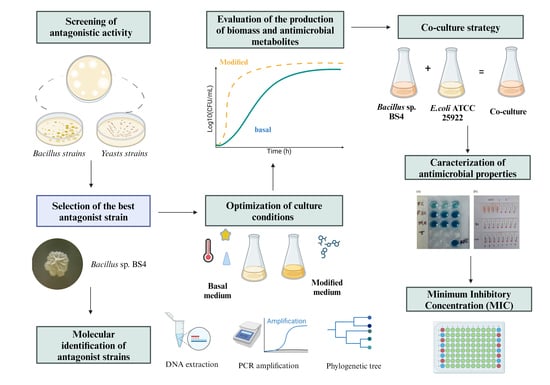
Antimicrobial Activity of Bacillus amyloliquefaciens BS4 against Gram-Negative Pathogenic Bacteria
Abstract
1. Introduction
2. Results
2.1. Screening of Antagonistic Yeast
2.2. Screening of Antagonistic Bacteria
2.3. Effect of Different Culture Conditions on Enhancing Antimicrobial Activity of the Bacillus sp. BS4 Strain
2.3.1. Selection of Fermentation Medium
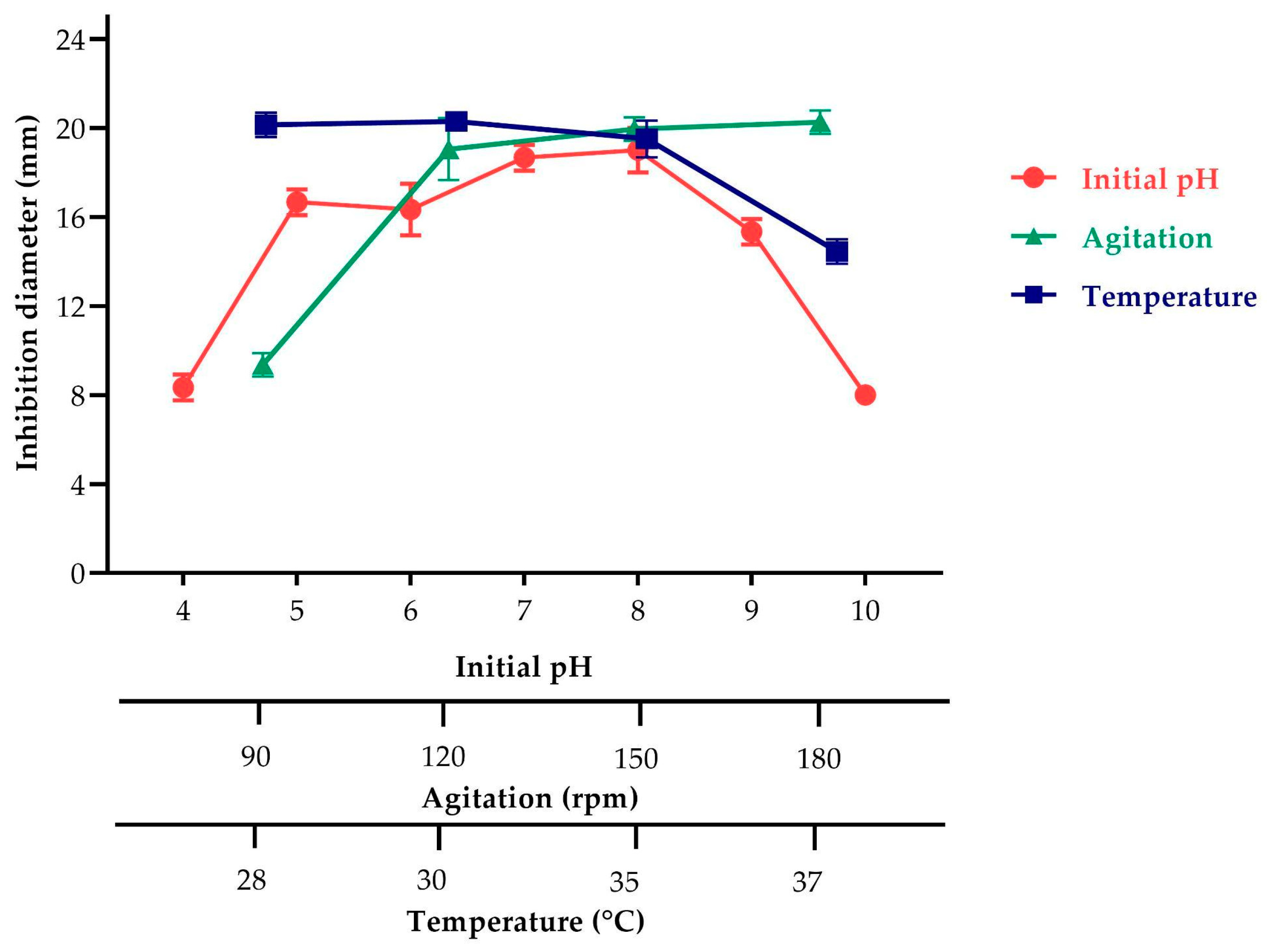
2.3.2. Effect of Agitation, pH, and Temperature on Antimicrobial Metabolite Production
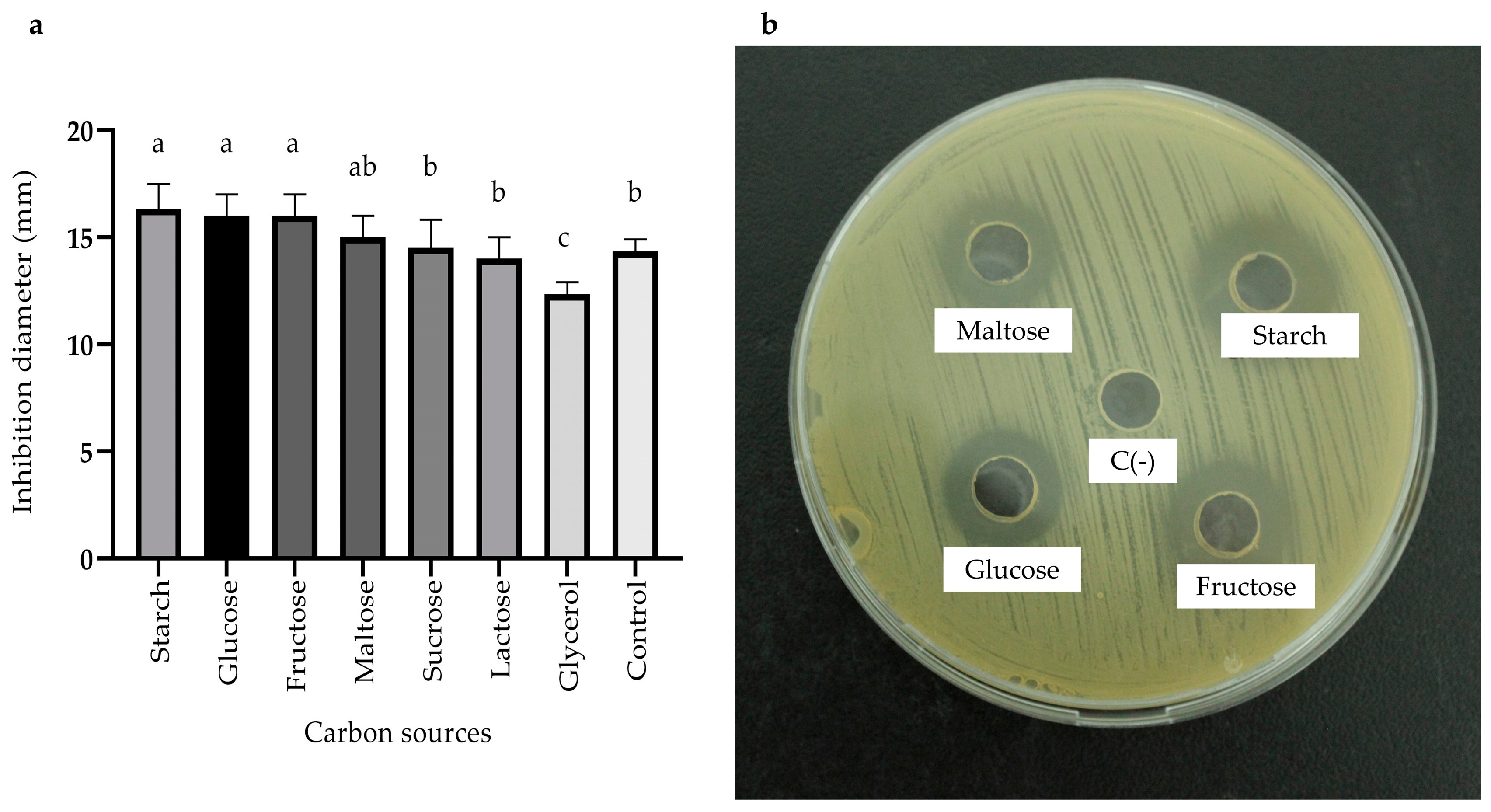
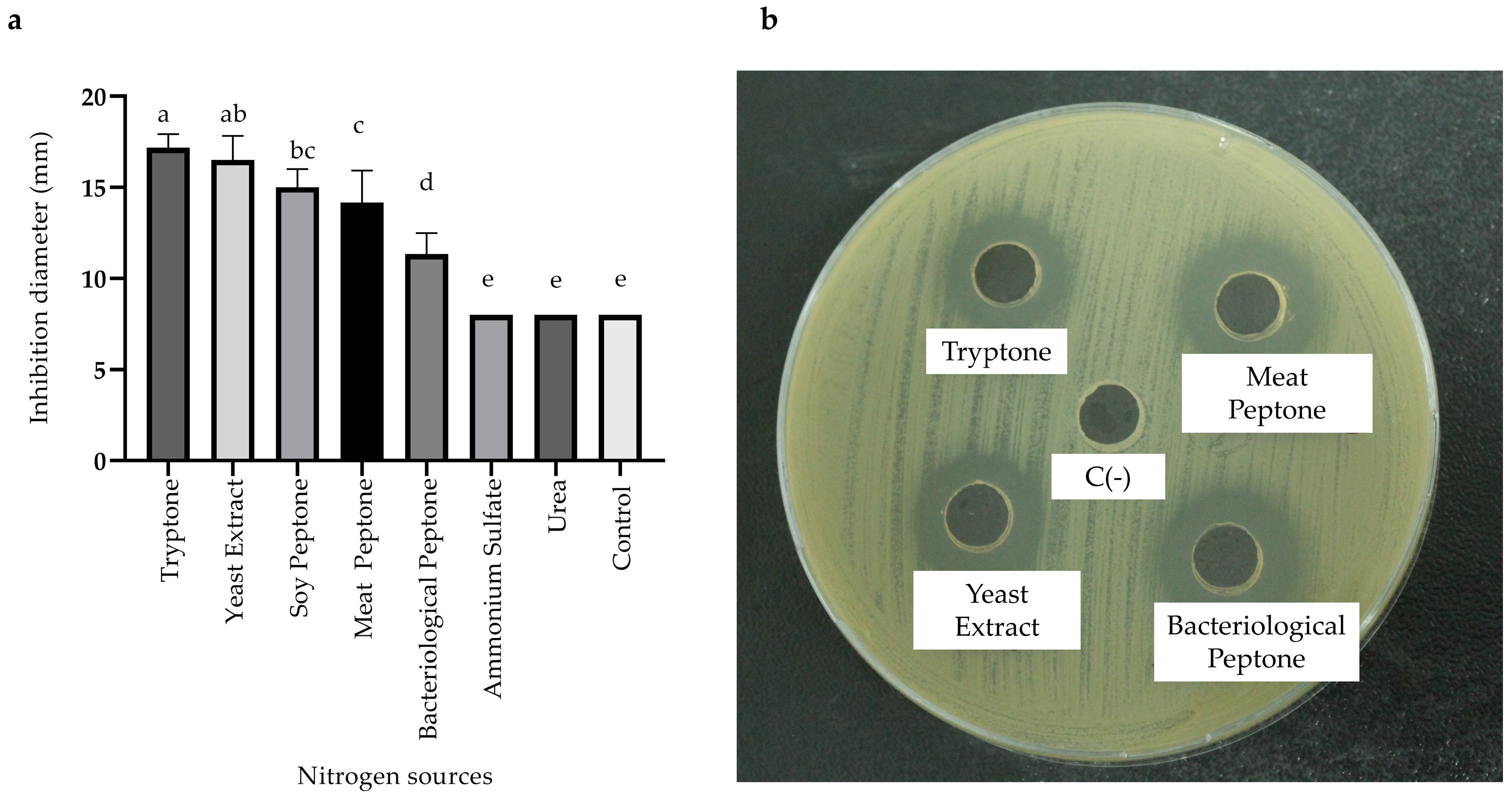
2.3.3. Effects of Carbon, Nitrogen, and Mineral Salts Sources on Metabolite Production
2.4. Evaluation of the Production of Biomass and Antimicrobial Metabolites Using Best Culture Conditions
2.5. Effect of the Antimicrobial Activity of BS4 Strain by Co-Culture Method
2.6. Characterization of the Properties of the Cell-Free Supernatant of the BS4 Strain
2.6.1. Evaluation of the Antimicrobial Spectrum of Selected Strain
2.6.2. Evaluation of Hemolytic and Antioxidant Activity
2.6.3. Effects of Enzymes, Temperature, Surfactants, and Metal Salts on the Cell-Free Supernatant of the Bacillus sp. BS4 Strain
2.7. Determination of the Minimum Inhibitory Concentration (MIC)
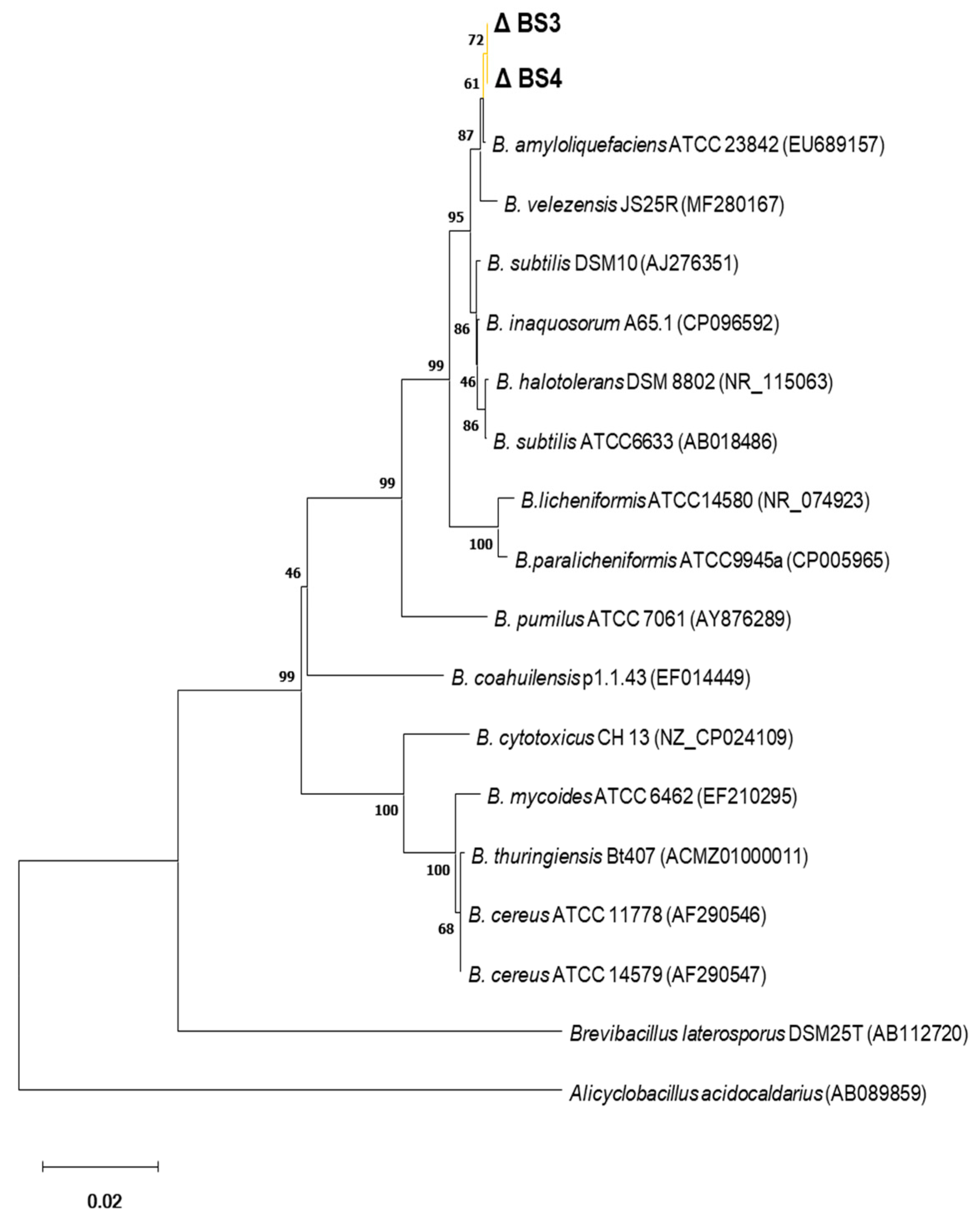
2.8. Phylogenetic Tree
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Strain and Growth Conditions
4.3. Screening of Antagonistic Yeast
4.4. Screening of Antagonistic Bacteria
4.5. Effect of Culture Conditions on Improving the Production of Antimicrobial Metabolites from the Bacillus sp. BS4 Strain
4.5.1. Selection of Fermentation Medium
4.5.2. Effect of Agitation, pH, and Temperature on Antimicrobial Metabolite Production Using the One-Factor-at-a-Time Method
4.5.3. Effect of Carbon, Nitrogen, and Mineral Salt Sources on Metabolite Production Using the One-Factor-at-A-Time Method
4.6. Evaluation of the Production of Biomass and Antimicrobial Metabolites Using Favorable Culture Conditions
4.7. Effect of the Antimicrobial Activity of the Bacillus sp BS4 Strain by Co-Culture Method
4.8. Characterization of the Antimicrobial Properties of the Cell-Free Supernatant of the Bacillus sp. BS4 Strain
4.8.1. Evaluation of the Antimicrobial Spectrum of the Selected Strain
4.8.2. Concentrated Fractions of the Supernatant
4.8.3. Evaluation of Hemolytic and Antioxidant Activity
4.8.4. Effects of Enzymes, Temperature, Surfactants, and Metal Salts on the Cell-Free Supernatant of the Bacillus sp. BS4 Strain
- Enzyme Effect
- Stability of Antimicrobial Activity
- Effect of Surfactants and Other Chemicals
- Effect of Metallic Salts
4.9. Determination of the Minimum Inhibitory Concentration (MIC)
4.10. Genomic DNA Extraction
4.11. 16S rRNA Gene Amplification and Nanopore Sequencing
4.12. Phylogenetic Analysis
4.13. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Review on Antimicrobial Resistance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. 2014. Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (accessed on 5 February 2024).
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef] [PubMed]
- Burow, E.; Käsbohrer, A. Risk Factors for Antimicrobial Resistance in Escherichia coli in Pigs Receiving Oral Antimicrobial Treatment: A Systematic Review. Microb. Drug Resist. 2017, 23, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [PubMed]
- Gulen, T.A.; Guner, R.; Celikbilek, N.; Keske, S.; Tasyaran, M. Clinical importance and cost of bacteremia caused by nosocomial multi drug resistant Acinetobacter baumannii. Int. J. Infect. Dis. 2015, 38, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, P.; Cooper, B.S.; Coast, J.; Oppong, R.; Do Thi Thuy, N.; Phodha, T.; Celhay, O.; Guerin, P.J.; Wertheim, H.; Lubell, H. Enumerating the economic cost of antimicrobial resistance per antibiotic consumed to inform the evaluation of interventions affecting their use. Antimicrob. Resist. Infect. Control 2018, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Coates, A.; Hu, Y.; Bax, R.; Page, C. The future challenges facing the development of new antimicrobial drugs. Nat. Rev. Drug Discov. 2002, 1, 895–910. [Google Scholar] [CrossRef] [PubMed]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar] [CrossRef]
- Suzuki, C.; Ando, Y.; Machida, S. Interaction of SMKT, a killer toxin produced by Pichia farinosa, with the yeast cell membranes. Yeast 2001, 18, 1471–1478. [Google Scholar] [CrossRef]
- Hatoum, R.; Labrie, S.; Fliss, I. Antimicrobial and probiotic properties of yeasts: From fundamental to novel applications. Front. Microbiol. 2012, 3, 421. [Google Scholar] [CrossRef]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the “one strain many compounds” (OSMAC) principle to marine microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Rammali, S.; Hilali, L.; Dari, K.; Bencharki, B.; Rahim, A.; Timinouni, M.; Gaboune, F.; Aalaoui, M.E.; Khattabi, A. Antimicrobial and antioxidant activities of Streptomyces species from soils of three different cold sites in the Fez-Meknes region Morocco. Sci. Rep. 2022, 12, 17233. [Google Scholar] [CrossRef]
- Acuña-Fontecilla, A.; Silva-Moreno, E.; Ganga, M.A.; Godoy, L. Evaluación de la actividad antimicrobiana de levaduras vínicas nativas contra microorganismos patógenos de la industria alimentaria. Food 2017, 15, 457–465. [Google Scholar] [CrossRef]
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. The microbial ecology of wine grape berries. Int. J. Food Microbiol. 2012, 153, 243–259. [Google Scholar] [CrossRef]
- Thompson, J.N. The Evolution of Species lnteractions. Science 1999, 284, 2116–2118. [Google Scholar] [CrossRef] [PubMed]
- Piskure, J.; Rozpedowska, E.; Polakova, S.; Merico, A.; Compagno, C. How did Saccharomyces evolve to become a good brewer? TRENDS Genet. 2006, 22, 2–5. [Google Scholar] [CrossRef]
- Zhou, N.; Katz, M.; Knecht, W.; Compagno, C.; Piškur, J. Genome dynamics and evolution in yeasts: A long-term yeast-bacteria competition experiment. PLoS ONE 2018, 13, e0194911. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.L. Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 2014, 32, 1180–1204. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, W.-C.; Shi, X.; Zheng, H.-Z.; Zheng, Z.-H.; Lu, X.-H.; Xing, Y.; Ji, K.; Liu, M.; Dong, Y.-S. Inducing secondary metabolite production of Aspergillus sydowii through microbial co-culture with Bacillus subtilis. Microb. Cell Fact. 2021, 20, 42. [Google Scholar] [CrossRef]
- Baruzzi, F.; Quintieri, L.; Morea, M.; Caputo, L. Antimicrobial compounds produced by Bacillus spp. and applications in food. In Science against Microbial Pathogens: Communicating Current Research and Technological Advances; Formatex Research Center: Badajoz, Spain, 2011; pp. 1102–1211. Available online: http://www.formatex.info/microbiology3/book/1102-1111.pdf (accessed on 7 March 2024).
- Sci, W.J. A Comparison of Two Methods Used for Measuring the Antagonistic Activity of Bacillus Species. Culture 2008, 5, 161–171. [Google Scholar]
- Ramachandran, R.; Chalasani, A.G.; Lal, R.; Roy, U. A broad-spectrum antimicrobial activity of Bacillus subtilis RLID 12.1. Sci. World J. 2014, 2014, 96848. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yilmaz, M.; Soran, H.; Beyatli, Y. Antimicrobial activities of some Bacillus spp. strains isolated from the soil. Microbiol. Res. 2006, 161, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Barale, S.S.; Ghane, S.G.; Sonawane, K.D. Purification and characterization of antibacterial surfactin isoforms produced by Bacillus velezensis SK. Amb. Express 2022, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Usta, A.; Demirkan, E. The Effect of Growth Parameters on the Antibiotic Activity and Sporulation in Bacillus spp. Isolated from Soil. J. Microbiol. Biotechnol. Food Sci. 2013, 2, 2310–2313. [Google Scholar]
- Pye, C.R.; Bertin, M.J.; Lokey, R.S.; Gerwick, W.H.; Linington, R.G. Retrospective analysis of natural products provides insights for future discovery trends. Proc. Natl. Acad. Sci. USA 2017, 114, 5601–5606. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Wang, Y.H.; Feng, J.T.; Zhang, Q.; Zhang, X. Optimization of fermentation condition for antibiotic production by Xenorhabdus nematophila with response surface methodology. J. Appl. Microbiol. 2008, 104, 735–744. [Google Scholar] [CrossRef]
- Murniasih, T.; Mardiana, N.A.; Untari, F.; Triwibowo, J.; Bayu, A. Optimization of Carbon and Nitrogen Source to Enhance Antibacterial Activity from a Sponge-derived Bacillus tequilensis. Turkish J. Fish. Aquat. Sci. 2024, 24, TRJFAS24222. [Google Scholar] [CrossRef]
- Li, T.; Fan, Y.; Nambou, K.; Hu, F.; Imanaka, T.; Wei, L.; Hua, Q. Improvement of Ansamitocin P-3 Production by Actinosynnema mirum with Fructose as the Sole Carbon Source. Appl. Biochem. Biotechnol. 2015, 175, 2845–2856. [Google Scholar] [CrossRef] [PubMed]
- Messis, A.; Bettache, A.; Brahami, A.; Kecha, M.; Benallaoua, S. Optimization of antifungal production from a novel strain Streptomyces sp. TKJ2 using response surface methodology. Med. Chem. Res. 2014, 23, 310–316. [Google Scholar] [CrossRef]
- Umezawa, H. Structures and Activities of Protease Inhibitors of Microbial Origin. Methods Enzymol. 1976, 45, 678–695. [Google Scholar]
- Saraniya, A.; Jeevaratnam, K. Optimization of nutritional and non-nutritional factors involved for production of antimicrobial compounds from Lactobacillus pentosus SJ65 using response surface methodology. Braz. J. Microbiol. 2014, 45, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Benitez, L.; Correa, A.; Daroit, D.; Brandelli, A. Antimicrobial activity of Bacillus amyloliquefaciens LBM 5006 is enhanced in the presence of Escherichia coli. Curr Microbiol. 2011, 62, 1017–1022. [Google Scholar] [CrossRef] [PubMed]
- Steinke, K.; Mohite, O.S.; Weber, T.; Kovács, Á.T. Phylogenetic Distribution of Secondary Metabolites in the Bacillus subtilis Species Complex. mSystems 2021, 6, 10–1128. [Google Scholar] [CrossRef]
- Choi, S.K.; Jeong, H.; Kloepper, J.W.; Ryu, C.M. Genome sequence of Bacillus amyloliquefaciens GB03, an active ingredient of the first commercial biological control product. Genome Announc. 2014, 2, 2–3. [Google Scholar] [CrossRef]
- Ji, S.H.; Paul, N.C.; Deng, J.X.; Kim, Y.S.; Yun, B.S.; Yu, S.H. Biocontrol activity of Bacillus amyloliquefaciens CNU114001 against fungal plant diseases. Mycobiology 2013, 41, 234–242. [Google Scholar] [CrossRef]
- Tran, C.; Cock, I.E.; Chen, X.; Feng, Y. Antimicrobial Bacillus: Metabolites and Their Mode of Action. Antibiotics 2022, 11, 88. [Google Scholar] [CrossRef]
- Fan, B.; Wang, C.; Song, X.; Ding, X.; Wu, L.; Wu, H.; Gao, X.; Borriss, R. Bacillus velezensis FZB42 in 2018: The gram-positive model strain for plant growth promotion and biocontrol. Front Microbiol. 2018, 9, 408393. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Heng, J.; Qin, S.; Bian, K. A comprehensive understanding of the biocontrol potential of Bacillus velezensis LM2303 against Fusarium head blight. PLoS ONE 2018, 13, e0198560. [Google Scholar] [CrossRef]
- Fazle Rabbee, M.; Baek, K.H. Antimicrobial Activities of Lipopeptides and Polyketides of Bacillus velezensis for Agricultural Applications. Molecules 2020, 25, 4973. [Google Scholar] [CrossRef]
- Lopes, C.A.; Sangorrín, M.P. Optimization of killer assays for yeast selection protocols. Rev. Argent. Microbiol. 2010, 42, 298–306. [Google Scholar]
- Sa-Uth, C.; Rattanasena, P.; Chandrapatya, A.; Bussaman, P. Modification of Medium Composition for Enhancing the Production of Antifungal Activity from Xenorhabdus stockiae PB09 by Using Response Surface Methodology. Int. J. Microbiol. 2018, 2018, 3965851. [Google Scholar] [CrossRef]
- Chen, X.H.; Lou, W.Y.; Zong, M.H.; Smith, T.J. Optimization of culture conditions to produce high yields of active Acetobacter sp. CCTCC M209061 cells for anti-Prelog reduction of prochiral ketones. BMC Biotechnol. 2011, 11, 110. [Google Scholar] [CrossRef]
- Latha, S.; Sivaranjani, G.; Dhanasekaran, D. Response surface methodology: A non-conventional statistical tool to maximize the throughput of Streptomyces species biomass and their bioactive metabolites. Crit. Rev. Microbiol. 2017, 43, 567–582. [Google Scholar] [CrossRef]
- Perez, K.J.; Viana, J.D.; Lopes, F.C.; Pereira, J.Q.; dos Santos, D.M.; Oliveira, J.S.; Velho, R.V.; Crispim, S.M.; Nicoli, J.R.; Brandelli, A.; et al. Bacillus spp. isolated from puba as a source of biosurfactants and antimicrobial lipopeptides. Front. Microbiol. 2017, 8, 61. [Google Scholar] [CrossRef]
- Pérez-Delgado, O.; Espinoza-Vergara, M.A.; Castro-Vega, N.A.; Reyes-Montaño, E.A. Evaluación preliminar de actividad antibacteriana in vitro del veneno de escorpión Hadruroides charcasus (Karsch, 1879) contra Pseudomonas aeruginosa y Staphylococcus aureus. Rev del Cuerpo Médico del HNAAA 2019, 12, 6–12. [Google Scholar] [CrossRef]
- Oddo, A.; Hansen, P.R. Hemolytic Activity of Antimicrobial Peptides; Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2017; pp. 427–435. [Google Scholar]
- Kayalvizhi, N.; Gunasekaran, P. Purification and characterization of a novel broad-spectrum bacteriocin from Bacillus licheniformis MKU3. Biotechnol. Bioprocess. Eng. 2010, 15, 365–370. [Google Scholar] [CrossRef]
- Hernández, D.; Cardell, E.; Zárate, V. Antimicrobial activity of lactic acid bacteria isolated from Tenerife cheese: Initial characterization of plantaricin TF711, a bacteriocin-like sufbstance produced by Lactobacillus plantarum TF711. J. Appl. Microbiol. 2005, 99, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Navarrete, P.; Magne, F.; Mardones, P.; Riveros, M.; Opazo, R.; Suau, A.; Pochart, P.; Romero, J. Molecular analysis of intestinal microbiota of rainbow trout (Oncorhynchus mykiss). FEMS Microbiol. Ecol. 2010, 71, 148–156. [Google Scholar] [CrossRef] [PubMed]
- de Koning, W.; Miladi, M.; Hiltemann, S.; Heikema, A.; Hays, J.P.; Flemming, S.; van den Beek, M.; Mustafa, D.A.; Backofen, R.; Grüning, B.; et al. NanoGalaxy: Nanopore long-read sequencing data analysis in Galaxy. GigaScience 2020, 9, giaa105. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]

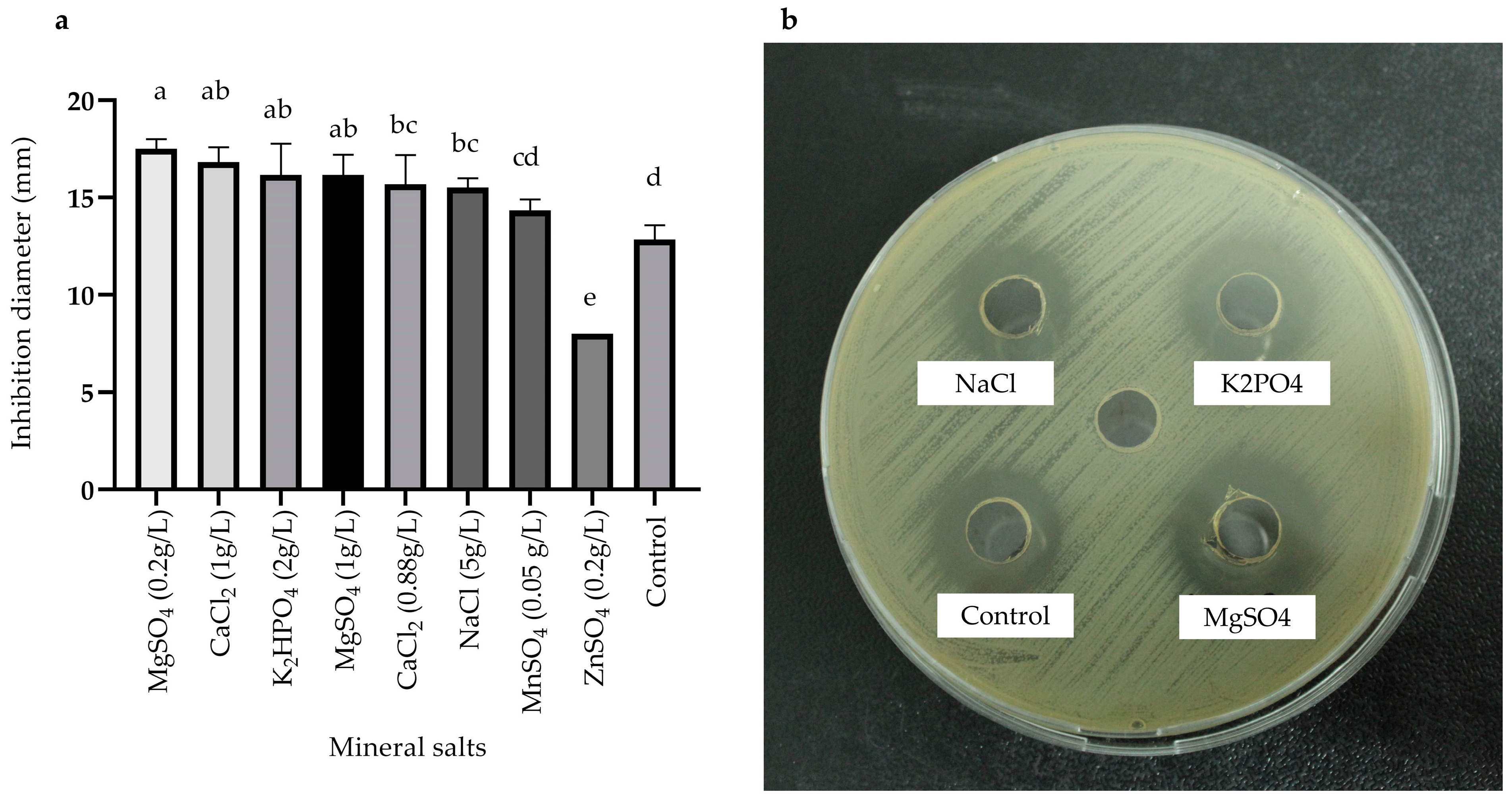
 ) and MOD broth (
) and MOD broth ( ). The error bars in the figure indicate the standard deviations from three replicates.
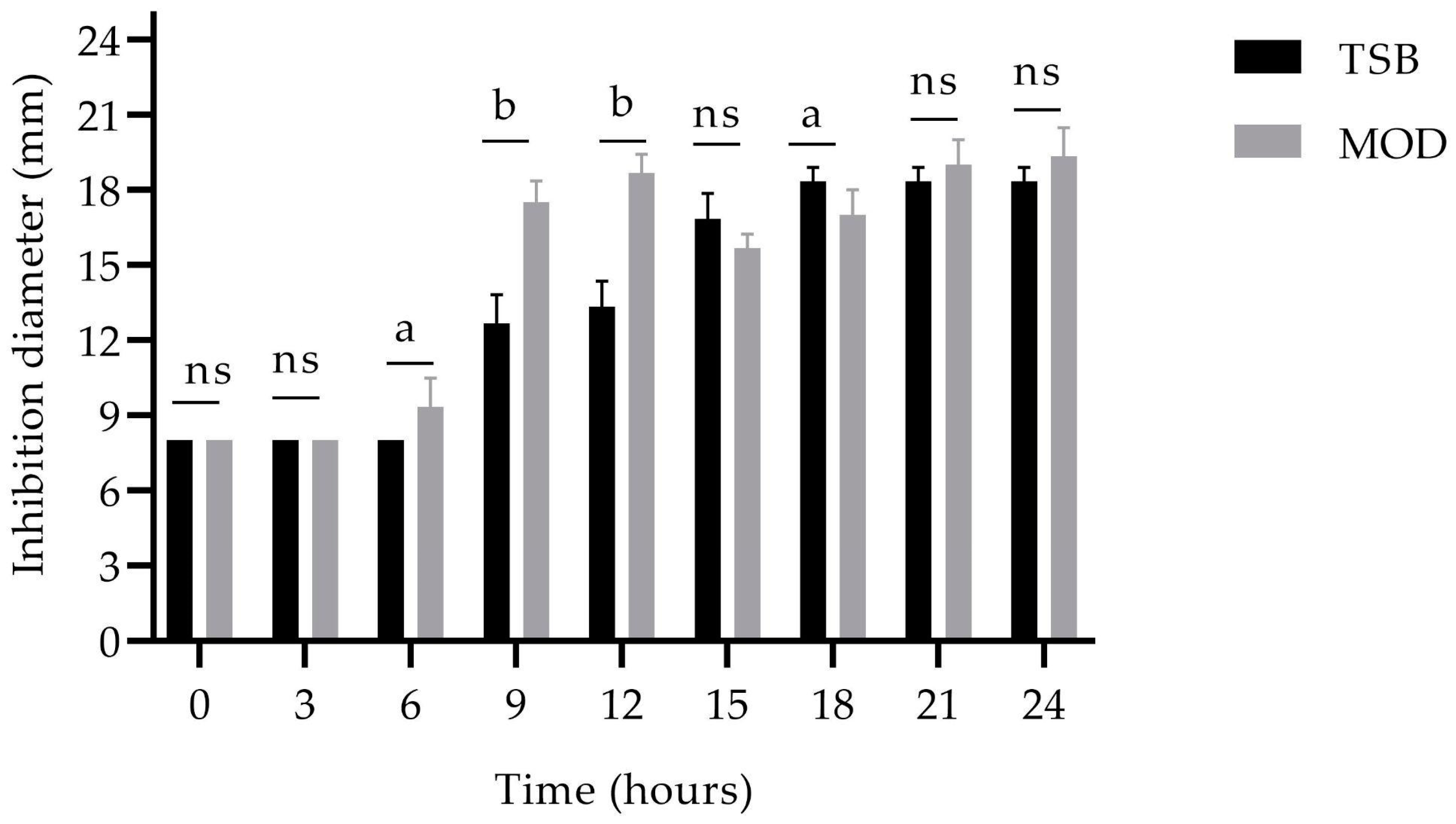
). The error bars in the figure indicate the standard deviations from three replicates.
 ) and MOD broth (
) and MOD broth ( ). The error bars in the figure indicate the standard deviations from three replicates.
). The error bars in the figure indicate the standard deviations from three replicates.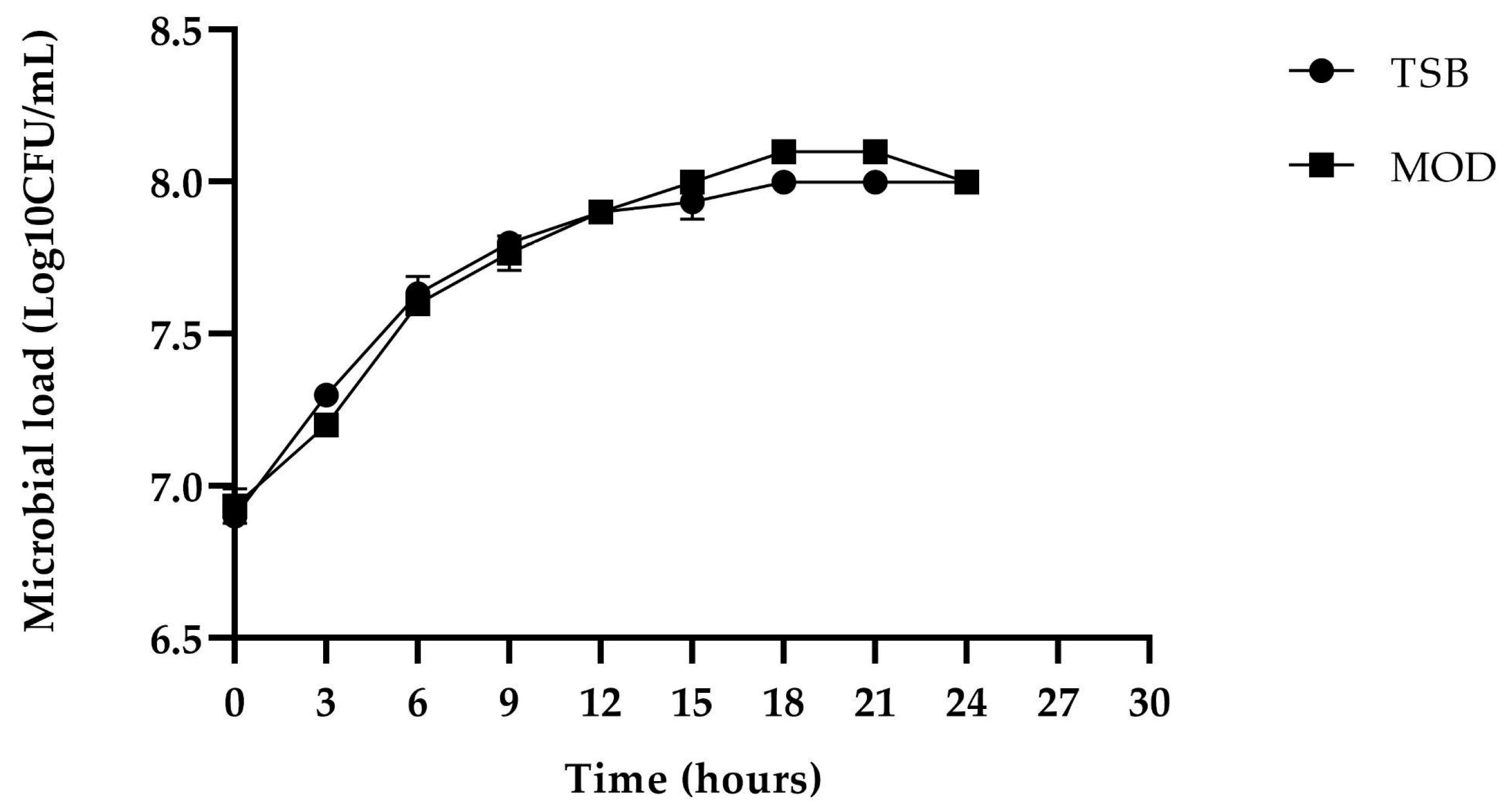


| Strains | Antimicrobial Activity (mm ± SD) | |||||
|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | ||||
| BC | CFS | BC | CFS | BC | CFS | |
| BS3 | 19.00 ± 1.41 a | 17.67 ± 0.58 a | 19.50 ± 1.41 a | 18.50 ± 1.53 a | 18.00 ± 0 b | 18.50 ± 1.00 b |
| BS4 | 20.50 ± 0.70 a | 19.67 ± 0.58 a | 20.00 ± 1.41 b | 20.00 ± 1.00 b | 18.50 ± 0.71 a | 19.33 ± 0.58 a |
| BS17 | 19.00 ± 2.83 a | 18.00 ± 1.80 a | 18.50 ± 2.12 c | 17.67 ± 1.15 c | 14.50 ± 2.12 a | 19.17 ± 0.76 a |
| BS21 | 18.50 ± 3.54 b | 17.00 ± 1.73 b | 18.30 ± 0.70 c | 17.33 ± 0.58 c | 17.33 ± 1.00 d | 16.50 ± 1.00 d |
| Control (+) | 26.00 ± 1.15 | 26.00 ± 1.15 | 26.00 ± 1.15 | 26.00 ± 1.15 | 26.00 ± 1.15 | 26.00 ± 1.15 |
| Control (−) | - | - | - | - | - | - |
| Culture Medium | Biomass (Log10 (CFU/mL ± SD)) | Diameter of Inhibition Zone (mm, Mean ± SD) | ||||
|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | |
| TSB | 8.86 ± 0.01 a | 8.79 ± 0.06 b | 8.77 ± 0.08 c | 19.66 ± 1.15 a | 19.88 ± 1.00 b | 19.33 ± 1.15 b |
| LB | 8.72 ± 0.04 c | 8.72 ± 0.01 a | 8.73 ± 0.05 b | 15.67 ± 0.58 a | 14.67 ± 0.58 b | 14.00 ± 1.00 a |
| YPG | 8.80 ± 0 b | 8.70 ± 0.07 c | 8.71 ± 0.08 a | 18.00 ± 1.00 a | 15.33 ± 0.58 a | 14.67 ± 1.53 a |
| Treatment | Zone of Inhibition (mm) | |
|---|---|---|
| 24 h | 48 h | |
| TSB + E. coli | 19.75 ± 0.96 b | 20.5 ± 1.29 b |
| MOD + E. coli | 21.85 ± 1.03 a | 21.12 ± 1.03 b |
| MOD + inactivated E. coli | 20.25 ± 0.50 b | 20.50 ± 0.58 b |
| Indicator Strains | TSB | MOD |
|---|---|---|
| Staphylococcus aureus ATCC 6538 | N.A. | N.A. |
| Staphylococcus aureus ATCC 25923 | N.A. | N.A. |
| Staphylococcus epidermidis ATCC 1228 | N.A. | N.A. |
| Staphylococcus saprophyticus ATCC 15305 | N.A. | 16.38 ± 0.38 |
| Citrobacter freundii ATCC 8090 | 16.00 ± 0 | 16.33 ± 0.58 |
| Escherichia coli ATCC 25922 | 19.67 ± 0.58 | 20.67 ± 0.58 |
| Proteus vulgaris ATCC 49132 | 19.33 ± 1.15 | 20.33 ± 0.58 |
| Enterobacter aerogenes | 17.67 ± 0.58 | 18.33 ± 0.58 |
| Salmonella enterica | 20.33 ± 0.58 | 23.67 ± 0.58 |
| Shigella flexneri ATCC 12022 | 15.00 ± 0 | 16.00 ± 0 |
| Pseudomonas aeruginosa | N.A. | N.A. |
| Yersinia enterocolitica ATCC 23715 | 21.67 ± 1.15 | 22.00 ± 1.00 |
| Serratia marcescens ATCC 14756 | 21.67 ± 0.58 | 22.33 ± 0.58 |
| Aeromonas sp. | 29.00 ± 0.15 | 30.00 ± 0.15 |
| Candida albicans ATCC 14053 | N.A. | 11.33 ± 1.15 |
| Candida tropicalis ATCC 1369 | N.A. | 13.33 ± 1.15 |
| Nosocomial Strains | ||
| Klebsiella pneumoniae 8636 (clinic isolation) | 24.50 ± 0.50 | 25.67 ± 0.58 |
| Escherichia coli (fistula, clinic isolation) | 19.00 ± 1.00 | 21.17 ± 0.29 |
| Escherichia coli BLEE 1705 (clinic isolation) | 16.00 ± 0 | 16.33 ± 1.15 |
| Escherichia coli BLEE 78 (clinic isolation) | 16.00 ± 1.00 | 16.00 ± 1.00 |
| Escherichia coli BLEE 79 (clinic isolation) | N.A. | N.A. |
| Pseudomonas aeruginosa (urine culture) | 13.33 ± 1.53 | 13.67 ± 0.30 |
| Pseudomonas aeruginosa (bronchial secretion) | 14.33 ± 0.58 | N.A. |
| Pseudomonas aeruginosa (clinic isolation) | 16.00 ± 0.00 | 15.67 ± 0.30 |
| Acinetobacter baumannii (clinic isolation) | N.A. | N.A. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palacios-Rodriguez, A.P.; Espinoza-Culupú, A.; Durán, Y.; Sánchez-Rojas, T. Antimicrobial Activity of Bacillus amyloliquefaciens BS4 against Gram-Negative Pathogenic Bacteria. Antibiotics 2024, 13, 304. https://doi.org/10.3390/antibiotics13040304
Palacios-Rodriguez AP, Espinoza-Culupú A, Durán Y, Sánchez-Rojas T. Antimicrobial Activity of Bacillus amyloliquefaciens BS4 against Gram-Negative Pathogenic Bacteria. Antibiotics. 2024; 13(4):304. https://doi.org/10.3390/antibiotics13040304
Chicago/Turabian StylePalacios-Rodriguez, Ana Paula, Abraham Espinoza-Culupú, Yerson Durán, and Tito Sánchez-Rojas. 2024. "Antimicrobial Activity of Bacillus amyloliquefaciens BS4 against Gram-Negative Pathogenic Bacteria" Antibiotics 13, no. 4: 304. https://doi.org/10.3390/antibiotics13040304
APA StylePalacios-Rodriguez, A. P., Espinoza-Culupú, A., Durán, Y., & Sánchez-Rojas, T. (2024). Antimicrobial Activity of Bacillus amyloliquefaciens BS4 against Gram-Negative Pathogenic Bacteria. Antibiotics, 13(4), 304. https://doi.org/10.3390/antibiotics13040304