Synthesis of 3-((4-Hydroxyphenyl)amino)propanoic Acid Derivatives as Promising Scaffolds for the Development of Antimicrobial Candidates Targeting Multidrug-Resistant Bacterial and Fungal Pathogens
Abstract
1. Introduction
2. Results
3. Discussion
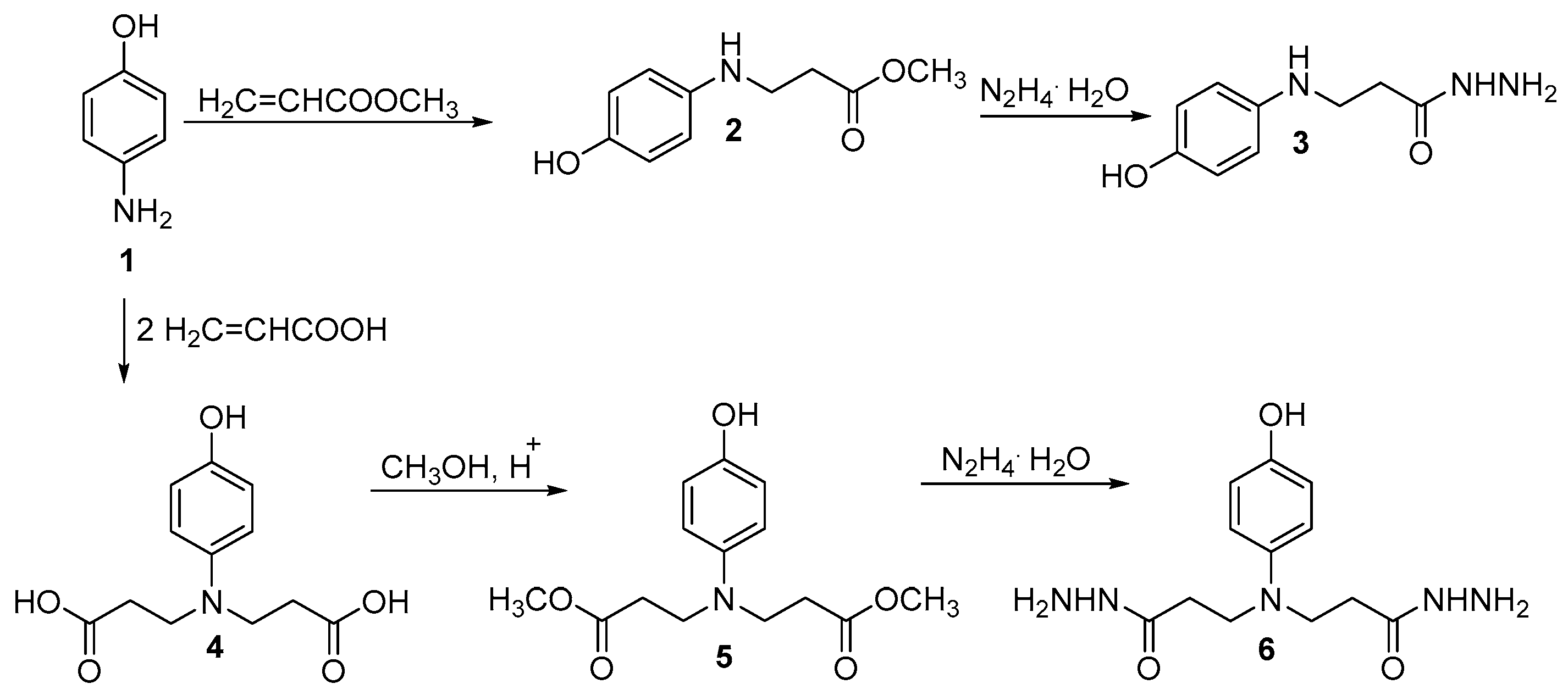
4. Materials and Methods
- 3,3′-((4-Hydroxyphenyl)azanediyl)di(propanehydrazide) (6)
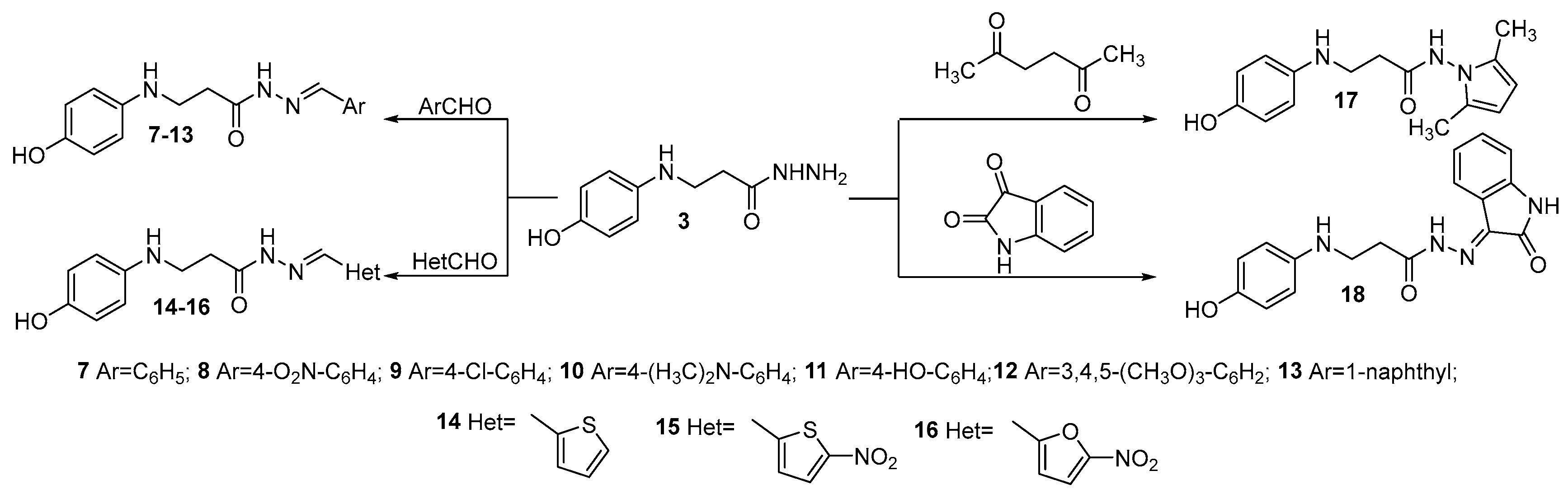
- General procedure for the preparation of hydrazones 7–13
- N′-benzylidene-3-((4-hydroxyphenyl)amino)propanehydrazide (7)
- 3-((4-Hydroxyphenyl)amino)-N′-(4-nitrobenzylidene)propanehydrazide (8)
- N′-(4-chlorobenzylidene)-3-((4-hydroxyphenyl)amino)propanehydrazide (9)
- N′-(4-(dimethylamino)benzylidene)-3-((4-hydroxyphenyl)amino)propanehydrazide (10)
- N′-(4-hydroxybenzylidene)-3-((4-hydroxyphenyl)amino)propanehydrazide (11)
- 3-((4-Hydroxyphenyl)amino)-N′-(3,4,5-trimethoxybenzylidene)propanehydrazide (12)
- 3-((4-Hydroxyphenyl)amino)-N′-(naphthalen-1-ylmethylene)propanehydrazide (13)
- General procedure for the preparation of hydrazones 14–16
- 3-((4-Hydroxyphenyl)amino)-N′-(thien-2-ylmethylene)propanehydrazide (14)
- 3-((4-Hydroxyphenyl)amino)-N′-((5-nitrothien-2-yl)methylene)propanehydrazide (15)
- 3-((4-Hydroxyphenyl)amino)-N′-((5-nitrofuryl-2-yl)methylene)propanehydrazide (16)
- N-(2,5-dimethyl-1H-pyrrol-1-yl)-3-((4-hydroxyphenyl)amino)propanamide (17)
- 3-((4-Hydroxyphenyl)amino)-N′-(2-oxoindolin-3-ylidene)propanehydrazide (18)
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N′-(-(2-oxoindolin-3-yl)methylene)propanehydrazide) (19)
- 5,5′-(((4-Hydroxyphenyl)azanediyl)bis(ethane-2,1-diyl))bis(1,3,4-oxadiazole-2(3H)-thione) (20)
- General procedure for the preparation of hydrazones 21–25.
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N′-(furan-2-ylmethylene)propanehydrazide) (21)
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N′-(thien-2-ylmethylene)propanehydrazide) (22)
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N′-(-(5-nitrothien-2yl)methylene)propanehydrazide) (23)
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N′-((5-nitrofuran-2-yl)methylene)propanehydrazide) (24)
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N′-(thien-3-ylmethylene)propanehydrazide) (25)
- General procedure for the preparation of hydrazones 26–28.
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N′-(propan-2-ylidene)propanehydrazide) (26)
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N′-(butan-2-ylidene)propanehydrazide) (27)
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N′-(1-phenylethylidene)propanehydrazide) (28)
- General procedure for the preparation of dihydrazones 29–35
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N′-(benzylidene)propanehydrazide) (29)
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N′-(4-nitrobenzylidene)propanehydrazide) (30)
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N′-(benzylidene)propanehydrazide) (31)
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N′-(4-(dimethylamino)benzylidene)propanehydrazide) (32)
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N′-(4-hydroxybenzylidene)propanehydrazide) (33)
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N′-(3,4,5-trimethoxybenzylidene)propanehydrazide) (34)
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N′-(naphthalen-1-ylmethylene)propanehydrazide) (35)
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(1-(3,5-dimethyl-1H-pyrazol-1-yl)propan-1-one) (36)
- 3,3′-((4-Hydroxyphenyl)azanediyl)bis(N-(2,5-dimethyl-1H-pyrrol-1-yl)propanamide) (37)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Septimus, E.J. Antimicrobial Resistance: An Antimicrobial/Diagnostic Stewardship and Infection Prevention Approach. Med. Clin. N. Am. 2018, 102, 819–829. [Google Scholar] [CrossRef]
- Rice, L.B. Antimicrobial Stewardship and Antimicrobial Resistance. Med. Clin. N. Am. 2018, 102, 805–818. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Roch, M.; Sierra, R.; Andrey, D.O. Antibiotic heteroresistance in ESKAPE pathogens, from bench to bedside. Clin. Microbiol. Infect. 2023, 29, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Bing, J.; Hu, T.; Ennis, C.L.; Nobile, C.J.; Huang, G. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020, 16, e1008921. [Google Scholar] [CrossRef]
- Pristov, K.E.; Ghannoum, M.A. Resistance of Candida to azoles and echinocandins worldwide. Clin. Microbiol. Infect. 2019, 25, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Nowak, M.G.; Skwarecki, A.S.; Milewska, M.J. Amino Acid Based Antimicrobial Agents—Synthesis and Properties. ChemMedChem 2021, 16, 3513–3544. [Google Scholar] [CrossRef]
- Li, X.; Meng, X.; Duan, H.; Wang, L.; Wang, S.; Zhang, Y.; Qin, D. Original and efficient synthesis of D-cycloserine. Arch. Pharm. 2010, 343, 473–475. [Google Scholar] [CrossRef]
- Kotnik, M.; Humljan, J.; Contreras-Martel, C.; Oblak, M.; Kristan, K.; Hervé, M.; Blanot, D.; Urleb, U.; Gobec, S.; Dessen, A.; et al. Structural and functional characterization of enantiomeric glutamic acid derivatives as potential transition state analogue inhibitors of MurD ligase. J. Mol. Biol. 2007, 370, 107–115. [Google Scholar] [CrossRef]
- Humljan, J.; Kotnik, M.; Contreras-Martel, C.; Blanot, D.; Urleb, U.; Dessen, A.; Solmajer, T.; Gobec, S. Novel naphthalene-N-sulfonyl-D-glutamic acid derivatives as inhibitors of MurD, a key peptidoglycan biosynthesis enzyme. J. Med. Chem. 2008, 51, 7486–7494. [Google Scholar] [CrossRef]
- Ellsworth, B.A.; Tom, N.J.; Bartlett, P.A. Synthesis and evaluation of inhibitors of bacterial D-alanine:D-alanine ligases. Chem. Biol. 1996, 3, 37–44. [Google Scholar] [CrossRef]
- Caselli, E.; Powers, R.A.; Blasczcak, L.C.; Wu, C.Y.; Prati, F.; Shoichet, B.K. Energetic, structural, and antimicrobial analyses of beta-lactam side chain recognition by beta-lactamases. Chem. Biol. 2001, 8, 17–31. [Google Scholar] [CrossRef]
- Wamp, S.; Rothe, P.; Stern, D.; Holland, G.; Döhling, J.; Halbedel, S. MurA escape mutations uncouple peptidoglycan biosynthesis from PrkA signaling. PLoS Pathog. 2022, 18, e1010406. [Google Scholar] [CrossRef]
- Hummels, K.R.; Berry, S.P.; Li, Z.; Taguchi, A.; Min, J.K.; Walker, S.; Marks, D.S.; Bernhardt, T.G. Coordination of bacterial cell wall and outer membrane biosynthesis. Nature 2023, 615, 300–304. [Google Scholar] [CrossRef]
- Weng, C.J.; Yen, G.C. Chemopreventive effects of dietary phytochemicals against cancer invasion and metastasis: Phenolic acids, monophenol, polyphenol, and their derivatives. Cancer Treat. Rev. 2012, 38, 76–87. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, Y.; Liu, X.; Zhao, C.; Yin, J.; Li, X.; Zhang, X.; Wang, J.; Wang, S. Distinctive anti-inflammatory effects of resveratrol, dihydroresveratrol, and 3-(4-hydroxyphenyl)-propionic acid on DSS-induced colitis in pseudo-germ-free mice. Food Chem. 2023, 400, 133904. [Google Scholar] [CrossRef]
- Bertašiūtė, M.; Kavaliauskas, P.; Vaickelionienė, R.; Grybaitė, B.; Petraitis, V.; Petraitienė, R.; Naing, E.; Garcia, A.; Šiugždaitė, J.; Lelešius, R.; et al. Synthesis of 1-(2-Hydroxyphenyl)- and (3,5-Dichloro-2-hydroxyphenyl)-5-oxopyrrolidine-3-carboxylic Acid Derivatives as Promising Scaffolds for the Development of Novel Antimicrobial and Anticancer Agents. Int. J. Mol. Sci. 2023, 24, 7966. [Google Scholar] [CrossRef]
- Phenol. IARC Monogr. Eval. Carcinog. Risks Hum. 1989, 47, 263–287. Available online: https://pubmed.ncbi.nlm.nih.gov/2699901/ (accessed on 5 January 2024).
- Scott, K.A.; Cox, P.B.; Njardarson, J.T. Phenols in Pharmaceuticals: Analysis of a Recurring Motif. J. Med. Chem. 2022, 65, 7044–7072. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, P.F.; Menini, L.A.P.; Bernardes, P.C.; Saraiva, S.H.; Carneiro, J.W.M.; Costa, A.V.; Arruda, T.R.; Lage, M.R.; Gonçalves, P.M.; Bernardes, C.O.; et al. Semisynthetic Phenol Derivatives Obtained from Natural Phenols: Antimicrobial Activity and Molecular Properties. J. Agric. Food Chem. 2018, 66, 323–330. [Google Scholar] [CrossRef]
- Baltrusis, R.S.; Beresnevičius, Z.J.; Mickevičius, V. Synthesis and transformations of N-(4-hydroxyphenyl)dihydrouracils. Chem. Heterocycl. Compd. 1982, 18, 1089–1095. [Google Scholar] [CrossRef]
- Beattie, J.K.; McErlean, C.S.P.; Phippen, C.B.W. On-water conjugate additions of anilines. Chem. Commun. 2010, 46, 8234–8236. [Google Scholar] [CrossRef]
- Tumosienė, I.; Jakienė, E.; Kantminienė, K.; Rutkauskas, K.; Beresnevičius, Z.J. Synthesis and plant growth regulating activity of halo derivatives of 3,3′-(arylimino)dipropanoic acids. Chemija 2010, 21, 139–144. [Google Scholar]
- Anusevičius, K.; Mickevičius, V.; Mikulskienė, G. Synthesis and structure of N-(4-bromophenyl)-N-carboxyethyl-β-alanine derivatives. Chemija 2010, 21, 127–134. [Google Scholar]
- Tumosienė, I.; Mikulskienė, G.; Kantminienė, K.; Beresnevičius, Z.J. Synthesis and structure of 3,3′-[(4-alkoxyphenyl)imino]bis(N’-phthaloyl- or N’-benzylidenepropanohydrazide) derivatives. Chemija 2011, 22, 65–72. [Google Scholar]
- Paulusma, C.C.; Lamers, W.H.; Broer, S.; van de Graaf, S.F.J. Amino acid metabolism, transport and signalling in the liver revisited. Biochem. Pharmacol. 2022, 201, 115074. [Google Scholar] [CrossRef]
- Ngo, D.H.; Vo, T.S. An Updated Review on Pharmaceutical Properties of Gamma-Aminobutyric Acid. Molecules 2019, 24, 2678. [Google Scholar] [CrossRef]
- Koksharova, O.A.; Safronova, N.A. Non-Proteinogenic Amino Acid β-N-Methylamino-L-Alanine (BMAA): Bioactivity and Ecological Significance. Toxins 2022, 14, 539. [Google Scholar] [CrossRef]
- López-López, L.I.; Rivera-Ávalos, E.; Villarreal-Reyes, C.; Martínez-Gutiérrez, F.; de Loera, D. Synthesis and Antimicrobial Evaluation of Amino Acid Naphthoquinone Derivatives as Potential Antibacterial Agents. Chemotherapy 2022, 67, 102–109. [Google Scholar] [CrossRef]
- Sajjad, F.; Sun, N.N.; Chen, T.; Yan, Y.J.; Margetić, D.; Chen, Z.L. Evaluation of antimicrobial photodynamic activities of 5-aminolevulinic acid derivatives. Photodermatol. Photoimmunol. Photomed. 2021, 37, 296–305. [Google Scholar] [CrossRef]
- Skwarecki, A.S.; Nowak, M.G.; Milewska, M.J. Amino Acid and Peptide-Based Antiviral Agents. ChemMedChem 2021, 16, 3106–3135. [Google Scholar] [CrossRef]
- Cui, J.; Ji, X.; Mi, Y.; Miao, Q.; Dong, F.; Tan, W.; Guo, Z. Antimicrobial and Antioxidant Activities of N-2-Hydroxypropyltrimethyl Ammonium Chitosan Derivatives Bearing Amino Acid Schiff Bases. Mar. Drugs 2022, 20, 86. [Google Scholar] [CrossRef]
- Chang, R.Y.K.; Nang, S.C.; Chan, H.K.; Li, J. Novel antimicrobial agents for combating antibiotic-resistant bacteria. Adv. Drug Deliv. Rev. 2022, 187, 114378. [Google Scholar] [CrossRef]
- Loyola-Cruz, M.Á.; Gonzalez-Avila, L.U.; Martínez-Trejo, A.; Saldaña-Padilla, A.; Hernández-Cortez, C.; Bello-López, J.M.; Castro-Escarpulli, G. ESKAPE and Beyond: The Burden of Coinfections in the COVID-19 Pandemic. Pathogens 2023, 12, 743. [Google Scholar] [CrossRef]

| Minimal Inhibitory Concentration (µg/mL) | ||||||
|---|---|---|---|---|---|---|
| Compound | S. aureus TCH 1516 1 | E. faecalis AR-0671 2 | E. coli AR-0001 3 | K. pneumoniae AR-0003 4 | P. aeruginosa AR-1114 5 | A. baumannii AR-0273 6 |
| 2 | 64 | 64 | >64 | >64 | >64 | >64 |
| 3 | >64 | >64 | >64 | >64 | >64 | >64 |
| 4 | >64 | >64 | >64 | >64 | >64 | >64 |
| 5 | >64 | >64 | >64 | >64 | >64 | >64 |
| 6 | >64 | >64 | 64 | 64 | 32 | 32 |
| 7 | 32 | >64 | 64 | 64 | >64 | >64 |
| 8 | 16 | 8 | 16 | >64 | >64 | >64 |
| 9 | 8 | 8 | 64 | >64 | >64 | 32 |
| 10 | >64 | >64 | >64 | >64 | >64 | >64 |
| 11 | >64 | >64 | >64 | >64 | >64 | >64 |
| 12 | >64 | >64 | >64 | >64 | >64 | >64 |
| 13 | 16 | 16 | 32 | 64 | >64 | 64 |
| 14 | 8 | 8 | 64 | 64 | 64 | 64 |
| 15 | 1 | <0.5 | 8 | 32 | 64 | 16 |
| 16 | 4 | 2 | 16 | >64 | >64 | 64 |
| 17 | >64 | >64 | >64 | >64 | >64 | 16 |
| 18 | >64 | >64 | >64 | >64 | >64 | >64 |
| 19 | >64 | >64 | >64 | >64 | >64 | >64 |
| 20 | 32 | 32 | >64 | >64 | >64 | >64 |
| 21 | 16 | 16 | 32 | 64 | >64 | 64 |
| 22 | 32 | 16 | 32 | 32 | 64 | 64 |
| 23 | >64 | >64 | >64 | >64 | >64 | >64 |
| 24 | 32 | 32 | 64 | 64 | 64 | 32 |
| 25 | 64 | 32 | >64 | >64 | >64 | >64 |
| 26 | >64 | >64 | >64 | >64 | >64 | >64 |
| 27 | >64 | >64 | >64 | >64 | >64 | >64 |
| 28 | >64 | >64 | 32 | 64 | >64 | 64 |
| 29 | 16 | >64 | 64 | 64 | >64 | >64 |
| 30 | 16 | 16 | 32 | 64 | >64 | >64 |
| 31 | 16 | 16 | 32 | 64 | 64 | 32 |
| 32 | >64 | >64 | >64 | >64 | >64 | 16 |
| 33 | 8 | 16 | 32 | 64 | 16 | 16 |
| 34 | >64 | >64 | >64 | >64 | >64 | >64 |
| 35 | >64 | >64 | >64 | >64 | >64 | >64 |
| 36 | 32 | 64 | >64 | >64 | >64 | >64 |
| 37 | 16 | 32 | 16 | >64 | >64 | >64 |
| Vancomycin | 2 | 4 | N/A | N/A | N/A | N/A |
| Cefazolin | 8 | 8 | >64 | >64 | >64 | >64 |
| Minimal inhibitory Concentration (µg/mL) | ||||||
|---|---|---|---|---|---|---|
| Compound | C. albicans AR-0761 1 | C. parapsilosis AR-0339 2 | C. auris AR-0381 3 | C. auris AR-0382 4 | C. auris AR-0383 5 | C. auris AR-0384 6 |
| 2 | >64 | >64 | >64 | >64 | >64 | >64 |
| 3 | >64 | >64 | >64 | >64 | >64 | >64 |
| 4 | >64 | >64 | >64 | >64 | >64 | >64 |
| 5 | >64 | >64 | >64 | >64 | >64 | >64 |
| 6 | >64 | >64 | >64 | >64 | >64 | >64 |
| 7 | >64 | >64 | >64 | >64 | >64 | >64 |
| 8 | >64 | >64 | >64 | >64 | >64 | >64 |
| 9 | >64 | >64 | >64 | >64 | >64 | >64 |
| 10 | >64 | >64 | >64 | >64 | >64 | >64 |
| 11 | >64 | >64 | >64 | >64 | >64 | >64 |
| 12 | >64 | >64 | >64 | >64 | >64 | >64 |
| 13 | >64 | >64 | >64 | >64 | >64 | >64 |
| 14 | 16 | 32 | 16 | 16 | 8 | 16 |
| 15 | 32 | >64 | >64 | >64 | >64 | >64 |
| 16 | 8 | 16 | 32 | 32 | 32 | 32 |
| 17 | 8 | 8 | 16 | 16 | 16 | 16 |
| 18 | >64 | >64 | >64 | >64 | >64 | >64 |
| 19 | >64 | >64 | >64 | >64 | >64 | >64 |
| 20 | >64 | >64 | >64 | >64 | >64 | >64 |
| 21 | >64 | >64 | >64 | >64 | >64 | >64 |
| 22 | >64 | >64 | >64 | >64 | >64 | >64 |
| 23 | >64 | >64 | >64 | >64 | >64 | >64 |
| 24 | >64 | >64 | >64 | >64 | >64 | >64 |
| 25 | >64 | >64 | >64 | >64 | >64 | >64 |
| 26 | >64 | >64 | >64 | >64 | >64 | >64 |
| 27 | >64 | >64 | >64 | >64 | >64 | >64 |
| 28 | >64 | >64 | >64 | >64 | >64 | >64 |
| 29 | >64 | >64 | >64 | >64 | >64 | >64 |
| 30 | >64 | >64 | >64 | >64 | >64 | >64 |
| 31 | >64 | >64 | >64 | >64 | >64 | >64 |
| 32 | >64 | >64 | >64 | >64 | >64 | >64 |
| 33 | >64 | >64 | >64 | >64 | >64 | >64 |
| 34 | >64 | >64 | >64 | >64 | >64 | >64 |
| 35 | >64 | >64 | >64 | >64 | >64 | >64 |
| 36 | 16 | >64 | >64 | >64 | >64 | >64 |
| 37 | >64 | >64 | >64 | >64 | >64 | >64 |
| Fluconazole | 8 | 16 | 32 | 32 | >64 | 64 |
| Amphotericin B | >0.5 | >0.5 | >0.5 | >0.5 | >0.5 | 0.5 |
| Molecule | Molecular Weight | No. Heavy Atoms | No. Aromatic Heavy Atoms | Fraction Csp3 | No. Rotatable Bonds | No. H Bond Acceptors | No. H Bond Donors |
|---|---|---|---|---|---|---|---|
| 9 | 317.77 | 22 | 12 | 0.12 | 7 | 3 | 3 |
| 13 | 333.38 | 25 | 16 | 0.1 | 7 | 3 | 3 |
| 14 | 289.35 | 20 | 11 | 0.14 | 7 | 3 | 3 |
| 15 | 334.35 | 23 | 11 | 0.14 | 8 | 5 | 3 |
| 16 | 318.28 | 23 | 11 | 0.14 | 8 | 6 | 3 |
| 20 | 365.43 | 24 | 16 | 0.29 | 7 | 5 | 3 |
| 21 | 437.45 | 32 | 16 | 0.18 | 13 | 7 | 3 |
| 22 | 469.58 | 32 | 16 | 0.18 | 13 | 5 | 3 |
| 24 | 527.44 | 38 | 16 | 0.18 | 15 | 11 | 3 |
| 29 | 457.52 | 34 | 18 | 0.15 | 13 | 5 | 3 |
| 30 | 547.52 | 40 | 18 | 0.15 | 15 | 9 | 3 |
| 31 | 526.41 | 36 | 18 | 0.15 | 13 | 5 | 3 |
| 33 | 489.52 | 36 | 18 | 0.15 | 13 | 7 | 5 |
| 36 | 409.48 | 30 | 16 | 0.36 | 9 | 5 | 1 |
| 37 | 437.53 | 32 | 16 | 0.33 | 11 | 3 | 3 |
| Ceftazidime | 454.51 | 29 | 10 | 0.43 | 8 | 9 | 2 |
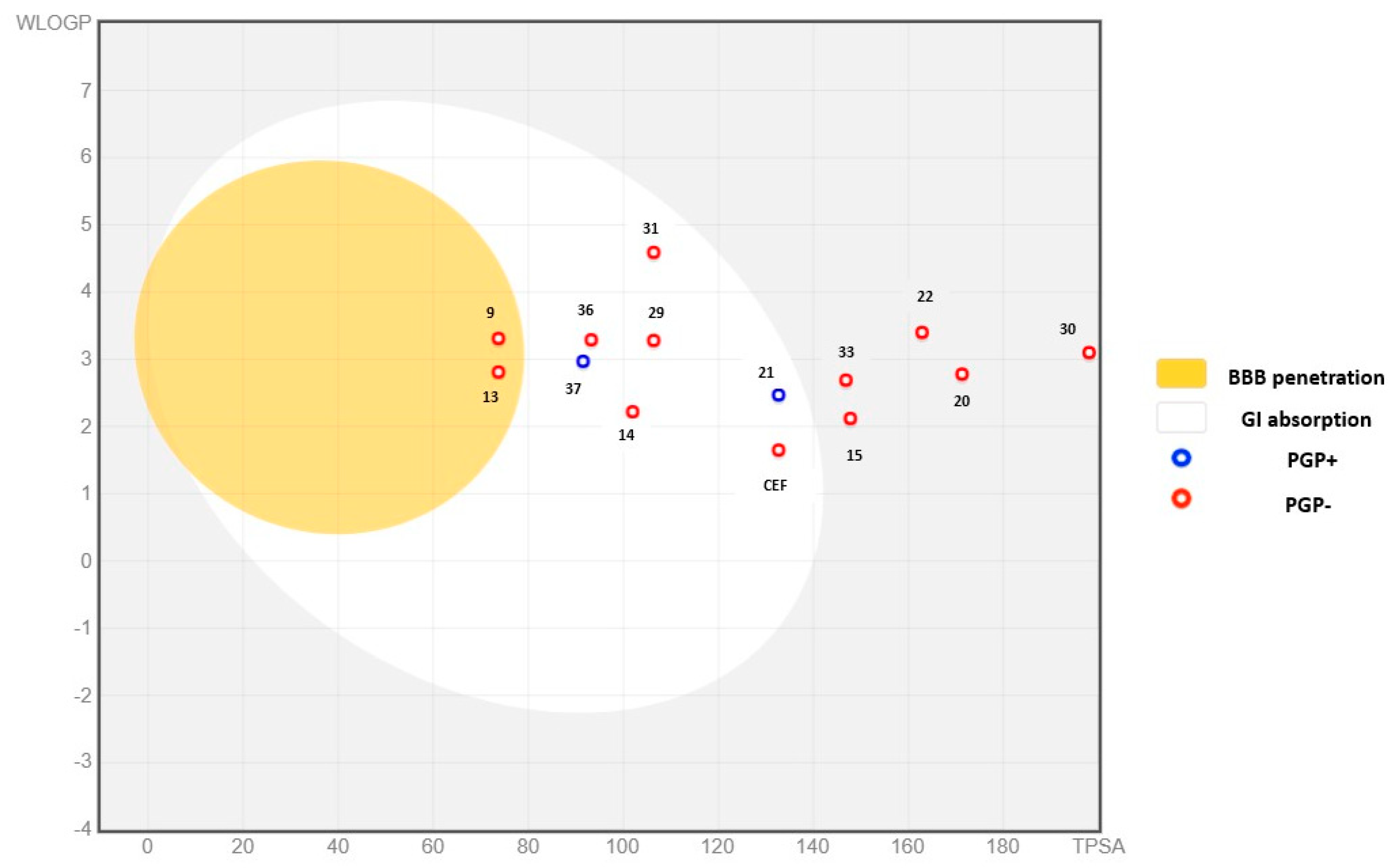
| Molecule | Lipophilicity | GI Absorption | BBB Permeant | P-gp Substrate | CYP1A2 Inhibitor | CYP2C19 Inhibitor | CYP2C9 Inhibitor | CYP2D6 Inhibitor | CYP3A4 Inhibitor |
|---|---|---|---|---|---|---|---|---|---|
| 9 | 2.85 | High | Yes | No | Yes | Yes | Yes | Yes | No |
| 13 | 3.26 | High | Yes | No | Yes | Yes | Yes | Yes | No |
| 14 | 2.27 | High | No | No | Yes | No | No | No | No |
| 15 | 1.55 | Low | No | No | Yes | No | No | No | No |
| 16 | 1.07 | High | No | No | No | No | No | No | No |
| 20 | 2.36 | Low | No | No | No | No | No | No | Yes |
| 21 | 1.96 | High | No | Yes | No | No | Yes | Yes | Yes |
| 22 | 3.27 | Low | No | No | No | Yes | Yes | Yes | Yes |
| 24 | 0.95 | Low | No | Yes | No | Yes | Yes | No | Yes |
| 29 | 3.24 | High | No | No | No | Yes | Yes | Yes | No |
| 30 | 1.86 | Low | No | No | No | Yes | Yes | No | No |
| 31 | 4.23 | High | No | No | No | Yes | Yes | Yes | No |
| 33 | 2.42 | Low | No | No | No | No | Yes | No | No |
| 36 | 2.74 | High | No | No | No | Yes | Yes | No | Yes |
| 37 | 2.82 | High | No | Yes | No | Yes | No | Yes | Yes |
| Ceftazidime | −0.15 | Low | No | Yes | No | No | No | No | No |
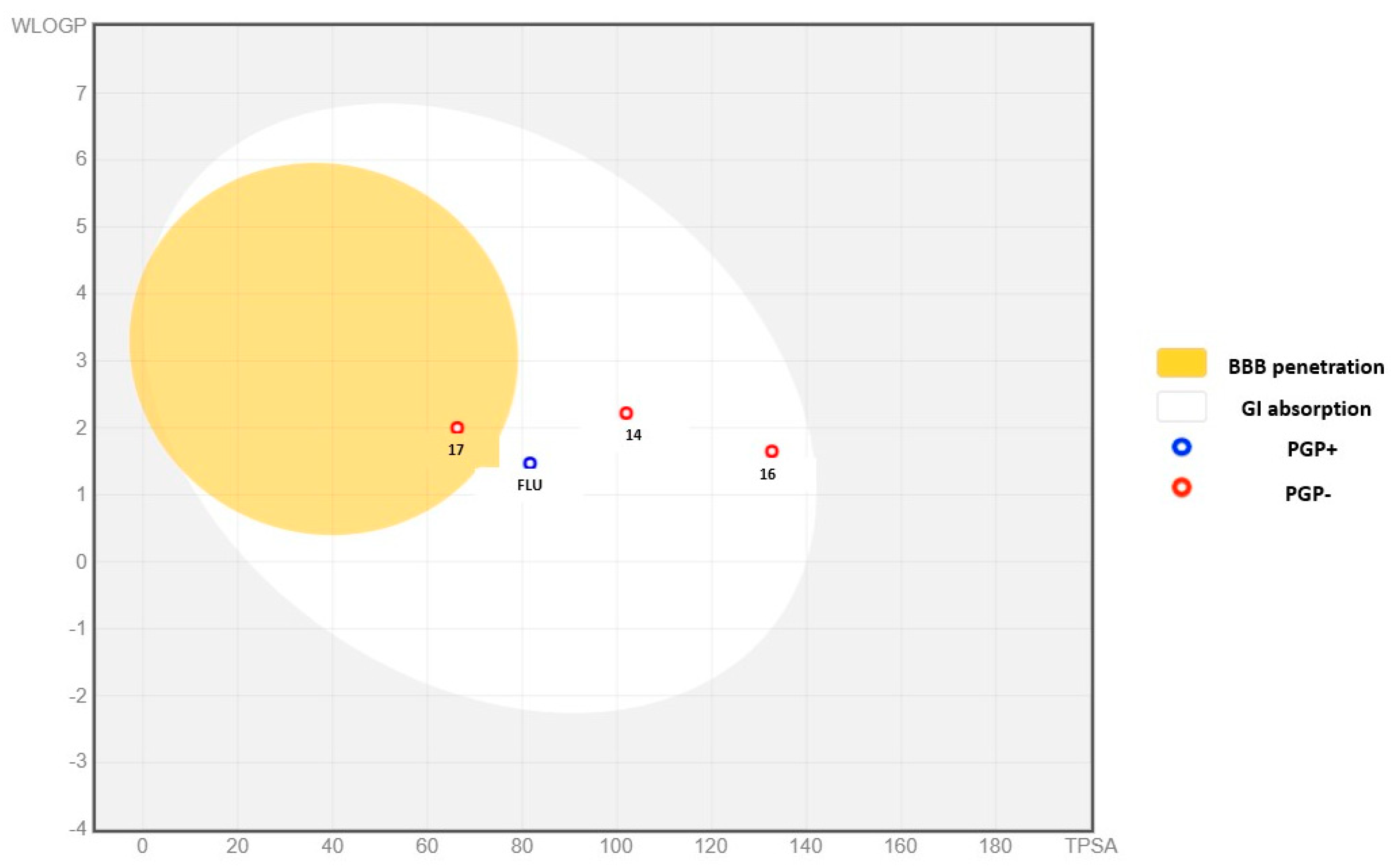
| Molecule | Lipophilicity | GI Absorption | BBB Permeant | P-Gp Substrate | CYP1A2 Inhibitor | CYP2C19 Inhibitor | CYP2C9 Inhibitor | CYP2D6 Inhibitor | CYP3A4 Inhibitor |
|---|---|---|---|---|---|---|---|---|---|
| 14 | 2.27 | High | No | No | Yes | No | No | No | No |
| 16 | 1.07 | High | No | No | No | No | No | No | No |
| 17 | 2.03 | High | Yes | No | Yes | No | No | Yes | No |
| FLU | 0.88 | High | No | Yes | No | Yes | No | No | No |
| Molecule | MW | No. Heavy Atoms | No. Aromatic Heavy Atoms | Fraction Csp3 | No. Rotatable Bonds | No. H Bond Acceptors | No. H Bond Donors |
|---|---|---|---|---|---|---|---|
| 14 | 289.35 | 20 | 11 | 0.14 | 7 | 3 | 3 |
| 16 | 318.28 | 23 | 11 | 0.14 | 8 | 6 | 3 |
| 17 | 273.33 | 20 | 11 | 0.27 | 6 | 2 | 3 |
| FLU | 306.27 | 22 | 16 | 0.23 | 5 | 7 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kavaliauskas, P.; Grybaitė, B.; Sapijanskaitė-Banevič, B.; Vaickelionienė, R.; Petraitis, V.; Petraitienė, R.; Naing, E.; Garcia, A.; Grigalevičiūtė, R.; Mickevičius, V. Synthesis of 3-((4-Hydroxyphenyl)amino)propanoic Acid Derivatives as Promising Scaffolds for the Development of Antimicrobial Candidates Targeting Multidrug-Resistant Bacterial and Fungal Pathogens. Antibiotics 2024, 13, 193. https://doi.org/10.3390/antibiotics13020193
Kavaliauskas P, Grybaitė B, Sapijanskaitė-Banevič B, Vaickelionienė R, Petraitis V, Petraitienė R, Naing E, Garcia A, Grigalevičiūtė R, Mickevičius V. Synthesis of 3-((4-Hydroxyphenyl)amino)propanoic Acid Derivatives as Promising Scaffolds for the Development of Antimicrobial Candidates Targeting Multidrug-Resistant Bacterial and Fungal Pathogens. Antibiotics. 2024; 13(2):193. https://doi.org/10.3390/antibiotics13020193
Chicago/Turabian StyleKavaliauskas, Povilas, Birutė Grybaitė, Birutė Sapijanskaitė-Banevič, Rita Vaickelionienė, Vidmantas Petraitis, Rūta Petraitienė, Ethan Naing, Andrew Garcia, Ramunė Grigalevičiūtė, and Vytautas Mickevičius. 2024. "Synthesis of 3-((4-Hydroxyphenyl)amino)propanoic Acid Derivatives as Promising Scaffolds for the Development of Antimicrobial Candidates Targeting Multidrug-Resistant Bacterial and Fungal Pathogens" Antibiotics 13, no. 2: 193. https://doi.org/10.3390/antibiotics13020193
APA StyleKavaliauskas, P., Grybaitė, B., Sapijanskaitė-Banevič, B., Vaickelionienė, R., Petraitis, V., Petraitienė, R., Naing, E., Garcia, A., Grigalevičiūtė, R., & Mickevičius, V. (2024). Synthesis of 3-((4-Hydroxyphenyl)amino)propanoic Acid Derivatives as Promising Scaffolds for the Development of Antimicrobial Candidates Targeting Multidrug-Resistant Bacterial and Fungal Pathogens. Antibiotics, 13(2), 193. https://doi.org/10.3390/antibiotics13020193








