Error in Figure and Table
In the original publication [1], there was a mistake in Figure 1 as published. The depictions of molecules 31 and 32 in Figure 1 erroneously contained a 2H-pyran ring instead of the correct 1,2-dihydropyridine ring. The corrected portion of Figure 1 appears as follows:
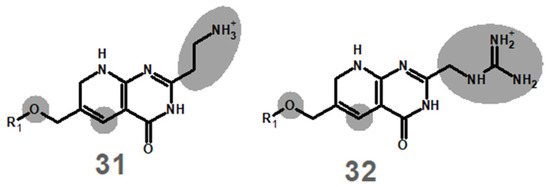
The protonation and reduction energies given in the original Table 1 referred to the 2H-pyran-containing analogues. The correct energies for the actual molecules 31 and 32 are given in Table 1 below. I apologize to the readers for this oversight. The results and conclusions of the original publication are not affected, since the protonation and reduction energies of these molecules are confirmed to render both its protonation and its reduction by NADH unfeasible, and therefore both 31 and 32 are (like the erroneously depicted counterparts) confirmed to be incapable of modification by plasmid-borne dihydrofolate reductase.

Table 1.
PBE0/6-311+G(2d,p)//PBE0/6-31+G(d) energies for the protonation (at position 5) and hydride transfer from NADH to each molecule (in its original protonation state). Bolded values show molecules significantly less basic than the pteridine core (>10 kcal·mol−1 difference) or with reduction energies above 22 kcal·mol−1.
The name of molecule 32 has been corrected in the abstract: “Molecule 32 ({[6-(hydroxymethyl)-4-oxo-3,4,7,8-tetrahydropyrido[2,3-d]pyrimidin-2-yl]methylguanidinium was shown by this methodology to afford extremely stable binding towards R67 DHFR and to prevent simultaneous binding to the substrate”.
No other portions of the text dealing with molecules 31 and 32 are affected, as both the docking and the molecular dynamics simulations described therein were performed with the correct 1,2-dihydropyridine-containing structures.
The authors state that the scientific conclusions are unaffected. This correction was approved by the Academic Editor. The original publication has also been updated.
Reference
- Silva, P.J. Computational Development of Inhibitors of Plasmid-Borne Bacterial Dihydrofolate Reductase. Antibiotics 2022, 11, 779. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).