Abstract
Clostridioides difficile infections (CDIs) continue to be a persistent healthcare concern despite newer antibiotic treatments, enhanced infection control practices, and preventive strategies focused on restoring the protective intestinal microbial barrier. Recent strides in gene sequencing research have identified many genes regulating diverse virulence factors for CDIs. These genes may be over- or under-expressed when triggered by various environmental and nutritional factors. The aims of this paper are to review the important genes involved in C. difficile pathogenesis and to identify modifiable environmental, nutritional, and other factors that may trigger the expression of these genes and thus offer new strategies to prevent CDIs.
1. Introduction
Clostridioides difficile infections (CDIs) have been a serious burden for healthcare systems for decades and continue to be a cause of outbreaks in hospitals. In the United States alone, CDI accounts for over 500,000 cases/year with 29,000 deaths [1,2]. Patients with CDIs typically have longer hospital stays (average 10 days) and higher intensive care unit admissions (up to 18%), resulting in higher healthcare costs and a heavier burden on healthcare workers [3]. Patients with an initial episode of CDI may also develop recurrent CDI (20–40%), which has a higher risk of developing sepsis and exposes patients to more antibiotic exposure for additional treatment [3].
Previous research initially focused on effective treatments for CDIs and, as more CDI outbreaks occurred, better strategies to control healthcare-associated outbreaks in hospitals [4]. During the last two decades, the roles of various toxins, risk factors, and the importance of the protective intestinal microbiome have been established [4]. The development of newer tools has expanded the scope of research into the field of genomic expression of virulence factors.
The pathogenesis of CDI involves a cascade of events: (1) disruption of the intestinal microbiome and metabolome, which acts as a protective barrier to the opportunistic invading C. difficile bacteria, (2) attachment of C. difficile to receptor sites on enterocyte cell surfaces, (3) reproduction of C. difficile reaching quorum sensing (QS) thresholds, which triggers the production of toxins, (4) toxin-mediated host cytoskeleton destruction, leading to enterocyte collapse, (5) opening of tight junctions, leading to fluid secretion, and (6) attraction of inflammatory cytokines to the site. A greater understanding of the pathogenesis cascade has led to strategies for the prevention and treatment of CDIs based on direct killing of the C. difficile bacterium with antibiotics or methods to restore the protective intestinal microbiome using probiotics or fecal microbial replacement. However, CDIs remain a global problem, and more innovative strategies to treat CDIs are urgently needed [4,5].
Recent research on the genome of the C. difficile chromosome has expanded our understanding of which genes are involved in the pathogenesis of CDI [6]. The emergence of a new “hypervirulent” strain (C. difficile NAP1/BI/027) was responsible for several outbreaks of CDIs in the 2000s [7]. The hypervirulent ribotype 027 strain has two distinguishing virulence determinants: the deletion of the tcdC gene, which is a negative regulator for toxin production, and a higher frequency of resistance to fluoroquinolones. The expansion of hypervirulent strains might have been due to genetic adaptations and environmental factors favoring the transmission of the virulence of C. difficile, but which environmental factors trigger genetic expression is still under investigation [6,8].
The aim of this paper is to review the most important genes involved in C. difficile pathogenesis, identify modifiable environmental, nutritional, and other factors that may trigger the expression of these genes, and thus offer new strategies to prevent CDIs.
2. Overview of Genes Associated with Virulence of C. difficile
A wide variety of genes have been found to be involved in the pathogenesis of C. difficile, including genes producing various toxins, regulator, and promoter genes, genes involved in QS cascades, and the production of important structural components of C. difficile cells (cell walls and flagella) and biofilm formation, as described in Table 1.

Table 1.
Genes involved in the pathogenesis of C. difficile and factors that influence their expression.
2.1. Production of Toxin A and Toxin B: tcdA and tcdB
The symptoms of CDIs result from the production of two exotoxins, which disrupt the cytoskeleton of the intestinal epithelial cells, and higher levels of toxins result in more severe CDIs [9,10]. Toxins A and B are encoded by the genes tcdA (308 kDa) and tcdB (270 kDa), respectively, and are located on the 19.6 Kb pathogenicity loci (PaLoc) on C. difficile’s chromosome (Figure 1A) [11]. The PaLoc possesses a mobile genetic element and is located at the same site in all toxigenic C. difficile strains [12]. Remarkable sequence diversity in genes that encode toxin B exists among different C. difficile strains and may contribute to differences in virulence [13]. Although non-pathogenic strains of C. difficile may lack of tcdA and tcdB genes, the PaLoc locus can be horizontally transferred, converting them into a pathogenic strains [14]. The toxin genes are transcribed when the accumulation of growth-inhibiting substances or nutrient limitation occurs, and QS systems upregulate toxin genes [9,15,16].
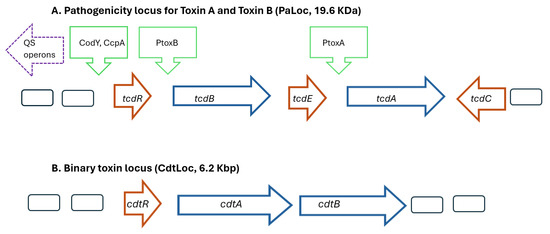
Figure 1.
Schematic models showing the major genes present in the pathogenicity locus regions for (A) Toxins A and B (PaLoc) and (B) binary toxin (CdtLoc) of Clostridioides difficile. Promoter proteins indicated by green arrows, regulatory genes indicated by red arrows, and toxin genes indicated by blue arrows. The agr1 operon located upstream of the PaLoc operon involves genes regulating quorum sensing. Abbreviations: PtoxA Promoter toxin A; PtoxB Promoter toxin B; QS, quorum sensing.
2.2. Regulatory Genes for Toxin A and Toxin B: tcdR, tcdC and tcdE
There are three important regulatory genes involved in the production of Toxin A and Toxin B. The first positive regulator gene (tcdR) codes for an RNA polymerase, TcdR (555 bp), which assists binding of promoters to the genes for Toxin A and Toxin B production [14]. The second positive regulator gene (tcdE) codes for a holin-like protein involved in the cell lysis and subsequent release of both Toxin A and Toxin B [6]. A negative regulator gene (tcdC) codes for an anti-sigma factor (TcdC) that downregulates tcdR expression and suppresses the production of Toxins A and B [17]. Increased virulence observed in the hypervirulent strains has been linked to mutations in the tcdC gene. A deletion of 18 bp in the tcdC gene enhances toxin production, resulting in more severe outcomes and higher mortality rates, highlighting the possible function of TcdC in regulating toxin expression in C. difficile [18].
2.3. Promoter Genes for Toxin A and Toxin B Production: ccpA, codY
Two promoter genes are involved in the production of Toxin A and Toxin B and are responsive to environmental factors. The gene ccpA produces a protein (CcpA), which binds to regulatory regions and reduces the production of these two toxins when sugar levels are low. CcpA is also involved in a diverse number of regulators involved in fermentation processes, especially for butyrate, which is known to increase Toxin A and B production. In addition, CcpA is involved in the sporulation process for C. difficile by repressing SpoOA and SigF, factors involved in the early development of spores. CcpA is also involved in the production of CodY.
The second promoter gene (codY) produces CodY, which is also involved in the expression of SpoOA. Like CcpA, CodY is involved in a wide range of transcription processes involving the production of Toxin A, Toxin B and the production of spores. CodY production is dependent upon nutrient levels in the environment [19,20,21].
2.4. Quorum Sensing Genes: agr1, agr2, luxS
Quorum sensing (QS) involves regulating gene expression in response to bacterial population densities. Different genes are involved in the C. difficile QS system depending upon whether the regulation occurs at an intraspecies level (agr1 and agr2) or at an interspecies level (luxS) [15,22]. Once a threshold of bacterial density is reached, these genes produce autoinducers, which, in turn, stimulate the production of Toxin A and Toxin B [23,24]. The Agr1 locus is found in all virulent C. difficile strains. Hypervirulent strains (such as NAP1/027 R20291) contain an additional Agr2 locus [15,25,26]. The luxS gene produces LuxS, an enzyme that produces auto-inducers that upregulate the expression of tcdA, tcdB, and tcdE, resulting in an increase in Toxin A and B production.
2.5. Binary Toxin Genes: cdtA and cdtB
A third type of toxin is produced by some strains of C. difficile called binary toxin (CDT), made of two polypeptides: CDTa (48 KDa) and CDTb (75 KDa). The cdtA gene encodes CDTa, a G-actin-specific ADP-ribosyl transferase that disrupts the actin cytoskeleton, leading to cell death. The cdtB gene encodes CDTb, which acts to internalize CDTa into the interior of the host cell [27]. The genes for these polypeptides are found on a separate locus (CdtLoc) distinct from the PaLoc locus (Figure 1B). A regulatory gene (cdtR, 747 bp) encodes a 30 KDa protein (CdtR) which results in higher levels of binary toxin [27]. Some strains producing binary toxin but not having the CdtLoc operon have a unique 68 bp sequence at the location but the function has not been well defined [28].
2.6. Adhesion Genes: cwp13, cwp66, cwp84, Etc
The C. difficile S-layer is composed of numerous proteins, and the surface layer protein A (SlpA) is essential for the attachment of C. difficile to intestinal epithelial cells. The different Slps, including SlpA, are part of a class of cell wall proteins (CWPs), including Cwp66, Cwp84, Cwp13, and CwpV, proteins that are involved in the assembly of the S-layer [29,30,31].
2.7. Biofilm Genes: spoOA
C. difficile colonizes the human gut and can persist through its ability to form biofilms, a complex process involving many factors [32,33,34]. Bacteria within biofilms can be more resistant to antibiotics than their planktonic counterparts via several mechanisms. However, the opposite has also been observed since antibiotic molecules can be more concentrated in biofilms [35]. An increased resistance has been observed for C. difficile strains within biofilms [36]. The protein Spo0A is an important contributor to the development of biofilms in C. difficile. It is also a critical factor in persistent infections, including CDI recurrences [36]. High concentrations of Spo0A are involved in sporulation, while intermediate levels activate genes shaping the biofilm matrix [37]. The presence of another protein, LuxS, has been linked with biofilm formation [38].
2.8. Motility Genes: fliC, fliE, filG
Some strains of C. difficile have flagella. These flagella allow movement from one site to another in the gut. The appendages are considered virulence factors and are involved in the modulation of toxin production [39]. FliC, a 39-kDa flagellin protein, and FliD, a 56-kDa flagellar cap protein, are critical structures for C. difficile adherence in gut cells. FliC and FliD have also been shown to trigger an inflammatory response [40,41].
2.9. Genes Involved in Spore Germination: cspA, cspC, spoOA
The formation of spores allows C. difficile to resist contact with antibiotics, oxygen, heat, and common disinfectants. The germination of C. difficile spores depends on the upregulation or downregulation of more than 500 genes on the cspBAC gene locus. Primary bile acids, such as taurocholate, bind to Cspc receptor sites on spore surfaces, followed by calcium and dipocolinic acid release and phosphorylation and activation of the transcription factor Spo0A, which directly regulates dozens of genes [42].
3. Modifiable Parameters of C. difficile Gene Expression
3.1. Antibiotics
Antibiotics are the most common risk factor for antibiotic-associated diarrhea and CDIs [43]. Historically, the elimination or reduction in protective bacterial species in the intestinal microbiome barrier by antibiotics was the accepted mechanism of this risk factor [17,24,43]. However, as our appreciation of the roles different genetic expressions have on C. difficile virulence has grown, the possibility that antibiotics may impact the host by other mechanisms is being explored.
3.1.1. Antibiotics and Toxin A and Toxin B Production
Recent studies have demonstrated the impact of various environmental factors on the expression of tcdA and tcdB genes in C. difficile [9]. Additionally, antibiotics have been shown to alter the pattern of toxin gene expression in C. difficile, as evidenced by in vitro experiments utilizing sub-minimal inhibitory concentrations (MIC) of different antibiotics. Metronidazole, vancomycin, clindamycin, and linezolid have been observed to enhance the transcription rate of genes for Toxin A and B during the exponential phase of bacterial growth in all tested antibiotics, except clindamycin, with C. difficile VPI 10463 (ATCC 43255), strain 2932 (which has a deletion in the PaLoc thus not able to produce Toxin A), strain 47, and strain 317 [44]. It has been demonstrated that the transcription of toxin genes varies depending on the C. difficile strain and the type of antibiotics [45].
C. difficile strain 039 is resistant to ciprofloxacin, and dose-dependent subinhibitory concentrations of ciprofloxacin have been found to significantly increase Toxin A gene expression and shift its expression to earlier in its growth cycle [46]. While a high concentration of ciprofloxacin increased tcdB gene expression, it displayed less sensitivity to low-dose ciprofloxacin. In clinical isolates of C. difficile strains 5325 and BI/NAP1/027, ciprofloxacin at 0.25 MIC markedly boosted both tcdA and tcdB gene expression, albeit their temporal dynamics are similar as compared to the control [46].
Tigecycline, a protein synthesis inhibitor, was studied in vitro in cultures of C. difficile strain 9689 and hypervirulent BI/NAP1/027 (strain 5325) isolates to verify the impact on Toxin A and B production. The antibiotic induced more expression of Toxin A and B genes, and this expression was observed faster during the culture of both strains [47].
During the stationary phase of C. difficile strains UK-1 and CD196, fidaxomicin inhibited the transcripts from the pathogenicity loci (tcdR, tcdA, and tcdB), while vancomycin did not [48].
3.1.2. Antibiotics and Adherence
The initial phase of infection hinges on the adherence to epithelial cells. In a comprehensive study, both toxigenic and non-toxigenic C. difficile isolates were exposed to a range of antibiotics [49]. Notably, exposure to clindamycin and ampicillin prompted a notable increase in the expression of genes encoding colonization factors across all six strains. However, the extent of gene regulation exhibited significant variability among the strains, with cwp84 having the most pronounced upregulation following antibiotic exposure. Conversely, the presence of ofloxacin, moxifloxacin, or kanamycin resulted in minimal alterations in the expression of these genes [49].
In another investigation, the impact of four antibiotics on the expression of genes encoding three colonization factors was examined in four NAP1/027 strains, including one moxifloxacin-susceptible and three moxifloxacin-resistant strains [50]. Interestingly, exposure to ampicillin or clindamycin substantially heightened the expression of cwp84 and slpA in NAP1/027 strains. However, following exposure to fluoroquinolones, increased expression of cwp84 and slpA was only observed in moxifloxacin-resistant strains. These findings underscore the potential of fluoroquinolones to selectively promote the expression of specific colonization factor-encoding genes in resistant C. difficile strains.
3.1.3. Antibiotics and Biofilm Formation
Fidaxomicin and its major metabolite OP-1118 can also be used to treat CDIs and has lower recurrence rates compared with vancomycin [51,52]. Fidaxomicin inhibited biofilm formation by influencing fliC transcription of C. difficile UK027, unlike vancomycin, which did not influence fliC mRNA expression [52].
3.1.4. Antibiotics and Spore Germination
Tigecycline induced a notable increase in spo0A transcription in C. difficile 9689. However, despite this transcriptional upregulation, viable spore formation was consistently reduced across all concentrations of tigecycline. Conversely, in the hypervirulent NAP1 5325 strain, tigecycline did not significantly affect the already elevated level of spo0A transcription at 48 h. Nevertheless, it significantly suppressed spore formation in this strain [47].
3.2. Nutritional Factors
The metabolic network and the nutritional status in the environment play important roles in the growth and expression of pathogenicity of C. difficile [53]. The production of toxins is dependent on the nutrient levels in the growth medium and environmental signals, such as low levels of phosphotransferase system sugars, biotin, and amino acids, especially cysteine [42,54]. The effects of nutritional limitation are varied; indeed, in a complex media, the presence of rapidly metabolizable carbon sources results in lower toxin production, while in contrast, the limitation in biotin leads to an increase in Toxin A and B production [55].
3.2.1. Amino Acids
There are several amino acids capable of regulating the production of C. difficile toxins. However, cysteine remains the most effective [56]. Cysteine negatively regulates several metabolic pathways, including the production of butyric acids and butanol, leading to a reduction in toxin production [54,56]. Cysteine affects many genes linked to NAP1/BI/027, as well as flagellar and ribosomal genes [56]. Moreover, in response to cysteine, the synthesis of toxins and the production of butyrate may be controlled in different ways [56].
3.2.2. Carbohydrates
Thanks to the activity of nutrient-sensitive regulators such as CcpA and CodY, certain sugars and peptides can suppress the expression of C. difficile toxin [9]. The CcpA regulon includes genes involved in fermentation, amino acid metabolism, and sugar absorption, confirming its role in linking carbon and nitrogen pathways [57,58,59]. The inhibition by CodY and CcpA of toxin and nutrient acquisition mechanisms is disrupted when these peptides and simple sugars are depleted. Glucose regulates approximately 20% of all C. difficile genes, with 50% tributary to CcpA for their regulation [57].
3.2.3. Green Tea
Epigallocatechin gallate, present in green tea, was found to downregulate the production of LuxS involved in QS cascades and cause a decrease in the amount of AI-2 and a lower expression of the tcdA and tcdR genes [60].
3.2.4. Bile Acids
The primary bile acid taurocholate can act as a co-germinant of the spores with the glycine, and another primary bile acid (deoxycholate) also supports germination and suppresses vegetative growth. In contrast, chenodeoxycholate inhibits spore germination by the action of taurocholate and suppresses vegetative growth at the same time [61]. The commensal microbiota plays a vital role in the production of secondary bile acids. Most probiotics have been shown to metabolize primary bile acids into secondary bile acids, which do not stimulate spore germination. In a randomized control trial, administration of secondary bile acid to 16 patients with recurrent CDIs resulted in a cure rate of 88% [62].
3.3. Environmental Stimuli
Bacteria must adapt to changes in temperature, nutrient deprivation, radiation, acidity, water scarcity, and more [63]. Variations in gene expression form the primary aspect of bacterial responses to stress and alterations in the environment, engaging diverse mechanisms [64]. Bacteria must constantly adapt physiologically via drastic changes in gene expression. Indeed, environmental conditions are constantly evolving.
3.3.1. Stress
Environmental stresses such as oxidative, nutritional, and osmotic stress can cause damage to C. difficile DNA compromising its genetic stability and ability to survive in adverse conditions. Proteins play a role in DNA repair when responding to stress. Mfd, a highly conserved bacterial protein, connects DNA repair with transcription. Mutations in the mfd gene of C. difficile result in a notable rise in the production of Toxins A and B and abnormal colony morphology. Enhanced transcription of the tcdA and tcdB genes is noted, implying the relaxation of transcriptional repression mediated by CcpA in the mfd mutant CodY. It is possible that Mfd facilitates the release of RNA polymerase molecules obstructed by barriers established by CodY and CcpA [65].
3.3.2. Temperature
Changes in temperature can affect gene expression in bacteria; these changes lead to upregulation of the expression of virulence determinants in many pathogens [66]. Increasing the temperature from 22 to 37 °C showed positive upregulation in tcdA and tcdB with an increase in toxins in the medium in certain strains of C. difficile [67]. These observations show the impact of temperature at the transcriptional level. Toxin production in C. difficile is temperature-dependent and is optimal at 37 °C. An alternative sigma factor, TcdD, encoded by a gene located in PaLoc immediately upstream of tcdB, is required for the temperature-dependent activity of the toxin promoters. Temperature was also found to affect twelve proteins that were previously regulated during biotin deficiency. They matched the criteria for having the highest expression at 37 °C and, thus, high toxin production. Upregulation of toxin synthesis with limitation of biotin and glucose-amino acids occurred at 37 °C but not at 22 °C or 42 °C, indicating that temperature regulation can operate at a basal level [67]. Biotin limitation also causes slower growth and high toxin production, but this can be reversed by adding amino acids such as asparagine, glutamine, or lysine in the medium. Levels of butyric acid production are also regulated by temperature. As for the toxins, expression of 3-hydroxybutyryl-CoA dehydrogenase, a key enzyme in butyric acid production, was at the highest at 37 °C, and the biggest difference (>10-fold) was found between the 22 °C and 37 °C. Other short-chain fatty acids like acetic or propionic did not demonstrate a temperature dependency.
3.3.3. pH
Low pH levels have been implicated in the upregulation of tcdA in some strains of C. difficile, like NAP1/BI/027, but not all C. difficile strains were sensitive to low pH levels in vitro [68]. Lower pH levels have also been found to result in lower sporulation rates [6]. Protein pump inhibitors are considered a risk factor for CDIs, thus highlighting the importance of pH levels, but the role has not been definitively proven [43,69].
3.4. Viral Factors: Prophages
Bacteriophages play a role in the development of most bacteria. Lysogenic phages have the ability to neutralize their host via the reproduction cycle and can create a stable parasitic connection with their host via a lysogenic cycle [70]. C. difficile may carry more than one prophage in most strains, some of which, such as ϕCD38-2 and ϕCD119, have the ability to modulate the expression of virulence genes [71]. The cpC6 phage can decrease the transcription of tcdA [72]. In addition, the lysogenization of C. difficile strains by the ϕCD119 phage results in a decrease in the production of Toxins A and B and the transcription of the PaLoc (tcdA, tcdB, tcdE, tcdR and tcdC). Indeed, a phage repressor, encoded by the repR gene, binds to the promoter region upstream of the tcdR gene, a positive transcriptional regulator gene of the pathogenicity locus. This interaction results in repressing the transcription of PaLoc genes and affects the production of Toxins A and B. Thus, phages are able to influence major bacterial phenotypes, such as toxin production [71].
Recently, a complete binary toxin locus (CdtLoc) was discovered in the genome of the phiSemix9P1 phage. This phage has been shown to carry a complete binary toxin locus. Additionally, several regulatory genes have been identified in the genomes of these phages, suggesting that their interactions with the host may be complex and often nuanced [73]. Phages can also be involved in cell-to-cell communication via QS. Research has shown that three homologs of bacterial genes involved in QS (agrB, agrC, and agrD) are encoded by the PhiCDHM1 prophage [74]. This implies that C. difficile prophages could express genes capable of influencing bacterial communities. The study of gene expression in biofilms of wild-type strains and luxS mutants indicated a negative regulation of prophage loci in luxS mutant biofilms [31]. The release of DNA by phage-mediated cell lysis likely contributes to the formation of C. difficile biofilms [38].
3.5. Microbial Factors: Probiotics
The treatment of patients with antibiotics is a standard treatment for CDIs, but probiotics have also been used to treat and prevent CDIs [75,76]. The most utilized types of probiotics are usually in the family Lactobacillaceae or within the genera Bifidobacterium spp. or yeast (Saccharomyces spp.), which show different efficiencies with CDIs. The efficacy and mechanisms of action of probiotics in regulating intestinal microbiota functions are both disease-specific and strain-specific [77]. All probiotic strains do not fight against all pathogens, as not all probiotic strains possess the same type of mechanisms, and one probiotic strain may be effective for different pathogens, depending on the type of pathogen-specific defenses it possesses. The mechanisms can be diverse, including the production of antimicrobial metabolites, bacteriocins, organic acids or short chain fatty acids (SCFAs) as well as restricting pathogenic growth by competing for the nutrients and adherence on the mucosal barrier in the gut, production of toxin-destructive proteases and the regulation of the immune response [69]. Lactic acid bacteria (LAB) such as lactobacilli are beneficial microorganisms present in healthy human microbiota and have been associated with immunomodulatory effects, reduction in pathogenic bacteria levels, reduction in antibiotic-associated gastrointestinal symptoms, reduction in acute diarrhea, inflammatory bowel disease, and allergy symptoms [78,79]. These mechanisms have been well-studied for a variety of probiotic types, but whether probiotics can affect C. difficile on a genetic level has not been well documented.
Technological advances allow us to study all messenger RNAs produced during the transcription process of the C. difficile genome, notably in contact with probiotic strains. The study of gene expression of C. difficile is essential to understanding the mechanisms of action of probiotics against C. difficile. At transcriptional levels for virulence-associated genes in C. difficile on ribotypes 027, 078, and 001, the luxS and txeR genes are decreased with the presence of Lactobacillus acidophilus GP1B [80]. These reduced levels of expression appeared to affect the transcription of tcdA and tcdB genes. Moreover, the downregulation of luxS induces the reduction in AI-2 levels. The addition of a cell extract of L. acidophilus GP1B resulted in the downregulation of virulence genes in C. difficile at the level of mRNA. L. acidophilus GP1B is not the only probiotic that can impact C. difficile gene expression.
L. acidophilus La-5 secretes bioactive molecules namely proteobiotics, which also causes a significant downregulation of tcdA and tcdB and cwp84 in C. difficile ribotypes 027, 078, and 001 [81].
Lactobacillus fermentum Lim2 isolated from kimchi can affect the QS and virulence factors of C. difficile 027. A gene expression analysis indicated that the presence of 100 mg per mL or heat-inactivated cell extract significantly suppressed the QS (luxS) and the virulence factors (tcdA, tcdB, and tcdE) while upregulating the negative regulator gene (tcdC) [82].
A three-strain probiotic blend (L. acidophilus CL1285, Lacticaseibacillus casei LBC80R, Lacticaseibacillus rhamnosus CLR2, Bio-K+) was found to alter genetic expression in C. difficile. A study detected 1156 differently expressed genes in C. difficile with this Lactobacilli mixture. The effect of the Lactobacilli probiotic included the overexpression of CDT binary toxin genes, underexpression of fliC gene and other flagella genes, up- and downregulation of different sporulation genes (spoOA), reduction in QS (luxS) genes, up- and downregulation of different cwp adhesion genes, but no effect on Toxin A or B gene expression [83].
Other genus of bacteria, such as Bifidobacterium spp., can modulate the expression of C. difficile genes. Bifidobacterium breve YH68 finds extensive application in food fermentation and biomedical fields. The cell-free supernatant (CFS) of B. breve YH68 notably decreased the gene expression levels of tcdA and tcdB in C. difficile ATCC 9689 [84]. Previous research has linked the expression of tcdA/B to the production of autoinducer-2 (AI-2) by C. difficile, an important structure of its quorum sensing (QS) system [80]. Consequently, B. breve YH68 CFS curbed C. difficile pathogenicity by inhibiting QS, subsequently leading to the downregulation of both tcdA and tcdB [84,85]. A more recent transcriptomic investigation explored the impact of varying doses of B. breve YH68 (108–1012) on C. difficile ATCC 9689 [86]. Both doses of B. breve YH68-CFS induced significant alterations in activities in these metabolic pathways [86]. Notably, the high dose of probiotics led to significant suppression of genes associated with QS and signal transduction while enhancing those encoding toxin production and sporulation factors. Conversely, the low dose of B. breve resulted in the suppression of genes related to flagellar assembly (e.g., fliD, fliC, and flgE) and biofilm formation, alongside significant upregulation of drug resistance-related genes.
Investigations of other types of Bifidobacterial and Lactobacilli probiotics on C. difficile gene expression involved in QS cascade pathways are also ongoing [78].
4. Discussion
Our review found several types of environmental and nutritional factors that act as triggers for upregulating or downregulating the genetic expression of genes involved in the virulence pathways of C. difficile. This is an important finding to clinicians and researchers, as these triggers may be amenable to modifying the risk of patients from developing CDIs and may provide new therapeutic strategies for the treatment of CDIs. However, it is apparent that further research is needed to understand how and when various genes are expressed and if there are additional triggers that can be identified. Future studies could also investigate if these diverse factors act synergistically or if there are significant interactions between them. The strengths of our review include an extensive search of the literature relating to the genetic expression of C. difficile virulence factors and associated factors that might trigger the transcription of these genes. In addition, this review had the original goal to link these triggers with the known roles of the genes involved in the pathogenesis of C. difficile. Limitations of this review include the relative scarcity of studies linking environmental triggers to gene expression. Also, we did not include small non-coding RNA regulators in our review (such as Hfq and KhpB), as we could not find a link with any environmental or nutritional factors [87,88]. Continued research into these fields would be most useful.
5. Conclusions
Recent investigations into the genetic expression for C. difficile virulence factors have revealed a deeper understanding of the pathogenesis of this infection. In addition to unraveling how and when C. difficile toxins are produced, our understanding of the importance of other types of genes involved in pathogenesis, such as genes for adhesion, biofilm formation, motility, and spore germination, has increased. A wider appreciation for the diversity of triggers for gene expression, including antibiotic exposure, nutritional factors, environmental factors, prophage, and probiotic strains, has been gained by recent studies. Translating these results into clinical strategies for CDIs will require future studies but offers enticing alternatives to current therapies.
Author Contributions
Conceptualization, Z.M., M.M. and M.L.; Data curation, S.G. and M.M.; Formal analysis, Z.M., S.G., M.M. and M.L.; Investigation, Z.M., S.G., M.M. and M.L.; Methodology, Z.M. and S.G.; Project administration, M.M.; Resources, Z.M. and M.M.; Software, M.M.; Supervision, M.M. and M.L.; Validation, S.G. and M.M.; Visualization, L.V.M.; Writing—original draft, Z.M. and M.M.; Writing—review and editing, Z.M., S.G., M.M., L.V.M. and M.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Engineering Research Council of Canada (NSERC), grant number “CRDPJ 505365-16”, Mitacs and Bio-K+/Kerry.
Conflicts of Interest
M.M. and S.G. are paid employees of Bio-K+/Kerry. L.V.M. is a member of the advisory board of Bio-K+/Kerry. The remaining authors (Z.M., M.L.) declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Centers for Communicable Diseases (CDC). National and State Healthcare-Associated Infections Progress Report. 2020. Available online: https://www.cdc.gov/hai/data/portal/progress-report.html (accessed on 2 February 2023).
- Wiuff, C.; Banks, A.L.; Fitzpatrick, F.; Cottom, L. The need for European surveillance of CDI. In Updates on Clostridium difficile in Europe. Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2018; Volume 1050, pp. 13–25. [Google Scholar]
- Feuerstadt, P.; Boules, M.; Strong, L.; Dahdal, D.N.; Sacks, N.C.; Lang, K.; Nelson, W.W. Clinical complications in patients with primary and recurrent Clostridioides difficile infection: A real-world data analysis. SAGE Open Med. 2021, 9, 2050312120986733. [Google Scholar] [CrossRef] [PubMed]
- McFarland, L.V.; Goldstein, E.J.C.; Kullar, R. Microbiome-related and infection control approaches to primary and secondary prevention of Clostridioides difficile infections. Microorganisms 2023, 11, 1534. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Monaghan, T.; Yadegar, A.; Louie, T.; Kao, D. Insights into the evolving epidemiology of Clostridioides difficile infection and treatment: A global perspective. Antibiotics 2023, 12, 1141. [Google Scholar] [CrossRef] [PubMed]
- Buddle, J.E.; Fagan, R.P. Pathogenicity and virulence of Clostridioides difficile. Virulence 2023, 14, 2150452. [Google Scholar] [CrossRef] [PubMed]
- Aldeyab, M.A.; Devine, M.J.; Flanagan, P.; Mannion, M.; Craig, A.; Scott, M.G.; Harbarth, S.; Vernaz, N.; Davies, E.; Brazier, J.S.; et al. Multihospital outbreak of Clostridium difficile ribotype 027 infection: Epidemiology and analysis of control measures. Infect. Cont. Hosp. Epidemiol. 2011, 32, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Paquette, I.M.; Stewart, D.B. Clostridium difficile Infection. In The ASCRS Textbook of Colon and Rectal Surgery; Springer: Berlin/Heidelberg, Germany, 2022; pp. 879–891. [Google Scholar]
- Majumdar, A.; Govind, R. Regulation of Clostridioides difficile toxin production. Curr. Opin. Microbiol. 2022, 65, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Fatima, R.; Aziz, M. The hypervirulent strain of Clostridium difficile: NAP1/B1/027—A brief overview. Cureus 2019, 11, e3977. [Google Scholar] [CrossRef] [PubMed]
- Vaishnavi, C. Virulence factors associated with Clostridiodes difficile: An overview. J. Gastro Infect. 2021, 11, 25. [Google Scholar] [CrossRef]
- Saund, K.; Pirani, A.; Lacy, B.; Hanna, P.C.; Snitkin, E.S. Strain variation in Clostridioides difficile toxin activity associated with genomic variation at both PaLoc and non-PaLoc loci. bioRxiv 2021. [Google Scholar] [CrossRef]
- Shen, J.; Mehrotra, D.V.; Dorr, M.B.; Zeng, Z.; Li, J.; Xu, X.; Nickle, D.; Holzinger, E.R.; Chhibber, A.; Wilcox, M.H. Genetic association reveals protection against recurrence of Clostridium difficile infection with bezlotoxumab treatment. Msphere 2020, 5, e00232-20. [Google Scholar] [CrossRef]
- Di Bella, S.; Ascenzi, P.; Siarakas, S.; Petrosillo, N.; Di Masi, A. Clostridium difficile toxins A and B: Insights into pathogenic properties and extraintestinal effects. Toxins 2016, 8, 134. [Google Scholar] [CrossRef] [PubMed]
- Gunaratnam, S.; Millette, M.; McFarland, L.V.; DuPont, H.L.; Lacroix, M. Potential role of probiotics in reducing Clostridioides difficile virulence: Interference with quorum sensing systems. Microb. Pathog. 2021, 153, 104798. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, R.; Lacy, D.B. The role of toxins in Clostridium difficile infection. FEMS Microbiol. Rev. 2017, 41, 723–750. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.R.; Pike, C.M.; Parsons, R.J.; Rivera, A.J.; Foley, M.H.; McLaren, M.R.; Montgomery, S.A.; Theriot, C.M. Clostridioides difficile exploits toxin-mediated inflammation to alter the host nutritional landscape and exclude competitors from the gut microbiota. Nat. Commun. 2021, 12, 462. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.S.; Li, W.G.; Zhang, W.Z.; Wu, Y.; Lu, J.X. Molecular characterization of Clostridium difficile isolates in China from 2010 to 2015. Front. Microbiol. 2018, 9, 845. [Google Scholar] [CrossRef] [PubMed]
- Neumann-Schaal, M.; Jahn, D.; Schmidt-Hohagen, K. Metabolism the difficile way: The key to the success of the pathogen Clostridioides difficile. Front. Microbiol. 2019, 10, 219. [Google Scholar] [CrossRef] [PubMed]
- Daou, N.; Wang, Y.; Levdikov, V.M.; Nandakumar, M.; Livny, J.; Bouillaut, L.; Blagova, E.; Zhang, K.; Belitsky, B.R.; Rhee, K. Impact of CodY protein on metabolism, sporulation and virulence in Clostridioides difficile ribotype 027. PLoS ONE 2019, 14, e0206896. [Google Scholar] [CrossRef] [PubMed]
- Dineen, S.S.; McBride, S.M.; Sonenshein, A.L. Integration of metabolism and virulence by Clostridium difficile CodY. J. Bacteriol. 2010, 192, 5350–5362. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, U.K.B.; Ballard, J.D. Autoinducing peptide-based quorum signaling systems in Clostridioides difficile. Curr. Opin. Microbiol. 2022, 65, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Boo, A.; Amaro, R.L.; Stan, G.B. Quorum sensing in synthetic biology: A review. Curr. Opin. Syst. Biol. 2021, 28, 100378. [Google Scholar] [CrossRef]
- Nag, M.; Lahiri, D.; Ghosh, A.; Das, D.; Ray, R.R. Quorum sensing. In Biofilm-Mediated Diseases: Causes and Controls; Springer: Singapore, 2021; pp. 21–45. [Google Scholar] [CrossRef]
- Darkoh, C.; Odo, C.; DuPont, H.L. Accessory gene regulator-1 locus is essential for virulence and pathogenesis of Clostridium difficile. Mbio 2016, 7, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.; Mawer, D.; Wilcox, M.H. Clostridium difficile: Biological therapies. Curr. Opin. Infect. Dis. 2013, 26, 454–460. [Google Scholar] [CrossRef]
- Young, M.; Leslie, J.L.; Madden, G.R.; Lyerly, D.M.; Carman, R.J.; Lyerly, M.W.; Stewart, D.B.; Abhyankar, M.M.; Petri, W.A., Jr. Binary toxin expression by Clostridioides difficile is associated with worse disease. Open For. Infect. Dis. 2022, 9, ofac001. [Google Scholar] [CrossRef] [PubMed]
- Monot, M.; Eckert, C.; Lemire, A.; Hamiot, A.; Dubois, T.; Tessier, C.; Dumoulard, B.; Hamel, B.; Petit, A.; Lalande, V. Clostridium difficile: New insights into the evolution of the pathogenicity locus. Sci. Rep. 2015, 5, 15023. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Rao, F.; Chen, Z.; Cheng, Y.; Zhang, Q.; Zhang, J.; Guan, Z.; He, Y.; Yu, W.; Cui, G. The cwp66 gene affects cell adhesion, stress tolerance, and antibiotic resistance in Clostridioides difficile. Microbiol. Spect. 2022, 10, e02704-21. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, Y.D.; Dupuy, B. The blueprint for building a biofilm the Clostridioides difficile way. Curr. Opin. Microbiol. 2022, 66, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Taggart, M.G.; Snelling, W.J.; Naughton, P.J.; La Ragione, R.M.; Dooley, J.S.; Ternan, N.G. Biofilm regulation in Clostridioides difficile: Novel systems linked to hypervirulence. PLoS Path 2021, 17, e1009817. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.B.; Emerson, J.E.; De la Riva, L.; Fagan, R.P.; Fairweather, N.F. The Clostridium difficile cell wall protein CwpV is antigenically variable between strains, but exhibits conserved aggregation-promoting function. PLoS Path 2011, 7, e1002024. [Google Scholar] [CrossRef] [PubMed]
- Vuotto, C.; Donelli, G.; Buckley, A.; Chilton, C. Clostridium difficile biofilm. In Updates on Clostridium difficile in Europe: Advances in Microbiology, Infectious Diseases and Public Health; Springer: Berlin/Heidelberg, Germany, 2018; Volume 8, pp. 97–115. [Google Scholar]
- Frost, L.R.; Cheng, J.K.; Unnikrishnan, M. Clostridioides difficile biofilms: A mechanism of persistence in the gut? PLoS Path 2021, 17, e1009348. [Google Scholar] [CrossRef] [PubMed]
- Arora, G.; Sajid, A.; Virmani, R.; Singhal, A.; Kumar, C.S.; Dhasmana, N.; Khanna, T.; Maji, A.; Misra, R.; Molle, V. Ser/Thr protein kinase PrkC-mediated regulation of GroEL is critical for biofilm formation in Bacillus anthracis. NPJ Biofilms Microbiomes 2017, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Dapa, T.; Unnikrishnan, M. Biofilm formation by Clostridium difficile. Gut Microbiol. 2013, 4, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, O.A. Characterisation of Reduced Susceptibility to Metronidazole in Epidemic Clostridium difficile Clinical Isolates. Ph.D. Thesis, University of Hertfordshire, Hatfield, UK, 2018. [Google Scholar] [CrossRef]
- Slater, R.T.; Frost, L.R.; Jossi, S.E.; Millard, A.D.; Unnikrishnan, M. Clostridioides difficile LuxS mediates inter-bacterial interactions within biofilms. Sci. Rep. 2019, 9, 9903. [Google Scholar] [CrossRef] [PubMed]
- Aubry, A.; Hussack, G.; Chen, W.; KuoLee, R.; Twine, S.M.; Fulton, K.M.; Foote, S.; Carrillo, C.D.; Tanha, J.; Logan, S.M. Modulation of toxin production by the flagellar regulon in Clostridium difficile. Infect. Immun. 2012, 80, 3521–3532. [Google Scholar] [CrossRef] [PubMed]
- Batah, J.; Kansau, I. Intestinal epithelial cell response to Clostridium difficile flagella. In Clostridium difficile: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2016; pp. 103–116. [Google Scholar]
- Dingle, T.; Mulvey, G.L.; Armstrong, G.D. Mutagenic analysis of the Clostridium difficile flagellar proteins, FliC and FliD, and their contribution to virulence in hamsters. Infect. Immun. 2011, 79, 4061–4067. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.N.; Willams, C.L.; Pareek, N.; McBride, S.M.; Tamayo, R. c-di-GMP inhibits early sporulation in Clostridioides difficile. Msphere 2021, 6, e00919–e00921. [Google Scholar] [CrossRef] [PubMed]
- McFarland, L.V.; Ozen, M.; Dinleyici, E.C.; Goh, S. Comparison of pediatric and adult antibiotic-associated diarrhea and Clostridium difficile infections. World J. Gastroenterol. 2016, 22, 3078–3104. [Google Scholar] [CrossRef] [PubMed]
- Gerber, M.; Walch, C.; Löffler, B.; Tischendorf, K.; Reischl, U.; Ackermann, G. Effect of sub-MIC concentrations of metronidazole, vancomycin, clindamycin and linezolid on toxin gene transcription and production in Clostridium difficile. J. Med. Microbiol. 2008, 57, 776–783. [Google Scholar] [CrossRef] [PubMed]
- Moradi, M.; Mansouri, S.; Nakhaee, N.; Sarafzadeh, F.; Zarandi, E.R. Toxin A and B genes expression of Clostridium difficile in the sub-minimum inhibitory concentration of clindamycin, vancomycin and in combination with ceftazidime. Iran. J. Microbiol. 2020, 12, 18. [Google Scholar] [CrossRef]
- Aldape, M.J.; Packham, A.E.; Nute, D.W.; Bryant, A.E.; Stevens, D.L. Effects of ciprofloxacin on the expression and production of exotoxins by Clostridium difficile. J. Med. Microbiol. 2013, 62 Pt 5, 741. [Google Scholar] [CrossRef] [PubMed]
- Aldape, M.J.; Heeney, D.D.; Bryant, A.E.; Stevens, D.L. Tigecycline suppresses toxin A and B production and sporulation in Clostridium difficile. J. Antimicrob. Chemother. 2015, 70, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Babakhani, F.; Bouillaut, L.; Sears, P.; Sims, C.; Gomez, A.; Sonenshein, A.L. Fidaxomicin inhibits toxin production in Clostridium difficile. J. Antimicrob. Chemother. 2013, 68, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Deneve, C.; Delomenie, C.; Barc, M.C.; Collignon, A.; Janoir, C. Antibiotics involved in Clostridium difficile-associated disease increase colonization factor gene expression. J. Med. Microbiol. 2008, 57, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Denève, C.; Bouttier, S.; Dupuy, B.; Barbut, F.; Collignon, A.; Janoir, C. Effects of subinhibitory concentrations of antibiotics on colonization factor expression by moxifloxacin-susceptible and moxifloxacin-resistant Clostridium difficile strains. Antimicrob. Agents Chemother. 2009, 53, 5155–5162. [Google Scholar] [CrossRef] [PubMed]
- Mikamo, H.; Tateda, K.; Yanagihara, K.; Kusachi, S.; Takesue, Y.; Miki, T.; Oizumi, Y.; Gamo, K.; Hashimoto, A.; Toyoshima, J. Efficacy and safety of fidaxomicin for the treatment of Clostridioides (Clostridium) difficile infection in a randomized, double-blind, comparative Phase III study in Japan. J. Infect. Chemother. 2018, 24, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.; Yamaguchi, T.; Ishii, Y.; Chono, K.; Tateda, K. Inhibitory effect of fidaxomicin on biofilm formation in Clostridioides difficile. J. Infect. Chemother. 2020, 26, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, J.D.; Otto, A.; Berges, M.; Biedendieck, R.; Michel, A.M.; Becher, D.; Jahn, D.; Neumann-Schaal, M. Metabolic reprogramming of Clostridioides difficile during the stationary phase with the induction of toxin production. Front. Microbiol. 2018, 9, 1970. [Google Scholar] [CrossRef] [PubMed]
- Dubois, T.; Dancer-Thibonnier, M.; Monot, M.; Hamiot, A.; Bouillaut, L.; Soutourina, O.; Martin-Verstraete, I.; Dupuy, B. Control of Clostridium difficile physiopathology in response to cysteine availability. Infect. Immun. 2016, 84, 2389–2405. [Google Scholar] [CrossRef] [PubMed]
- Sommermeyer, H.; Piątek, J. Pathophysiology of C. difficile. In Clostridioides difficile: Infections, Risk Factors, Prevention and Treatment; Springer International Publisher: New York, NY, USA, 2021; pp. 19–33. [Google Scholar]
- Gu, H.; Shi, K.; Liao, Z.; Qi, H.; Chen, S.; Wang, H.; Li, S.; Ma, Y.; Wang, J. Time-resolved transcriptome analysis of Clostridium difficile R20291 response to cysteine. Microbiol. Res. 2018, 215, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, K.L.; Wetzel, D.; Jones, J.B.; Woods, E.C.; McBride, S.M. Ethanolamine is a valuable nutrient source that impacts Clostridium difficile pathogenesis. Environ. Microbiol. 2018, 20, 1419–1435. [Google Scholar] [CrossRef] [PubMed]
- Antunes, A.; Martin-Verstraete, I.; Dupuy, B. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol. Microbiol. 2011, 79, 882–899. [Google Scholar] [CrossRef]
- Antunes, A.; Camiade, E.; Monot, M.; Courtois, E.; Barbut, F.; Sernova, N.V.; Rodionov, D.A.; Martin-Verstraete, I.; Dupuy, B. Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucl. Acids Res. 2012, 40, 10701–10718. [Google Scholar] [CrossRef] [PubMed]
- Yun, B.; Oh, S.; Song, M.; Hong, Y.S.; Park, S.; Park, D.J.; Griffiths, M.W.; Oh, S. Inhibitory effect of epigallocatechin gallate on the virulence of Clostridium difficile PCR ribotype 027. J. Food Sci. 2015, 80, M2925–M2931. [Google Scholar] [CrossRef] [PubMed]
- Budi, N.; Safdar, N.; Rose, W.E. Treatment issues in recurrent Clostridioides difficile and the possible role of germinants. FEMS Microbes 2020, 1, xtaa001. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.J.; Brunner, A.; Lewis, J.; Ford, C.D.; Lopansri, B.K. Repurposing an old drug for a new epidemic: Ursodeoxycholic acid to prevent recurrent Clostridioides difficile infection. Clin. Infect. Dis. 2019, 68, 498–500. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Rahal, A.; Sohal, J.S.; Gupta, V.K. Bacterial stress response: Understanding the molecular mechanics to identify possible therapeutic targets. Expert Rev. Anti-Infect. Ther. 2021, 19, 121–127. [Google Scholar] [CrossRef] [PubMed]
- De Bruijn, F.J. Introduction. In Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria; De Bruijn, F.J., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2016; Volume 1, pp. 1–2. [Google Scholar]
- Willing, S.E.; Richards, E.J.; Sempere, L.; Dale, A.G.; Cutting, S.M.; Fairweather, N.F. Increased toxin expression in a Clostridium difficile mfd mutant. BMC Microbiol. 2015, 15, 280. [Google Scholar] [CrossRef] [PubMed]
- Konkel, M.E.; Tilly, K. Temperature-regulated expression of bacterial virulence genes. Microb. Infect. 2000, 2, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, S.; Dupuy, B.; Mukherjee, K.; Norin, E.; Burman, L.G.; Akerlund, T. Expression of Clostridium difficile toxins A and B and their sigma factor TcdD is controlled by temperature. Infect. Immun. 2003, 71, 1784–1793. [Google Scholar] [CrossRef] [PubMed]
- Stewart, D.; Hegarty, J.P. Correlation between virulence gene expression and proton pump inhibitors and ambient pH in Clostridium difficile: Results of an in vitro study. J. Med. Microbiol. 2013, 62, 1517–1523. [Google Scholar] [CrossRef] [PubMed]
- Mekonnen, S.A.; Merenstein, D.; Fraser, C.M.; Marco, M.L. Molecular mechanisms of probiotic prevention of antibiotic-associated diarrhea. Curr. Opin. Biotechnol. 2020, 61, 226–234. [Google Scholar] [CrossRef]
- Schroven, K.; Aertsen, A.; Lavigne, R. Bacteriophages as drivers of bacterial virulence and their potential for biotechnological exploitation. FEMS Microbiol. Rev. 2021, 45, fuaa041. [Google Scholar] [CrossRef] [PubMed]
- Fortier, L.C. Bacteriophages contribute to shaping Clostridioides (Clostridium) difficile species. Front. Microbiol. 2018, 9, 2033. [Google Scholar] [CrossRef] [PubMed]
- Govind, R.; Vediyappan, G.; Rolfe, R.D.; Dupuy, B.; Fralick, J.A. Bacteriophage-mediated toxin gene regulation in Clostridium difficile. J. Virol. 2009, 83, 12037–12045. [Google Scholar] [CrossRef]
- Riedel, T.; Wittmann, J.; Bunk, B.; Schober, I.; Spröer, C.; Gronow, S.; Overmann, J. A Clostridioides difficile bacteriophage genome encodes functional binary toxin-associated genes. J. Biotechnol. 2017, 250, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, K.R.; Kropinski, A.M.; Clokie, M.R. What does the talking?: Quorum sensing signalling genes discovered in a bacteriophage genome. PLoS ONE 2014, 9, e85131. [Google Scholar] [CrossRef] [PubMed]
- Su, G.L.; Ko, C.W.; Bercik, P.; Falck-Ytter, Y.; Sultan, S.; Weizman, A.V.; Morgan, R.L. AGA clinical practice guidelines on the role of probiotics in the management of gastrointestinal disorders. Gastroenterology 2020, 159, 697–705. [Google Scholar] [CrossRef] [PubMed]
- McFarland, L.V.; Ship, N.; Auclair, J.; Millette, M. Primary prevention of Clostridium difficile infections with a specific probiotic combining Lactobacillus acidophilus, L. casei and L. rhamnosus strains: Assessing the evidence. J. Hosp. Infect. 2018, 99, 443–452. [Google Scholar] [CrossRef] [PubMed]
- McFarland, L.V.; Evans, C.T.; Goldstein, E.J.C. Strain-specificity and disease-specificity of probiotic efficacy: A systematic review and meta-analysis. Front. Med. 2018, 5, 124. [Google Scholar] [CrossRef]
- Guglielmetti, S.; Mora, D.; Gschwender, M.; Popp, K. Randomised clinical trial: Bifidobacterium bifidum MIMBb75 significantly alleviates irritable bowel syndrome and improves quality of life—A double-blind, placebo-controlled study. Aliment. Pharmacol. Ther. 2011, 33, 1123–1132. [Google Scholar] [CrossRef] [PubMed]
- Sniffen, J.C.; McFarland, L.V.; Evans, C.T.; Goldstein, E.J.C. Choosing an appropriate probiotic product for your patient: An evidence-based practical guide. PLoS ONE 2018, 13, e0209205. [Google Scholar] [CrossRef] [PubMed]
- Yun, B.; Oh, S.; Griffiths, M. Lactobacillus acidophilus modulates the virulence of Clostridium difficile. J. Dairy Sci. 2014, 97, 4745–4758. [Google Scholar] [CrossRef] [PubMed]
- Najarian, A.; Sharif, S.; Griffiths, M. Evaluation of protective effect of Lactobacillus acidophilus La-5 on toxicity and colonization of Clostridium difficile in human epithelial cells in vitro. Anaerobe 2019, 55, 142–151. [Google Scholar] [CrossRef]
- Yong, C.; Lim, J.; Kim, B.K.; Park, D.J.; Oh, S. Suppressive effect of Lactobacillus fermentum Lim2 on Clostridioides difficile 027 toxin production. Lett. Appl. Microbiol. 2019, 68, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Masset, Z.; Gunaratnam, S.; Millette, M.; McFarland, L.V.; Lacroix, M. Transcriptome analysis of the Clostridioides difficile response to a specific lactobacilli probiotic formulation: Explanations for its mechanisms of action. J. Appl. Microbiol. 2023, 134, lxad047. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yang, H. Antibacterial activity of Bifidobacterium breve against Clostridioides difficile. Front. Cell Infect. Microbiol. 2019, 9, 288. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Schmidt, D.; Liu, W.; Li, S.; Shi, L.; Sheng, J.; Chen, K.; Yu, H.; Tremblay, J.M.; Chen, X. A novel multivalent, single-domain antibody targeting TcdA and TcdB prevents fulminant Clostridium difficile infection in mice. J. Infect. Dis. 2014, 210, 964–972. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yang, H. Evaluation of the therapeutic effect and dose–effect of Bifidobacterium breve on the primary Clostridioides difficile infected mice. Appl. Microbiol. Biotechnol. 2021, 105, 9243–9260. [Google Scholar] [CrossRef] [PubMed]
- Lamm-Schmidt, V.; Fuchs, M.; Sulzer, J.; Gerovac, M.; Hör, J.; Dersch, P.; Vogel, J.; Faber, F. Grad-seq identifies KhpB as a global RNA-binding protein in Clostridioides difficile that regulates toxin production. Microlife 2021, 2, uqab004. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, M.; Lamm-Schmidt, V.; Sulzer, J.; Ponath, F.; Jenniches, L.; Kirk, J.A.; Fagan, R.P.; Barquist, L.; Vogel, J.; Faber, F. An RNA-centric global view of Clostridioides difficile reveals broad activity of Hfq in a clinically important gram-positive bacterium. Proc. Nat. Acad. Sci. USA 2021, 118, e2103579118. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).