PA-Win2: In Silico-Based Discovery of a Novel Peptide with Dual Antibacterial and Anti-Biofilm Activity
Abstract
1. Introduction
2. Results
2.1. Identification of PA-Win2 from the Transcriptome of Pardosa Astrigera Venom Gland via In Silico Method
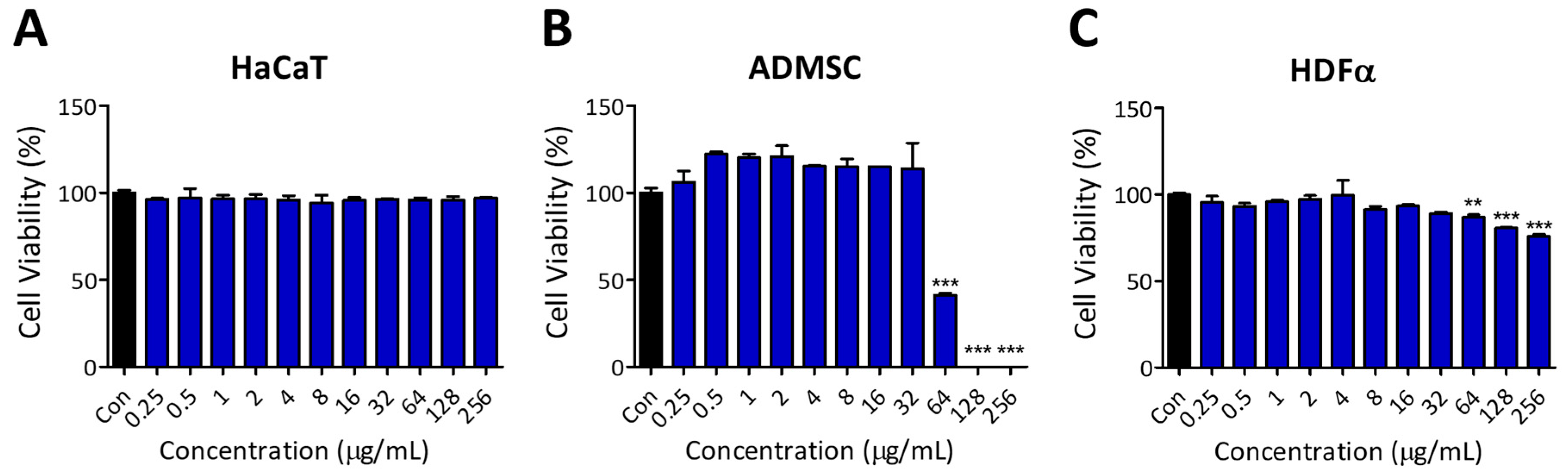
2.2. Evaluation of Antibacterial Activity and Cytocompatibility of PA-Win2
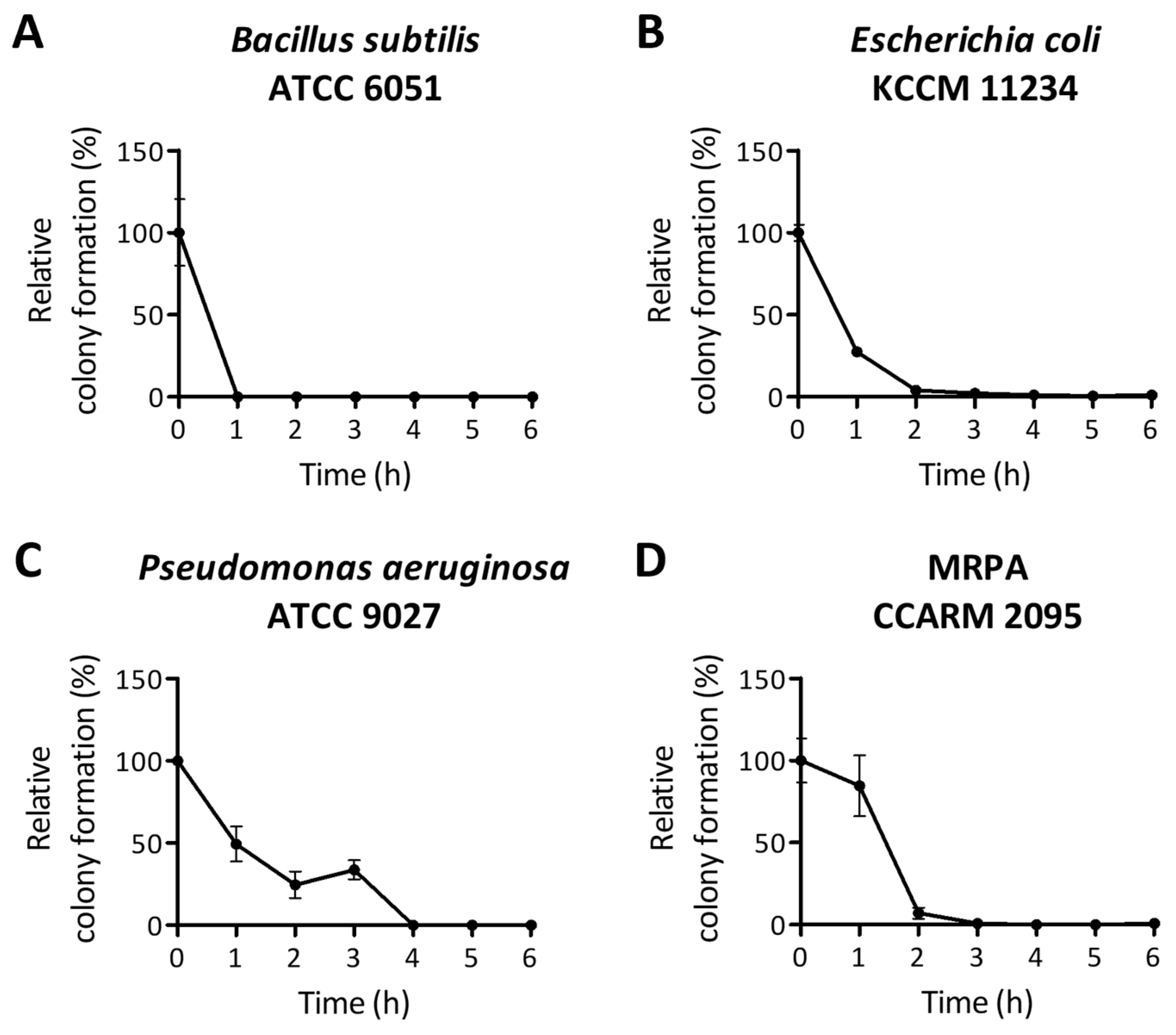
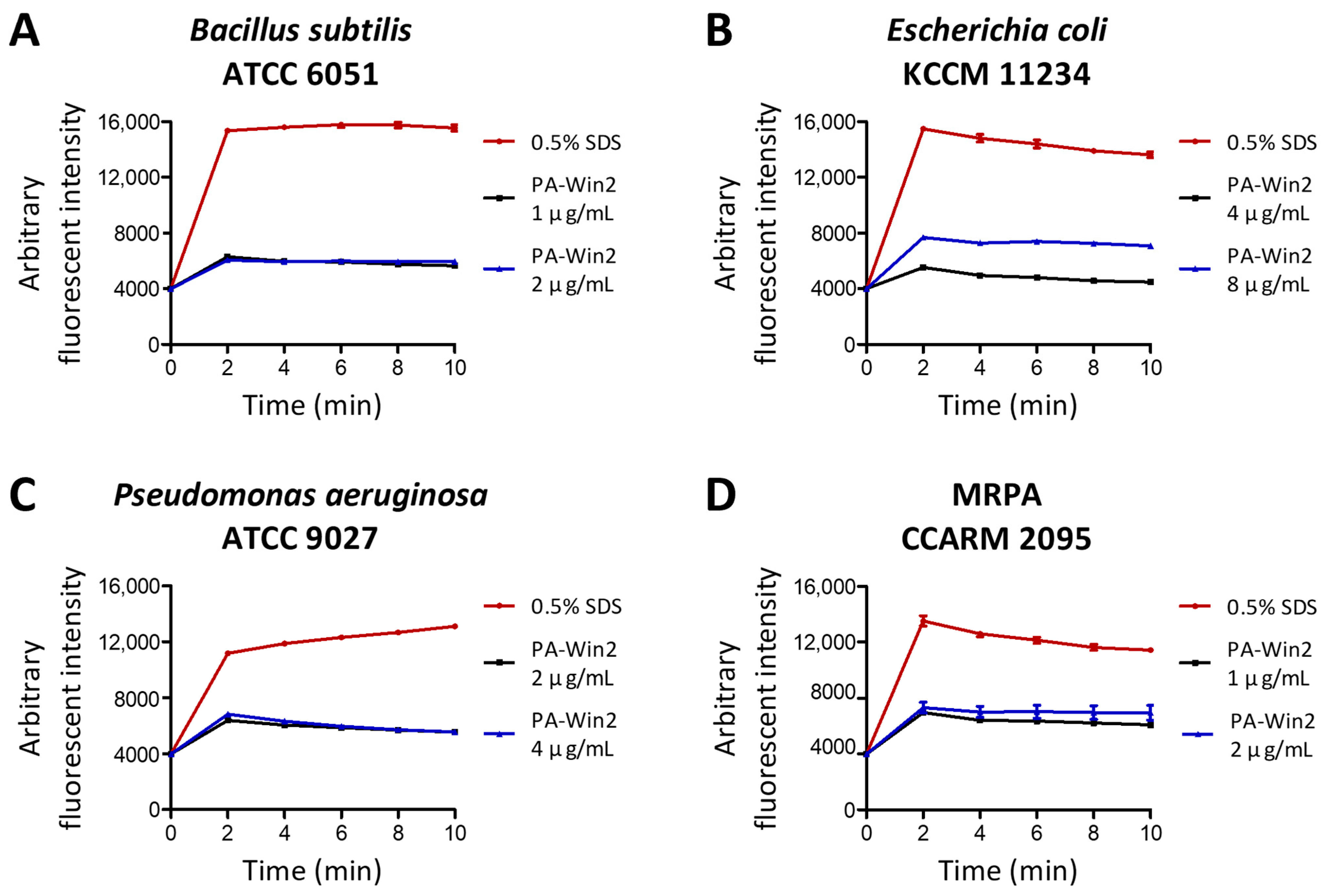
2.3. Bactericidal Activity of PA-Win2 by Membrane Depolarization
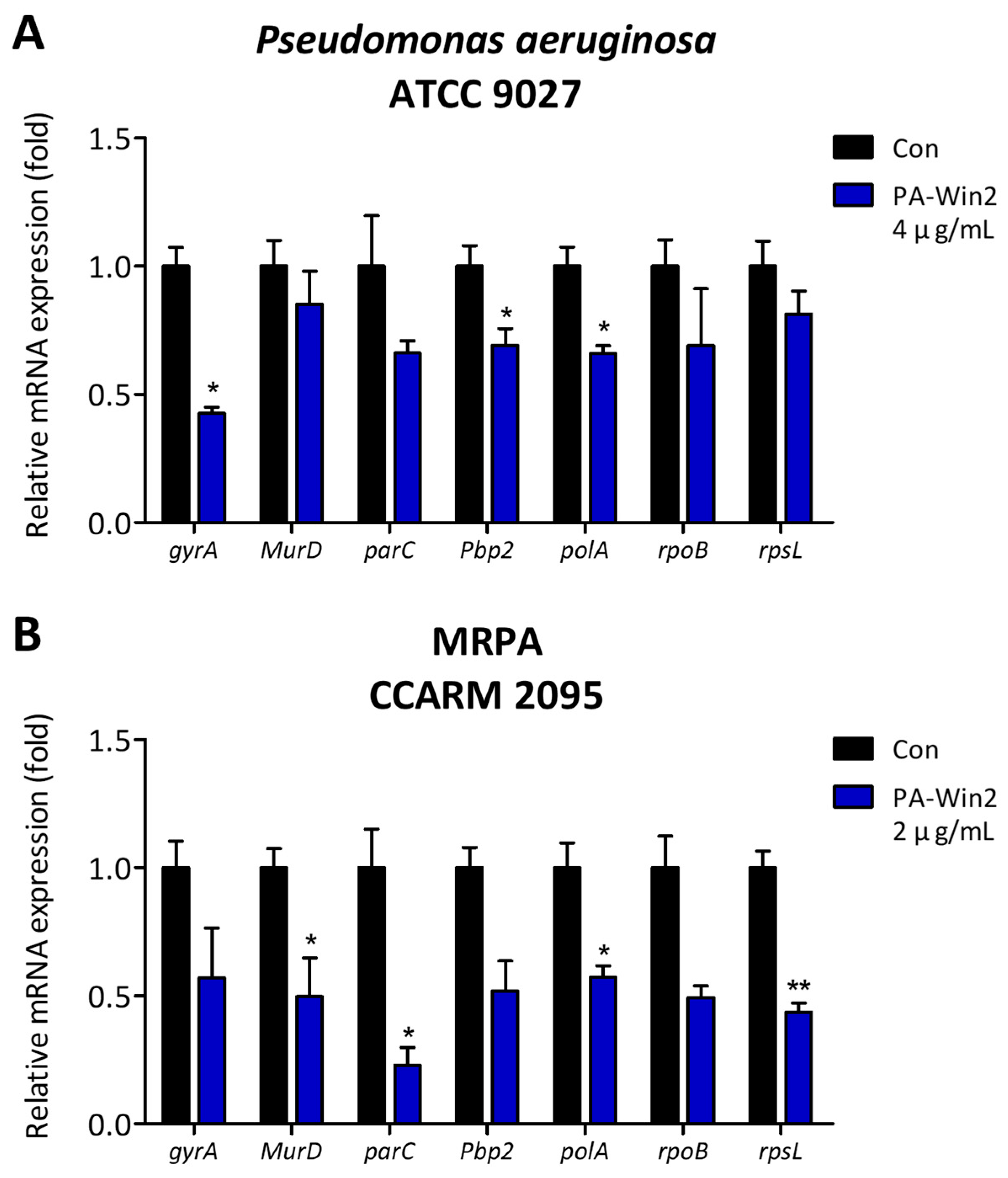
2.4. Changes in P. aeruginosa and MRPA mRNA Expression upon PA-Win2 Treatment
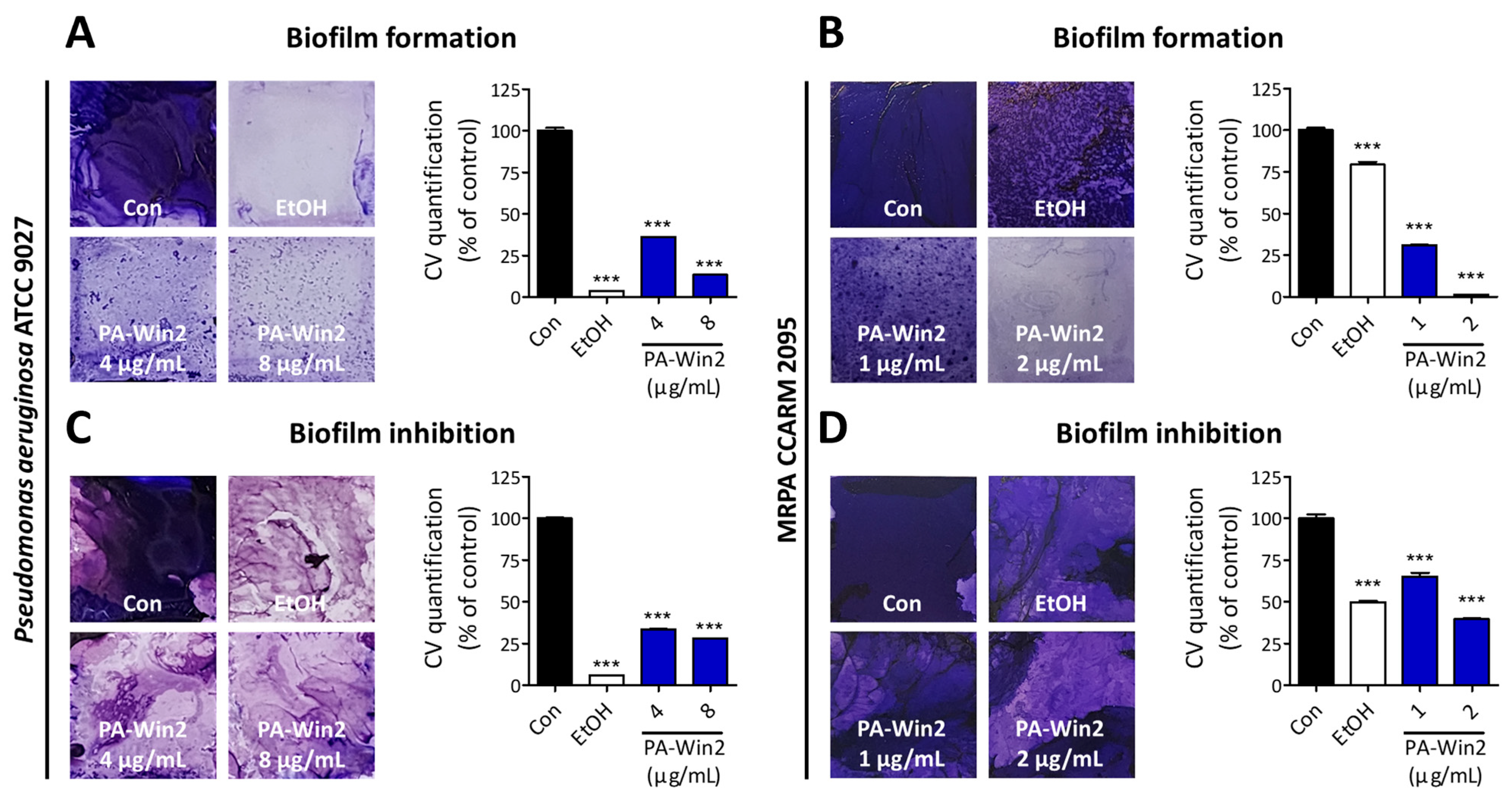
2.5. PA-Win2 Inhibits Biofilm and QS Gene Expressions in P. aeruginosa and MRPA
3. Discussion
4. Materials and Methods
4.1. In Silico Methods Used for AMP Discovery
4.2. Peptide Synthesis and Preparation
4.3. Peptide Stability Test
4.4. Bacterial Strains and Cell Lines
4.5. Antimicrobial Activity Assays
4.6. Cell Viability Assay
4.7. Time-Kill Curve Assay
4.8. Membrane Depolarization Measurement
4.9. RT-qPCR
4.10. Biofilm Formation and Inhibition Assay
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef] [PubMed]
- Gould, I.M.; Bal, A.M. New antibiotic agents in the pipeline and how they can help overcome microbial resistance. Virulence 2013, 4, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Tuon, F.F.; Dantas, L.R.; Suss, P.H.; Tasca Ribeiro, V.S. Pathogenesis of the Pseudomonas aeruginosa Biofilm: A Review. Pathogens 2022, 11, 300. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef]
- Mwangi, J.; Hao, X.; Lai, R.; Zhang, Z.Y. Antimicrobial peptides: New hope in the war against multidrug resistance. Zool. Res. 2019, 40, 488–505. [Google Scholar] [CrossRef]
- Bechinger, B.; Gorr, S.-U. Antimicrobial Peptides: Mechanisms of Action and Resistance. J. Dent. Res. 2017, 96, 254–260. [Google Scholar] [CrossRef]
- Giovagnorio, F.; de Vito, A.; Madeddu, G.; Parisi, S.G.; Geremia, N. Resistance in Pseudomonas aeruginosa: A Narrative Review of Antibiogram Interpretation and Emerging Treatments. Antibiotics 2023, 12, 1621. [Google Scholar] [CrossRef]
- Zhang, Y. Why do we study animal toxins? Dongwuxue Yanjiu 2015, 36, 183–222. [Google Scholar] [CrossRef]
- Vidya, V.; Achar, R.R.; Himathi, M.U.; Akshita, N.; Kameshwar, V.H.; Byrappa, K.; Ramadas, D. Venom peptides—A comprehensive translational perspective in pain management. Curr. Res. Toxicol. 2021, 2, 329–340. [Google Scholar] [CrossRef]
- Ageitos, L.; Torres, M.D.T.; de la Fuente-Nunez, C. Biologically Active Peptides from Venoms: Applications in Antibiotic Resistance, Cancer, and Beyond. Int. J. Mol. Sci. 2022, 23, 15437. [Google Scholar] [CrossRef]
- Escoubas, P.; Sollod, B.; King, G.F. Venom landscapes: Mining the complexity of spider venoms via a combined cDNA and mass spectrometric approach. Toxicon 2006, 47, 650–663. [Google Scholar] [CrossRef]
- Guo, R.; Guo, G.; Wang, A.; Xu, G.; Lai, R.; Jin, H. Spider-Venom Peptides: Structure, Bioactivity, Strategy, and Research Applications. Molecules 2023, 29, 35. [Google Scholar] [CrossRef] [PubMed]
- Munhoz, J.; Thomé, R.; Rostami, A.; Ishikawa, L.L.W.; Verinaud, L.; Rapôso, C. The SNX-482 peptide from Hysterocrates gigas spider acts as an immunomodulatory molecule activating macrophages. Peptides 2021, 146, 170648. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Ma, T.; Yang, L.; Peng, S.; Li, L.; Wang, Z.; Xiao, Z.; Zhang, Q.; Wang, L.; Huang, Y.; et al. Spider venom-derived peptide induces hyperalgesia in Nav1.7 knockout mice by activating Nav1.9 channels. Nat. Commun. 2020, 11, 2293. [Google Scholar] [CrossRef]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M.; et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef]
- Porto, W.F.; Irazazabal, L.; Alves, E.S.F.; Ribeiro, S.M.; Matos, C.O.; Pires, Á.S.; Fensterseifer, I.C.M.; Miranda, V.J.; Haney, E.F.; Humblot, V.; et al. In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design. Nat. Commun. 2018, 9, 1490. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.K.; Hwang, I.-W.; Jang, B.-Y.; Bu, K.-B.; Yoo, J.S.; Sung, J.-S. In silico identification of novel antimicrobial peptides from the venom gland transcriptome of the spider Argiope bruennichi (Scopoli, 1772). Front. Microbiol. 2023, 14, 1249175. [Google Scholar] [CrossRef]
- Shin, M.K.; Park, H.R.; Hwang, I.W.; Bu, K.B.; Jang, B.Y.; Lee, S.H.; Oh, J.W.; Yoo, J.S.; Sung, J.S. In Silico-Based Design of a Hybrid Peptide with Antimicrobial Activity against Multidrug-Resistant Pseudomonas aeruginosa Using a Spider Toxin Peptide. Toxins 2023, 15, 668. [Google Scholar] [CrossRef]
- Gupta, R.; Srivastava, D.; Sahu, M.; Tiwari, S.; Ambasta, R.K.; Kumar, P. Artificial intelligence to deep learning: Machine intelligence approach for drug discovery. Mol. Divers. 2021, 25, 1315–1360. [Google Scholar] [CrossRef]
- Wang, G.; Vaisman, I.I.; van Hoek, M.L. Machine Learning Prediction of Antimicrobial Peptides. Methods Mol. Biol. 2022, 2405, 1–37. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.K.; Hwang, I.-W.; Kim, Y.; Kim, S.T.; Jang, W.; Lee, S.; Bang, W.Y.; Bae, C.-H.; Sung, J.-S. Antibacterial and Anti-Inflammatory Effects of Novel Peptide Toxin from the Spider Pardosa astrigera. Antibiotics 2020, 9, 422. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Shin, M.K.; Yoo, J.S.; Jang, W.; Sung, J.S. Identifying novel antimicrobial peptides from venom gland of spider Pardosa astrigera by deep multi-task learning. Front. Microbiol. 2022, 13, 971503. [Google Scholar] [CrossRef]
- Zhang, J.; Li, W.; Chen, J.; Qi, W.; Wang, F.; Zhou, Y. Impact of biofilm formation and detachment on the transmission of bacterial antibiotic resistance in drinking water distribution systems. Chemosphere 2018, 203, 368–380. [Google Scholar] [CrossRef]
- Nikaido, H. Multidrug resistance in bacteria. Annu. Rev. Biochem. 2009, 78, 119–146. [Google Scholar] [CrossRef]
- Siegel, J.D.; Rhinehart, E.; Jackson, M.; Chiarello, L. Management of multidrug-resistant organisms in health care settings, 2006. Am. J. Infect. Control 2007, 35, S165–S193. [Google Scholar] [CrossRef] [PubMed]
- Ben, Y.; Fu, C.; Hu, M.; Liu, L.; Wong, M.H.; Zheng, C. Human health risk assessment of antibiotic resistance associated with antibiotic residues in the environment: A review. Environ. Res. 2019, 169, 483–493. [Google Scholar] [CrossRef]
- Parmanik, A.; Das, S.; Kar, B.; Bose, A.; Dwivedi, G.R.; Pandey, M.M. Current Treatment Strategies Against Multidrug-Resistant Bacteria: A Review. Curr. Microbiol. 2022, 79, 388. [Google Scholar] [CrossRef]
- Lee, H.; Lee, S.; Lee, I.; Nam, H. AMP-BERT: Prediction of antimicrobial peptide function based on a BERT model. Protein Sci. 2023, 32, e4529. [Google Scholar] [CrossRef]
- Veltri, D.; Kamath, U.; Shehu, A. Deep learning improves antimicrobial peptide recognition. Bioinformatics 2018, 34, 2740–2747. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Espada, R.; Shahrour, H.; Pitts, B.; Stewart, P.S.; Sánchez-Gómez, S.; Martínez-de-Tejada, G. A permeability-increasing drug synergizes with bacterial efflux pump inhibitors and restores susceptibility to antibiotics in multi-drug resistant Pseudomonas aeruginosa strains. Sci. Rep. 2019, 9, 3452. [Google Scholar] [CrossRef] [PubMed]
- Mba, I.E.; Nweze, E.I. Antimicrobial Peptides Therapy: An Emerging Alternative for Treating Drug-Resistant Bacteria. Yale J. Biol. Med. 2022, 95, 445–463. [Google Scholar] [PubMed]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef]
- Rázquin-Olazarán, I.; Shahrour, H.; Martínez-de-Tejada, G. A synthetic peptide sensitizes multi-drug resistant Pseudomonas aeruginosa to antibiotics for more than two hours and permeabilizes its envelope for twenty hours. J. Biomed. Sci. 2020, 27, 85. [Google Scholar] [CrossRef]
- Luo, Y.; Song, Y. Mechanism of Antimicrobial Peptides: Antimicrobial, Anti-Inflammatory and Antibiofilm Activities. Int. J. Mol. Sci. 2021, 22, 1401. [Google Scholar] [CrossRef]
- Bombaywala, S.; Bajaj, A.; Dafale, N.A. Deterministic effect of oxygen level variation on shaping antibiotic resistome. J. Hazard Mater. 2024, 465, 133047. [Google Scholar] [CrossRef]
- Sanders Laurie, H.; Rockel, A.; Lu, H.; Wozniak Daniel, J.; Sutton Mark, D. Role of Pseudomonas aeruginosa dinB-Encoded DNA Polymerase IV in Mutagenesis. J. Bacteriol. 2006, 188, 8573–8585. [Google Scholar] [CrossRef] [PubMed]
- Spinnato, M.C.; Lo Sciuto, A.; Mercolino, J.; Lucidi, M.; Leoni, L.; Rampioni, G.; Visca, P.; Imperi, F. Effect of a Defective Clamp Loader Complex of DNA Polymerase III on Growth and SOS Response in Pseudomonas aeruginosa. Microorganisms 2022, 10, 423. [Google Scholar] [CrossRef]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Stewart, P.S. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 2002, 292, 107–113. [Google Scholar] [CrossRef]
- Wilder, C.N.; Diggle, S.P.; Schuster, M. Cooperation and cheating in Pseudomonas aeruginosa: The roles of the las, rhl and pqs quorum-sensing systems. ISME J. 2011, 5, 1332–1343. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Mao, S.; Wang, H.; Ye, X. The Molecular Architecture of Pseudomonas aeruginosa Quorum-Sensing Inhibitors. Mar. Drugs 2022, 20, 488. [Google Scholar] [CrossRef]
- Yan, S.; Wu, G. Can Biofilm Be Reversed Through Quorum Sensing in Pseudomonas aeruginosa? Front. Microbiol. 2019, 10, 1582. [Google Scholar] [CrossRef] [PubMed]
- Rey, J.; Murail, S.; de Vries, S.; Derreumaux, P.; Tuffery, P. PEP-FOLD4: A pH-dependent force field for peptide structure prediction in aqueous solution. Nucleic Acids Res. 2023, 51, W432–W437. [Google Scholar] [CrossRef]


| Transcript ID | 20-Mer Sequence | Antibacterial Activity Prediction (%) | Net Charge | Water Solubility | ||||
|---|---|---|---|---|---|---|---|---|
| Bacillus subtilis | Escherichia coli | Pseudomonas aeruginosa | Staphylococcus aureus | Staphylococcus epidermidis | ||||
| TBIU038741 | IILLIIIILVVIYYRRRLRR | 0.994 | 0.990 | 0.991 | 0.995 | 0.981 | +5 | Poor |
| TBIU034561 | LILFRFLGYIVLRYVRKPK | 0.994 | 0.989 | 0.990 | 0.995 | 0.979 | +5 | Poor |
| TBIU038389 | LFLLIFCLWKLGFFKRRKPG | 0.994 | 0.988 | 0.990 | 0.995 | 0.979 | +4.9 | Poor |
| TBIU038647 | LLRGLRYLCLKILYILKLRK | 0.994 | 0.988 | 0.990 | 0.994 | 0.979 | +5.9 | Good |
| TBIU038959 | LILLIIILWKCGFFKRKKPG | 0.994 | 0.987 | 0.990 | 0.994 | 0.979 | +4.9 | Poor |
| Concentration (μg/mL) | Bacillus subtilis ATCC 6051 | Escherichia coli KCCM 11234 | Pseudomonas aeruginosa ATCC 9027 | Staphylococcus aureus KCCM 11335 | Staphylococcus epidermidis ATCC 12228 | MRPA CCARM 2095 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| Ampicillin | 8 | 8 | 64 | 128 | 256 | >256 | 0.125 | 1 | 32 | 64 | >512 | >512 |
| Streptomycin | 128 | >256 | 8 | 16 | 32 | 32 | 8 | 16 | >512 | >512 | >512 | >512 |
| Tetracycline | 2 | 32 | 0.5 | 32 | 32 | >256 | 0.125 | >32 | 8 | 64 | >512 | >512 |
| Rifampicin | 1 | 8 | 8 | 16 | 16 | >16 | 0.125 | 0.5 | 0.125 | 0.125 | 16 | 16 |
| PA-Win2 | 2 | 2 | 8 | 32 | 4 | 4 | 256 | >256 | 64 | >256 | 2 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, J.W.; Shin, M.K.; Park, H.-R.; Kim, S.; Lee, B.; Yoo, J.S.; Chi, W.-J.; Sung, J.-S. PA-Win2: In Silico-Based Discovery of a Novel Peptide with Dual Antibacterial and Anti-Biofilm Activity. Antibiotics 2024, 13, 1113. https://doi.org/10.3390/antibiotics13121113
Oh JW, Shin MK, Park H-R, Kim S, Lee B, Yoo JS, Chi W-J, Sung J-S. PA-Win2: In Silico-Based Discovery of a Novel Peptide with Dual Antibacterial and Anti-Biofilm Activity. Antibiotics. 2024; 13(12):1113. https://doi.org/10.3390/antibiotics13121113
Chicago/Turabian StyleOh, Jin Wook, Min Kyoung Shin, Hye-Ran Park, Sejun Kim, Byungjo Lee, Jung Sun Yoo, Won-Jae Chi, and Jung-Suk Sung. 2024. "PA-Win2: In Silico-Based Discovery of a Novel Peptide with Dual Antibacterial and Anti-Biofilm Activity" Antibiotics 13, no. 12: 1113. https://doi.org/10.3390/antibiotics13121113
APA StyleOh, J. W., Shin, M. K., Park, H.-R., Kim, S., Lee, B., Yoo, J. S., Chi, W.-J., & Sung, J.-S. (2024). PA-Win2: In Silico-Based Discovery of a Novel Peptide with Dual Antibacterial and Anti-Biofilm Activity. Antibiotics, 13(12), 1113. https://doi.org/10.3390/antibiotics13121113






