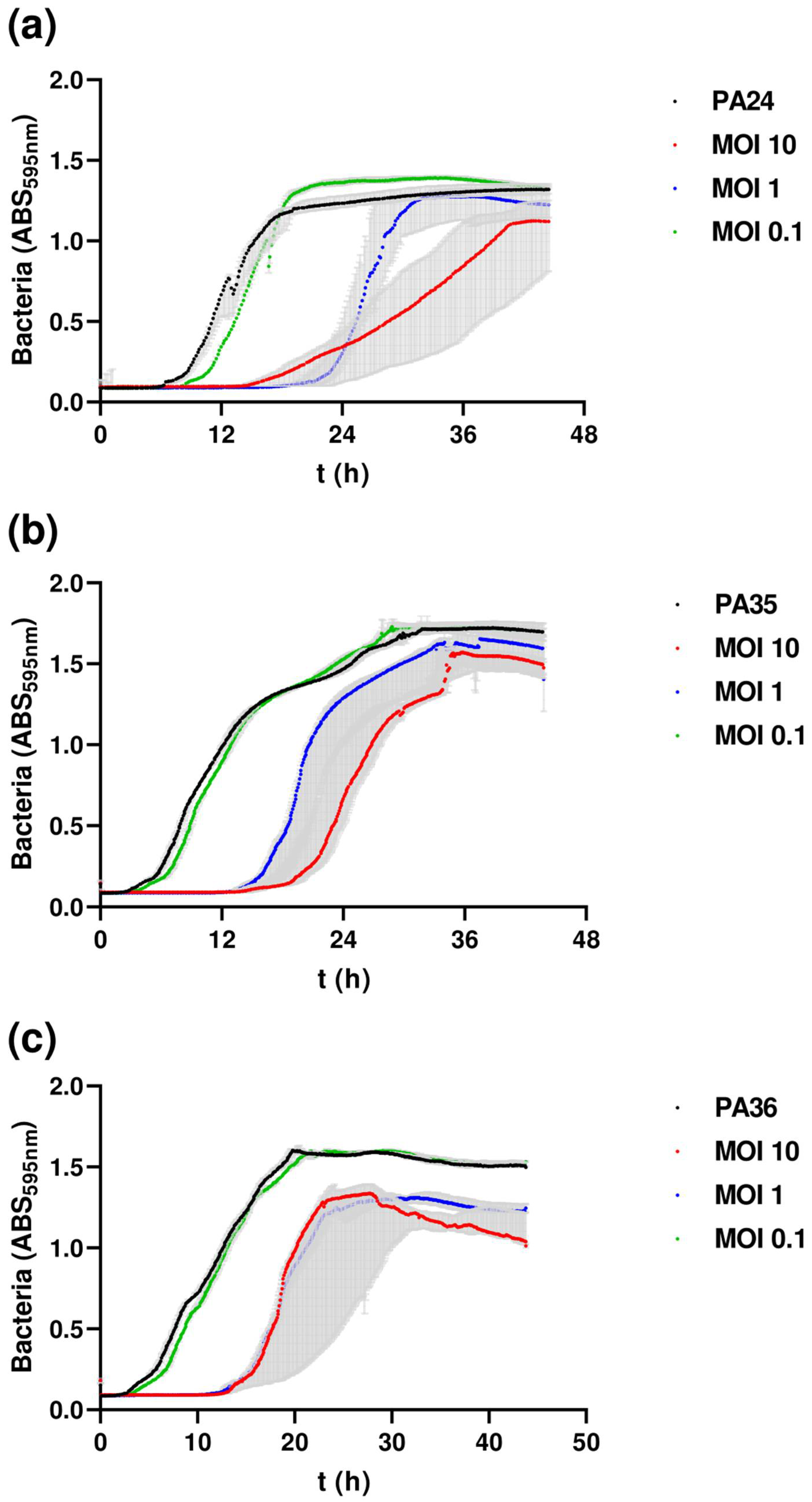
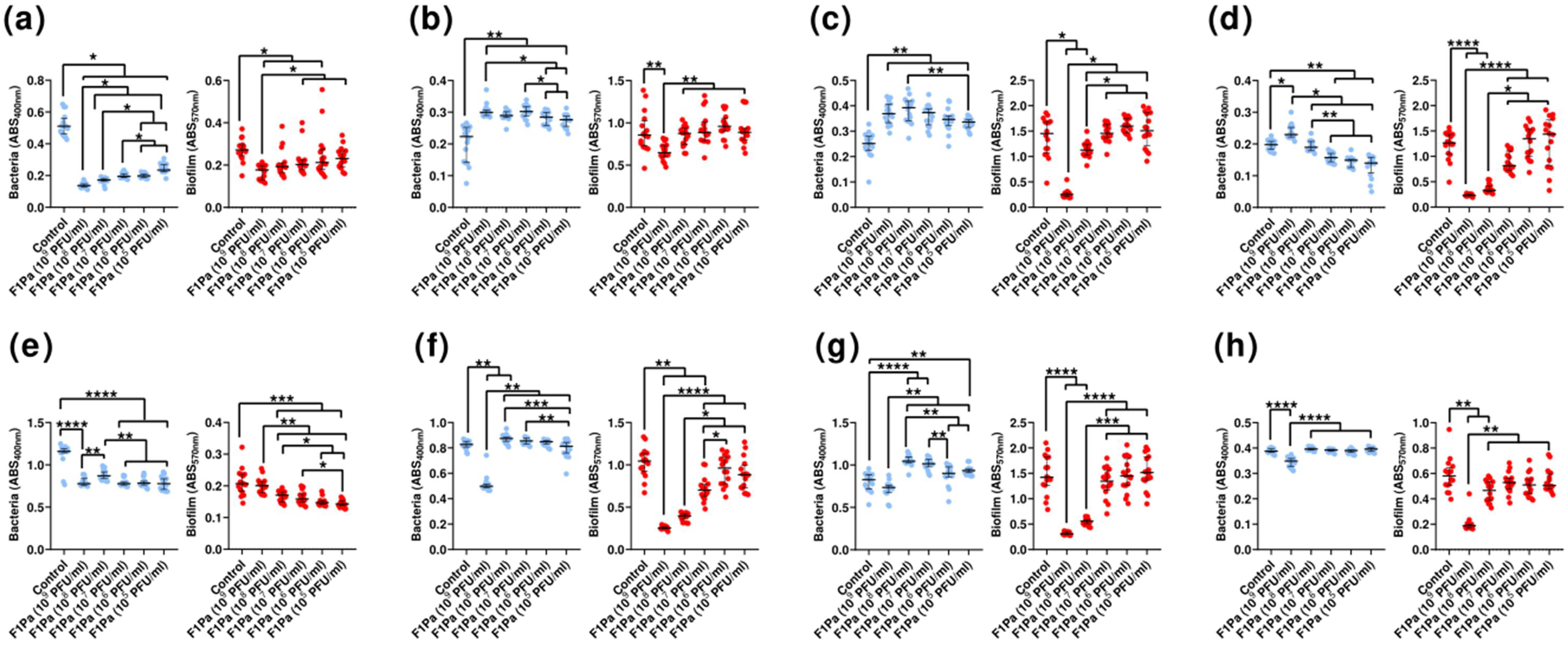
3.6. Inhibition Assays
3.7. Effect of Phage on Preformed Biofilm
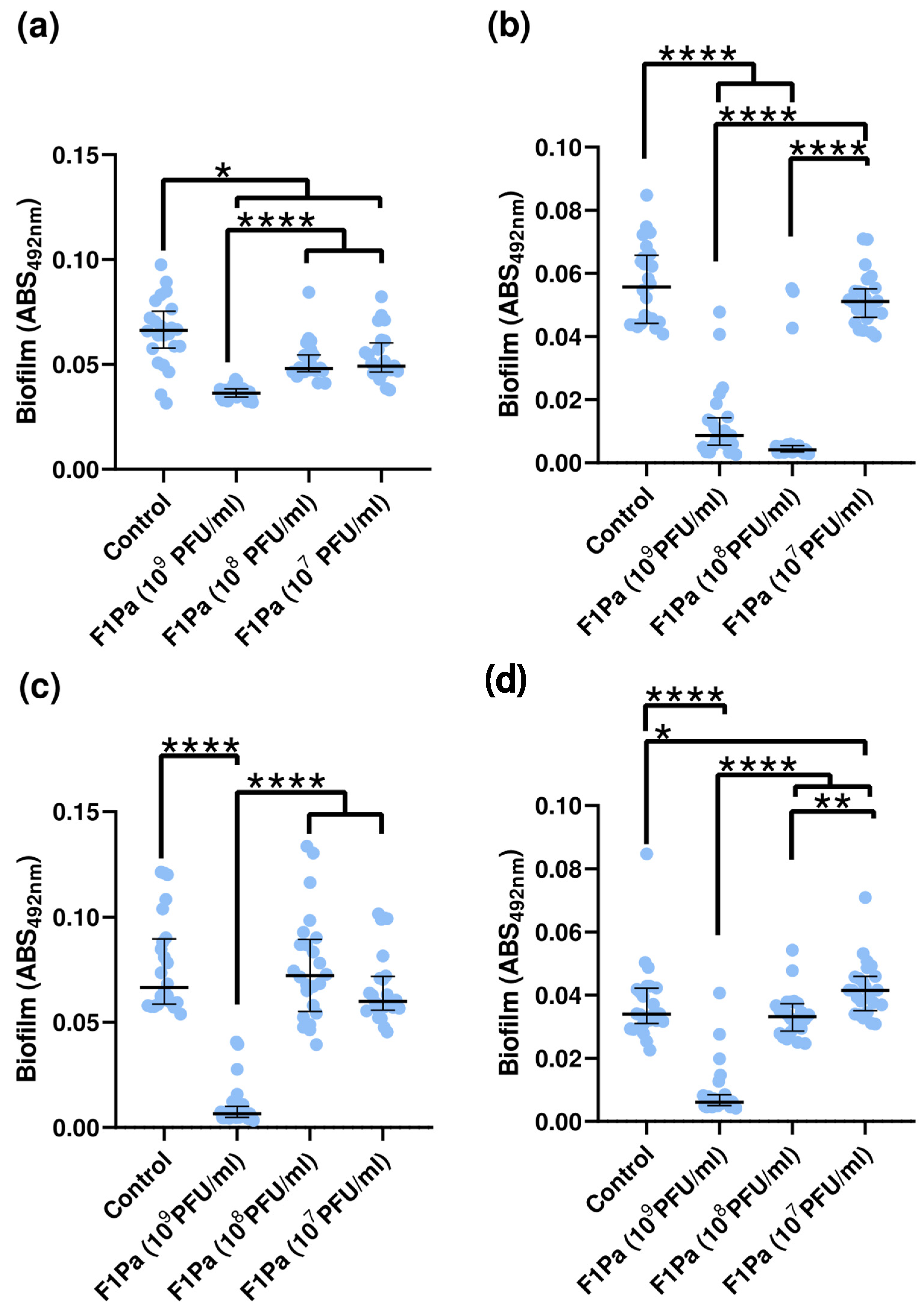
3.8. Inhibition of Biofilm Formation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lachiewicz, A.M.; Hauck, C.G.; Weber, D.J.; Cairns, B.A.; Van Duin, D. Bacterial infections after burn injuries: Impact of multidrug resistance. Clin. Infect. Dis. 2017, 65, 2130. [Google Scholar] [CrossRef]
- Weber, D.J.; Rutala, W.A.; Sickbert-Bennett, E.E.; Samsa, G.P.; Brown, V.; Niederman, M.S. Microbiology of ventilator-associated pneumonia compared with that of hospital-acquired pneumonia. Infect. Control Hosp. Epidemiol. 2007, 28, 825–831. [Google Scholar] [CrossRef]
- Rello, J.; Borgatta, B.; Lisboa, T. Risk factors for Pseudomonas aeruginosa pneumonia in the early twenty-first century. Intensive Care Med. 2013, 39, 2204–2206. [Google Scholar] [CrossRef]
- Al-Hasan, M.N.; Wilson, J.W.; Lahr, B.D.; Eckel-Passow, J.E.; Baddour, L.M. Incidence of Pseudomonas aeruginosa bacteremia: A population-based study. Am. J. Med. 2008, 121, 702. [Google Scholar] [CrossRef]
- Zarza, V.M.P.; Mordani, S.M.; Maldonado, A.M.; Hernández, D.Á.; Georgina, S.G.S.; Vázquez-López, R. Pseudomonas aeruginosa: Pathogenicity and antimicrobial resistance in urinary tract infection. Rev. Chilena Infectol. 2019, 36, 180–189. [Google Scholar] [CrossRef]
- Estahbanati, H.K.; Kashani, P.P.; Ghanaatpisheh, F. Frequency of Pseudomonas aeruginosa serotypes in burn wound infections and their resistance to antibiotics. Burns 2002, 28, 340–348. [Google Scholar] [CrossRef]
- McManus, A.T.; Mason, A.D.; McManus, W.F.; Pruitt, B.A. Twenty-five year review of Pseudomonas aeruginosa bacteremia in a burn center. Eur. J. Clin. Microbiol. 1985, 4, 219–223. [Google Scholar] [CrossRef]
- Gang, R.K.; Bang, R.L.; Sanyal, S.C.; Mokaddas, E.; Lari, A.R. Pseudomonas aeruginosa septicaemia in burns. Burns 1999, 25, 611–616. [Google Scholar] [CrossRef]
- Kwiecinska-Piróg, J.; Przekwas, J.; Majkut, M.; Skowron, K.; Gospodarek-Komkowska, E. Biofilm formation reducing properties of manuka honey and propolis in proteus mirabilis rods isolated from chronic wounds. Microorganisms 2020, 8, 1823. [Google Scholar] [CrossRef]
- Saltoglu, N.; Ergonul, O.; Tulek, N.; Yemisen, M.; Kadanali, A.; Karagoz, G.; Batirel, A.; Ak, O.; Sonmezer, C.; Eraksoy, H.; et al. Influence of multidrug resistant organisms on the outcome of diabetic foot infection. Int. J. Infect. Dis. 2018, 70, 10–14. [Google Scholar] [CrossRef]
- Saltoglu, N.; Surme, S.; Ezirmik, E.; Kadanali, A.; Kurt, A.F.; Sahin Ozdemir, M.; Ak, O.; Altay, F.A.; Acar, A.; Cakar, Z.S.; et al. The effects of antimicrobial resistance and the compatibility of initial antibiotic treatment on clinical outcomes in patients with diabetic foot infection. Int. J. Low. Extrem. Wounds 2023, 22, 283–290. [Google Scholar] [CrossRef]
- Saltoglu, N.; Yemisen, M.; Ergonul, O.; Kadanali, A.; Karagoz, G.; Batirel, A.; Ak, O.; Eraksoy, H.; Cagatay, A.; Vatan, A.; et al. Predictors for limb loss among patient with diabetic foot infections: An observational retrospective multicentric study in Turkey. Clin. Microbiol. Infect. 2015, 21, 659–664. [Google Scholar] [CrossRef]
- Pires, D.P.; Vilas Boas, D.; Sillankorva, S.; Azeredo, J. Phage therapy: A step forward in the treatment of Pseudomonas aeruginosa infections. J. Virol. 2015, 89, 7449–7456. [Google Scholar] [CrossRef]
- Hraiech, S.; Brégeon, F.; Rolain, J. Bacteriophage-based therapy in cystic infections: Rationale and current status. Drug Des. Devel Ther. 2015, 9, 3653–3663. [Google Scholar]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 284, 1318–1322. [Google Scholar] [CrossRef]
- Lewis, K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 2001, 45, 999–1007. [Google Scholar] [CrossRef]
- Breidenstein, E.B.M.; de la Fuente-Núñez, C.; Hancock, R.E.W. Pseudomonas Aeruginosa: All Roads Lead to Resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef]
- López-Causapé, C.; Cabot, G.; del Barrio-Tofiño, E.; Oliver, A. The versatile mutational resistome of pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 685. [Google Scholar] [CrossRef]
- Hill, C.; Mills, S.; Ross, R.P. Phages & antibiotic resistance: Are the most abundant entities on earth ready for a comeback? Future Microbiol. 2018, 13, 711–726. [Google Scholar]
- Potron, A.; Poirel, L.; Nordmann, P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int. J. Antimicrob. Agents 2015, 45, 568–585. [Google Scholar] [CrossRef]
- Eichenberger, E.M.; Thaden, J.T. Epidemiology and mechanisms of resistance of extensively drug resistant Gram-negative bacteria. Antibiotics 2019, 8, 37. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016, 4, 481–511. [Google Scholar] [CrossRef]
- Pérez, A.; Gato, E.; Pérez-Llarena, J.; Fernández-Cuenca, F.; José Gude, M.; Ovia, M.; Eugenia Pachón, M.; Garnacho, J.; Gonzá lez, V.; Pascual, L.; et al. High incidence of MDR and XDR Pseudomonas aeruginosa isolates obtained from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J. Antimicrob. Chemother. 2019, 74, 1244–1252. [Google Scholar] [CrossRef]
- del Barrio-Tofiño, E.; Zamorano, L.; Cortes-Lara, S.; López-Causapé, C.; Sá nchez-Diener, I.; Cabot, G.; Bou, G.; Martínez-Martínez, L.; Oliver, A. Spanish nationwide survey on Pseudomonas aeruginosa antimicrobial resistance mechanisms and epidemiology. J. Antimicrob. Chemother. 2019, 74, 1825–1835. [Google Scholar] [CrossRef]
- Tomczyk, S.; Zanichelli, V.; Grayson, M.L.; Twyman, A.; Abbas, M.; Pires, D.; Allegranzi, B.; Harbarth, S. Clinical infectious diseases control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii, and Pseudomonas aeruginosa in healthcare facilities: A systematic review and reanalysis of quasi-experimental studies. Clin. Infect. Dis. 2019, 68, 873–884. [Google Scholar] [CrossRef]
- Fong, S.A.; Drilling, A.J.; Ooi, M.L.; Paramasivan, S.; Finnie, J.W.; Morales, S.; Psaltis, A.J.; Vreugde, S.; Wormald, P.J. Safety and efficacy of a bacteriophage cocktail in an in vivo model of Pseudomonas aeruginosa sinusitis. Transl. Res. 2019, 206, 41–56. [Google Scholar] [CrossRef]
- Holloway, B.W.; Egan, J.B.; Monk, M. Lysogeny in Pseudomonas aeruginosa. Aust. J. Exp. Biol. Med. Sci. 1960, 38, 321–329. [Google Scholar] [CrossRef]
- Kellenberger, E.; Séchaud, J. Electron microscopical studies of phage multiplication. Virology 1957, 3, 256–274. [Google Scholar] [CrossRef]
- Drilling, A.; Morales, S.; Jardeleza, C.; Vreugde, S.; Speck, P.; Wormald, P.J. Bacteriophage reduces biofilm of Staphylococcus aureus ex vivo isolates from chronic rhinosinusitis patients. Am. J. Rhinol. Allergy 2014, 28, 3–11. [Google Scholar] [CrossRef]
- Clokie, M.R.J.; Kropinski, A.M.; Lavigne, R. (Eds.) Bacteriophages: Methods and protocols-Volume III. In Methods in Molecular Biology; Springer Science + Business Media: New York, NY, USA, 2018; Volume 1681. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates Europe PMC funders group. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Chibeu, A.; Ceyssens, P.J.; Hertveldt, K.; Volckaert, G.; Cornelis, P.; Matthijs, S.; Lavigne, R. The adsorption of pseudomonas aeruginosa bacteriophage PhiKMV is dependent on expression regulation of type IV pili genes. FEMS Microbiol. Lett. 2009, 296, 210–218. [Google Scholar] [CrossRef]
- Zurabov, F.; Zhilenkov, E. Characterization of four virulent Klebsiella pneumoniae bacteriophages, and evaluation of their potential use in complex phage preparation. Virol. J. 2021, 18, 9. [Google Scholar] [CrossRef]
- Okoliegbe, I.N.; Hijazi, K.; Cooper, K.; Ironside, C.; Gould, I.M. Antimicrobial synergy testing: Comparing the tobramycin and ceftazidime gradient diffusion methodology used in assessing synergy in cystic fibrosis-derived multidrug-resistant Pseudomonas aeruginosa. Antibiotics 2021, 10, 967. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by Staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A roadmap for genome-based phage taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef]
- Hyman, P.; Abedon, S.T. Practical methods for determining phage growth parameters. Methods Mol. Biol. 2009, 501, 175–202. [Google Scholar] [CrossRef]
- Hyman, P.; Abedon, S.T. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 2010, 70, 217–248. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, X.; Mou, Z.; Ren, H.; Zhang, C.; Zou, L.; Liu, H.; Liu, W.; Liu, Z. Characterization and genome analysis of Pseudomonas aeruginosa phage VB_PaeP_Lx18 and the antibacterial activity of its lysozyme. Arch. Virol. 2022, 167, 1805–1817. [Google Scholar] [CrossRef]
- Zhang, Y.; Meng, B.; Wei, X.; Li, Y.; Wang, X.; Zheng, Y.; Wang, C.; Cui, L.; Zhao, X. Evaluation of phage therapy for pulmonary infection of mouse by liquid aerosol-exposure Pseudomonas aeruginosa. Infect. Drug Resist. 2021, 14, 4457–4469. [Google Scholar] [CrossRef]
- Knezevic, P.; Obreht, D.; Curcin, S.; Petrusic, M.; Aleksic, V.; Kostanjsek, R.; Petrovic, O. Phages of Pseudomonas aeruginosa: Response to environmental factors and in vitro ability to inhibit bacterial growth and biofilm formation. J. Appl. Microbiol. 2011, 111, 245–254. [Google Scholar] [CrossRef]
- Alvi, I.A.; Asif, M.; Tabassum, R.; Aslam, R.; Abbas, Z.; Rehman, S. ur RLP, a bacteriophage of the family Podoviridae, rescues mice from bacteremia caused by multi-drug-resistant Pseudomonas aeruginosa. Arch. Virol. 2020, 165, 1289–1297. [Google Scholar] [CrossRef]
- Quirós, P.; Colomer-Lluch, M.; Martínez-Castillo, A.; Miró, E.; Argente, M.; Jofre, J.; Navarro, F.; Muniesa, M. Antibiotic resistance genes in the bacteriophage DNA fraction of human fecal samples. Antimicrob. Agents Chemother. 2014, 58, 606–609. [Google Scholar] [CrossRef]
- McCullor, K.; Postoak, B.; Rahman, M.; King, C.; Michael McShana, W. Genomic sequencing of high-efficiency transducing streptococcal bacteriophage A25: Consequences of escape from lysogeny. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef]
- Chaturongakul, S.; Ounjai, P. Phage-host interplay: Examples from tailed phages and Gram-negative bacterial pathogens. Front. Microbiol. 2014, 5, 442. [Google Scholar] [CrossRef]
- Mahdavi, S.; Sadeghi, M.; Shokri, R.; Sadegh, B. The role of bacteriophages as important reservoirs of extended-spectrum beta-lactamase genes in Azerbaijan hospitals. Microb. Drug Resist. 2022, 28, 436–443. [Google Scholar] [CrossRef]
- Anomaly, J. The future of phage: Ethical challenges of using phage therapy to treat bacterial infections. Public. Health Ethics 2020, 13, 82–88. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Alfenas-Zerbini, P.; Dempsey, D.M.; Dutilh, B.E.; García, M.L.; Curtis Hendrickson, R.; et al. Recent changes to virus taxonomy ratified by the International Committee on Taxonomy of Viruses (2022). Arch. Virol. 2022, 167, 2429–2440. [Google Scholar] [CrossRef]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; McVeigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef]
- Campbell, R.A.; Farlow, J.; Freyberger, H.R.; He, Y.; Ward, A.M.; Ellison, D.W.; Getnet, D.; Swierczewski, B.E.; Nikolich, M.P.; Filippov, A.A. Genome sequences of 17 diverse Pseudomonas aeruginosa phages. Microbiol. Resour. Announc. 2021, 10. [Google Scholar] [CrossRef]
- Liu, X.; Feng, Z.; Fan, X.; Nie, Y.; Wu, X.L. Isolation and characterization of the novel Pseudomonas stutzeri bacteriophage 8P. Arch. Virol. 2021, 166, 601–606. [Google Scholar] [CrossRef]
- Dąbrowska, K. Phage therapy: What factors shape phage pharmacokinetics and bioavailability? Systematic and critical review. Med. Res. Rev. 2019, 39, 2000. [Google Scholar] [CrossRef]
- Khanal, D.; Chang, R.Y.K.; Hick, C.; Morales, S.; Chan, H.K. Enteric-coated bacteriophage tablets for oral administration against gastrointestinal infections. Int. J. Pharm. 2021, 609, 121206. [Google Scholar] [CrossRef]
- Richards, K.; Malik, D.J. Bacteriophage encapsulation in Ph-responsive core-shell capsules as an animal feed additive. Viruses 2021, 13, 1131. [Google Scholar] [CrossRef]
- Lorenzo-Rebenaque, L.; Malik, D.J.; Catalá-Gregori, P.; Marin, C.; Sevilla-Navarro, S. In Vitro and in vivo gastrointestinal survival of non-encapsulated and microencapsulated salmonella bacteriophages: Implications for bacteriophage therapy in poultry. Pharmaceuticals 2021, 14, 434. [Google Scholar] [CrossRef]
- Kwiatek, M.; Parasion, S.; Rutyna, P.; Mizak, L.; Gryko, R.; Niemcewicz, M.; Olender, A.; Łobocka, M. Isolation of bacteriophages and their application to control pseudomonas aeruginosa in planktonic and biofilm models. Res. Microbiol. 2017, 168, 194–207. [Google Scholar] [CrossRef]
- Yu, X.; Xu, Y.; Gu, Y.; Zhu, Y.; Liu, X. Characterization and genomic study of “PhiKMV-Like” phage PAXYB1 infecting Pseudomonas aeruginosa. Sci. Rep. 2017, 7, 13068. [Google Scholar] [CrossRef]
- Shi, X.; Zhao, F.; Sun, H.; Yu, X.; Zhang, C.; Liu, W.; Pan, Q.; Ren, H. Characterization and complete genome analysis of Pseudomonas aeruginosa bacteriophage VB_PaeP_LP14 belonging to genus Litunavirus. Curr. Microbiol. 2020, 77, 2465–2474. [Google Scholar] [CrossRef]
- Namonyo, S.; Carvalho, G.; Guo, J.; Weynberg, K.D. Novel bacteriophages show activity against selected Australian clinical strains of Pseudomonas aeruginosa. Microorganisms 2022, 10, 210. [Google Scholar] [CrossRef]
- Barazandeh, M.; Shahin, K.; Hedayatkhah, A.; Komijani, M.; Mansoorianfar, M. Characterization of a novel bullet-shaped lytic bacteriophage against extensively drug-resistant Pseudomonas aeruginosa isolated from human and domestic sources. Vet. Res. Forum 2021, 12, 401. [Google Scholar] [CrossRef]
- Jerne, N.K. The presence in normal serum of specific antibody against bacteriophage T4 and its increase during the earliest stages of immunization. J. Immunol. 1956, 76, 209–216. [Google Scholar] [CrossRef]
- Jerne, N.K. Bacteriophage inactivation by antiphage serum diluted in distilled water. Nature 1952, 169, 117–118. [Google Scholar] [CrossRef]
- Egido, J.E.; Dekker, S.O.; Toner-Bartelds, C.; Lood, C.; Rooijakkers, S.H.M.; Bardoel, B.W.; Haas, P.-J. Human complement inhibits myophages against Pseudomonas aeruginosa. Viruses 2023, 15, 2211. [Google Scholar] [CrossRef]
- Van Nieuwenhuyse, B.; Van der Linden, D.; Chatzis, O.; Lood, C.; Wagemans, J.; Lavigne, R.; Schroven, K.; Paeshuyse, J.; de Magnée, C.; Sokal, E.; et al. Bacteriophage-antibiotic combination therapy against extensively drug-resistant Pseudomonas aeruginosa infection to allow liver transplantation in a toddler. Nat. Commun. 2022, 13, 5725. [Google Scholar] [CrossRef]
- Landa, K.J.; Mossman, L.M.; Whitaker, R.J.; Rapti, Z.; Clifton, S.M. Phage-antibiotic synergy inhibited by temperate and chronic virus competition. Bull. Math. Biol. 2022, 84, 54. [Google Scholar] [CrossRef]
- Kasman, L.M.; Porter, L.D. Bacteriophages. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 280–283. [Google Scholar] [CrossRef]
- Gibson, S.B.; Green, S.I.; Liu, C.G.; Salazar, K.C.; Clark, J.R.; Terwilliger, A.L.; Kaplan, H.B.; Maresso, A.W.; Trautner, B.W.; Ramig, R.F. Constructing and characterizing bacteriophage libraries for phage therapy of human infections. Front. Microbiol. 2019, 10, 2537. [Google Scholar] [CrossRef]
- Putzeys, L.; Poppeliers, J.; Boon, M.; Lood, C.; Vallino, M.; Lavigne, R. Transcriptomics-driven characterization of LUZ100, a T7-like Pseudomonas phage with temperate features. mSystems 2023, 8, e01189-22. [Google Scholar] [CrossRef]
- Yuanyuan, N.; Xiaobo, Y.; Shang, W.; Yutong, Y.; Hongrui, Z.; Chenyu, L.; Bin, X.; Xi, Z.; Chen, Z.; Zhiqiang, S.; et al. Isolation and characterization of two homolog phages infecting Pseudomonas aeruginosa. Front. Microbiol. 2022, 13, 946251. [Google Scholar] [CrossRef]
- Zhang, F.; Huang, K.; Yang, X.; Sun, L.; You, J.; Pan, X.; Cui, X.; Yang, H. Characterization of a novel lytic Podovirus O4 of Pseudomonas aeruginosa. Arch. Virol. 2018, 163, 2377–2383. [Google Scholar] [CrossRef]
- Xu, Y.; Yu, X.; Gu, Y.; Huang, X.; Liu, G.; Liu, X. Characterization and genomic study of phage VB_EcoS-B2 infecting multidrug-resistant Escherichia coli. Front. Microbiol. 2018, 9, 793. [Google Scholar] [CrossRef]
- Ghafoor, A.; Hay, I.D.; Rehm, B.H.A. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 2011, 77, 5238–5246. [Google Scholar] [CrossRef]
- Tam, M.; Thi, T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Chan, B.; Abedon, S. Bacteriophages and their enzymes in biofilm control. Curr. Pharm. Des. 2015, 21, 85–99. [Google Scholar] [CrossRef]
- Amgarten, D.; Martins, L.F.; Lombardi, K.C.; Antunes, L.P.; Paula, A.; De Souza, S.; Gonçalves Nicastro, G.; Watanabe Kitajima, E.; Bento Quaggio, R.; Upton, C.; et al. Three novel Pseudomonas phages isolated from composting provide insights into the evolution and diversity of tailed phages. BMC Genom. 2017, 18, 346. [Google Scholar] [CrossRef]
- Mendes, J.J.; Leandro, C.; Mottola, C.; Barbosa, R.; Silva, F.A.; Oliveira, M.; Vilela, C.L.; Melo-Cristino, J.; Górski, A.; Pimentel, M.; et al. In vitro design of a novel lytic bacteriophage cocktail with therapeutic potential against organisms causing diabetic foot infections. J. Med. Microbiol. 2014, 63, 1055–1065. [Google Scholar] [CrossRef]
- Ahiwale, S.; Tamboli, N.; Thorat, K.; Kulkarni, R.; Ackermann, H.; Kapadnis, B. In vitro management of hospital Pseudomonas aeruginosa biofilm using indigenous T7-like lytic phage. Curr. Microbiol. 2011, 62, 335–340. [Google Scholar] [CrossRef]
- Pei, R.; Lamas-Samanamud, G.R. Inhibition of biofilm formation by T7 bacteriophages producing quorum-quenching enzymes. Appl. Environ. Microbiol. 2014, 80, 5340–5348. [Google Scholar] [CrossRef]

| Strain | Origin | MDR/XDR | AMK | GE | TOB | AZ | P/T | CFT | CEF | C/T | IM | MP | CIP | CO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PA1 | Blood | - | S | S | S | S | S | S | S | S | S | S | S | S |
| PA2 | Blood | - | S | S | S | S | S | S | S | S | S | S | S | S |
| PA3 | Prosthetics | - | S | S | S | S | S | S | S | S | S | S | S | S |
| PA4 | Blood | - | S | S | S | S | S | S | S | S | R | S | R | S |
| PA5 | Peritoneum | - | S | S | S | R | S | S | S | S | R | R | S | S |
| PA6 | Ulcer | - | S | S | S | S | S | S | S | S | S | S | S | S |
| PA7 | Wound | - | S | S | S | S | S | S | S | S | S | S | S | S |
| PA8 | Wound | - | S | S | S | S | S | S | S | S | S | S | S | S |
| PA9 | Wound | MDR | S | R | S | R | R | R | R | R | S | R | R | S |
| PA10 | Ulcer | S | R | S | S | S | S | S | - | S | R | S | S | |
| PA11 | Ulcer | - | S | S | S | S | S | S | S | S | S | S | S | S |
| PA12 | Wound | - | R | S | S | S | S | S | S | R | S | S | S | S |
| PA13 | Wound | - | S | S | S | S | R | S | R | S | S | S | R | S |
| PA14 | Sputum | MDR | S | R | S | R | R | R | R | R | S | S | R | S |
| PA16 | Sputum | - | S | R | - | S | R | R | R | R | R | R | S | S |
| PA17 | Wound | - | S | S | S | R | R | S | S | S | S | S | R | S |
| PA18 | Bronchial | XDR | S | R | - | S | R | S | S | R | R | R | R | R |
| PA19 | Wound | - | S | S | S | S | S | S | S | S | R | S | S | R |
| PA20 | Bronchial | MDR | S | R | S | R | R | R | R | S | R | R | R | S |
| PA21 | Bronchial | MDR | S | R | S | R | R | R | R | R | R | - | R | S |
| PA22 | Wound | - | S | S | S | S | S | S | S | - | R | S | S | S |
| PA23 | Bronchial | XDR | S | R | - | S | S | S | S | R | R | R | R | R |
| PA24 | Sputum | MDR | S | R | R | S | S | S | S | S | R | S | R | S |
| PA25 | Sputum | MDR | S | R | R | S | R | S | S | S | R | S | R | S |
| PA26 | Sputum | - | S | S | S | S | S | S | S | S | R | S | S | S |
| PA27 | Bronchial | - | S | S | S | R | S | S | S | S | R | R | S | S |
| PA28 | Sputum | XDR | S | S | - | R | R | R | R | - | R | R | R | S |
| PA29 | Wound | - | S | S | S | - | R | S | S | S | R | R | R | S |
| PA30 | Wound | - | S | S | S | R | S | S | S | S | R | R | R | S |
| PA31 | Bronchial | - | S | S | S | R | R | S | S | S | R | R | S | S |
| PA32 | Bronchial | - | S | S | S | S | S | S | S | S | R | S | S | S |
| PA33 | Perianal | XDR | R | R | - | R | R | R | R | R | R | R | R | S |
| PA34 | Otic | - | S | S | S | - | S | S | S | S | S | S | S | S |
| PA35 | - | XDR | S | R | R | R | R | R | R | R | R | R | R | S |
| PA36 | Bronchial | MDR | S | R | R | S | R | R | S | S | R | S | R | S |
| PA37 | Urine | XDR | R | R | R | S | R | R | R | R | R | S | R | S |
| PA38 | Urine | XDR | R | R | R | R | R | R | R | R | R | S | R | S |
| PA39 | Urine | MDR | R | R | R | S | R | R | R | R | R | R | R | S |
| Strain | Biofilm Formation (Q1 to Q3) |
|---|---|
| PA1 | 25.79 (22.02 to 31.64) |
| PA2 | 23.95 (20.97 to 27.51) |
| PA3 | 2.01 (1.66 to 3.58) |
| PA4 | 1.59 (1.45 to 1.85) |
| PA5 | 0.85 (0.65 to 0.95) |
| PA6 | 6.88 (6.56 to 7.09) |
| PA7 | 1.24 (1.18 to 1.39) |
| PA8 | 9.54 (9.20 to 9.91) |
| PA9 | 2.32 (2.16 to 3.00) |
| PA10 | 1.65 (1.51 to 1.78) |
| PA11 | 2.40 (1.55 to 3.16) |
| PA12 | 4.31 (3.65 to 4.54) |
| PA13 | 4.61 (3.99 to 4.87) |
| PA14 | 2.55 (2.25 to 2.75) |
| PA16 | 4.91 (4.45 to 5.67) |
| PA17 | 1.55 (1.35 to 1.66) |
| PA18 | 1.89 (1.64 to 2.49) |
| PA19 | 7.39 (6.28 to 8.62) |
| PA20 | 0.83 (0.68 to 0.92) |
| PA21 | 0.84 (0.79 to 1.02) |
| PA22 | 9.73 (8.48 to 10.55) |
| PA23 | 3.85 (3.48 to 4.14) |
| PA24 | 7.50 (5.54 to 9.19) |
| PA25 | 4.59 (4.04 to 5.22) |
| PA26 | 16.69 (16.06 to 17.08) |
| PA27 | 5.82 (5.55 to 6.13) |
| PA28 | 1.21 (1.05 to 1.49) |
| PA29 | 25.40 (22.39 to 27.52) |
| PA30 | 5.42 (4.98 to 5.85) |
| PA31 | 0.64 (0.55 to 0.95) |
| PA32 | 1.04 (0.86 to 2.38) |
| PA33 | 5.59 (3.69 to 6.69) |
| PA34 | 9.96 (8.91 to 11.23) |
| PA35 | 15.26 (13.39 to 16.78) |
| PA36 | 12.30 (10.79 to 14.43) |
| PA37 | 1.68 (1.15 to 3.07) |
| PA38 | 1.24 (0.81 to 1.68) |
| PA39 | 8.03 (6.56 to 9.08) |
| PAO1 | 20.93 (15.62 to 26.86) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santamaría-Corral, G.; Pagán, I.; Aguilera-Correa, J.J.; Esteban, J.; García-Quintanilla, M. A Novel Bacteriophage Infecting Multi-Drug- and Extended-Drug-Resistant Pseudomonas aeruginosa Strains. Antibiotics 2024, 13, 523. https://doi.org/10.3390/antibiotics13060523
Santamaría-Corral G, Pagán I, Aguilera-Correa JJ, Esteban J, García-Quintanilla M. A Novel Bacteriophage Infecting Multi-Drug- and Extended-Drug-Resistant Pseudomonas aeruginosa Strains. Antibiotics. 2024; 13(6):523. https://doi.org/10.3390/antibiotics13060523
Chicago/Turabian StyleSantamaría-Corral, Guillermo, Israel Pagán, John Jairo Aguilera-Correa, Jaime Esteban, and Meritxell García-Quintanilla. 2024. "A Novel Bacteriophage Infecting Multi-Drug- and Extended-Drug-Resistant Pseudomonas aeruginosa Strains" Antibiotics 13, no. 6: 523. https://doi.org/10.3390/antibiotics13060523
APA StyleSantamaría-Corral, G., Pagán, I., Aguilera-Correa, J. J., Esteban, J., & García-Quintanilla, M. (2024). A Novel Bacteriophage Infecting Multi-Drug- and Extended-Drug-Resistant Pseudomonas aeruginosa Strains. Antibiotics, 13(6), 523. https://doi.org/10.3390/antibiotics13060523








