The Synergy of Machine Learning and Epidemiology in Addressing Carbapenem Resistance: A Comprehensive Review
Abstract
1. Introduction
- Comprehensive overview of synergy. A detailed analysis of how ML and epidemiological methods can complement each other in addressing carbapenem resistance is provided.
- Identification of gaps in traditional approaches. The review outlines the limitations of traditional epidemiological methods in capturing the complexity of resistance mechanisms and transmission patterns and discusses how ML can fill these gaps.
- Evaluation of ML applications. It examines the current state of ML applications in antimicrobial resistance, particularly in predicting CR, and the potential effectiveness of these models in clinical and public health settings.
- Proposals for future research. The review identifies key areas for future research, including the need for more robust data integration, model validation, and the development of real-time surveillance systems.
- Clinical and public health implications. The review emphasizes the clinical and public health benefits of integrating ML and epidemiology to improve predictions, patient outcomes, and intervention strategies.
2. Specific Focus on Carbapenem Resistance
2.1. Mechanisms of Resistance
2.2. Epidemiology and Incidence of Carbapenem-Resistant Organisms
2.3. Clinical Implications
3. Epidemiological Methods
3.1. Introduction to Epidemiology
3.2. Traditional Epidemiological Approaches to Studying AMR
3.3. Strengths and Limitations of Epidemiological Approaches in the Context of Rapidly Evolving Resistance Patterns
- Data lag. The time required to collect, process, and analyze data can result in delays, making it challenging to respond promptly to emerging resistance threats [35].
- Data completeness. Incomplete data collection and reporting can lead to gaps in understanding the full scope of AMR. Variability in laboratory capacities and surveillance systems across regions further complicates this issue [36].
- Complexity of AMR. AMR is influenced by a multitude of factors, including antibiotic usage, infection control practices, and genetic mechanisms. Traditional methods may struggle to account for these complex, multifactorial influences without integrating more advanced analytical techniques [37].
- Predictive limitations. Traditional epidemiological methods often focus on descriptive and retrospective analyses, which may not be sufficient for predicting future resistance trends or for real-time surveillance [38].
4. Machine Learning in Healthcare
4.1. Introduction to Machine Learning
- Supervised learning, which involves training an algorithm on a labeled dataset, where the input–output pairs are known. The algorithm learns to map inputs to the correct output. Common algorithms include linear regression, decision trees, and support vector machines [41].
- Unsupervised learning, where the algorithm is trained on data without labeled responses and aims to find hidden patterns or intrinsic structures in the input data. Key techniques include clustering (e.g., k-means and hierarchical clustering) and association (e.g., Apriori algorithm) [42].
- Reinforcement learning, where the algorithm learns by interacting with an environment, receiving rewards or penalties based on the actions it takes. It aims to maximize cumulative rewards over time. Examples include Q-learning and deep reinforcement learning [43].
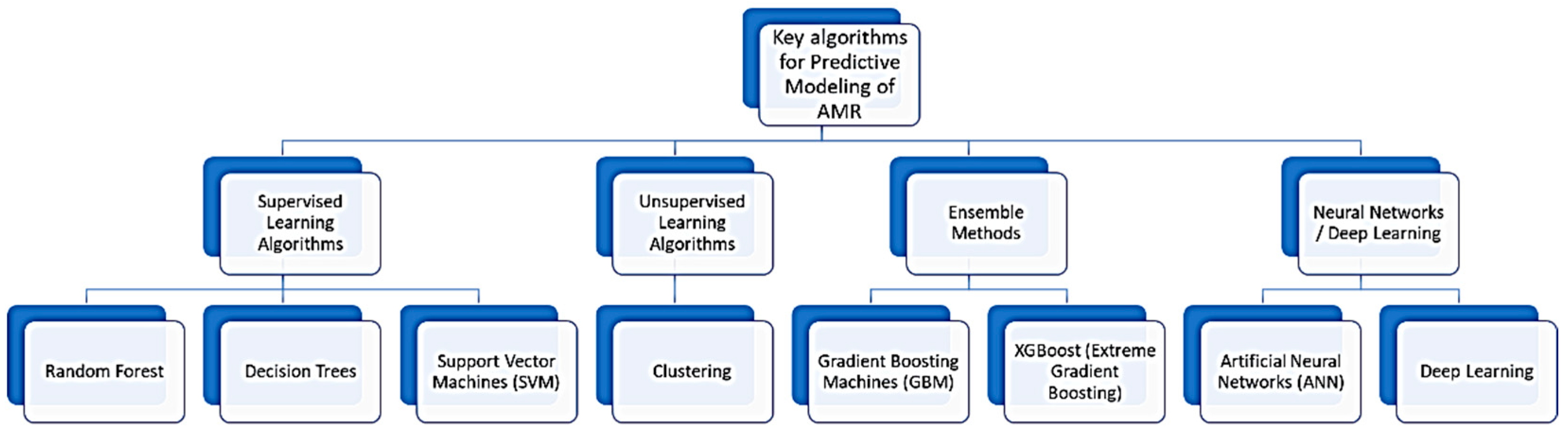
4.2. Key Algorithms and Applications
- Linear regression, which is used for predicting a continuous target variable based on one or more predictor variables [44].
- Decision trees, with a flowchart-like structure, where each internal node represents a decision based on an attribute, and each leaf node represents an outcome [45].
- Support vector machine (SVM), which is a classification method that finds the hyperplane that best separates the data into classes [46].
- Neural networks and deep learning models are inspired by the human brain’s structure, capable of learning complex patterns from large datasets, used extensively in image and speech recognition. In the context of deep learning, an artificial neural network with more than one hidden layer is referred to as deep learning, distinguishing it from simpler models with fewer layers [47].
4.3. Bridging Terminology: Aligning Epidemiology and Machine Learning Concepts
4.4. Benefits of Machine Learning in Analyzing Complex Biological Data and Predicting Trends
5. Integration of Machine Learning and Epidemiology
5.1. Data Sources and Preprocessing Techniques
- Genomic data. Genomic sequences, which comprise DNA or RNA of both pathogens and hosts, are helpful in the identification of genetic markers responsible for specific traits, such as drug resistance and virulence, which become indispensable for full comprehension of infectious disease mechanisms and epidemiology [52].
- Clinical data. Patients’ electronic health records (EHRs) are a rich source of vital information, such as demographics, diagnosis, treatment, and results/outcomes. This dataset captures detailed patient histories that can be used to track disease progression and treatment responses [53].
- Environmental data. Environmental factors, such as air quality, water quality, and climatic variables, may affect dissemination of infectious diseases. Such information may even indicate changing environmental conditions and, thus, the impact on disease transmission [54].
- Sociodemographic data. Information on aspects such as the population’s economic status, density, and education level is critical for understanding disease transmission within populations. More so, such elements can bring to light health-related disparities and susceptibilities [55].
- Data cleaning. Identification and correction of errors, inconsistencies, or incompleteness. Cleaning the data assures dependability and quality within the data and, hence, validation of ML models [56].
- Normalization. When several datasets are combined, normalization is necessary to standardize their scale. Certain algorithms are sensitive to the range of the data; thus, normalization ensures that no feature dominates the model because of a difference in its scale [57].
- Feature selection, which determines the most relevant variables. This, in turn, aids model performance by reducing dimensionality and weeding out insignificant or redundant data. It is totally focused on the most important part of the data and yields a higher performance with low computational complexity [58]. Figure 2 outlines the steps for a machine learning workflow for predictive modeling of AMR. This workflow illustrates how machine learning models handle diverse datasets for predicting AMR trends and providing clinical decision support.
5.2. Predictive Modeling
- Accuracy. The proportion of true results (both true positives and true negatives) among the total number of cases examined. It indicates the overall correctness of the model [74].
- Precision. The proportion of true-positive results among all positive results predicted by the model. It measures the model’s ability to correctly identify true resistance cases without including false positives [75].
- Recall (sensitivity). The proportion of true-positive results among all actual positive cases. It assesses the model’s ability to detect true resistance cases [76].
- F1 score. The harmonic mean of precision and recall, providing a single metric that balances both. It is particularly useful when the data are imbalanced, meaning the number of positive cases is much smaller than the number of negative cases [77].
- Area under the receiver operating characteristic (ROC) curve (AUROC). A plot of the true-positive rate against the false-positive rate at various threshold settings. The AUROC provides a single measure of the model’s ability to discriminate between positive and negative cases [78]. In the context of predicting antibiotic resistance, it measures how effectively the model can differentiate between cases where bacteria are resistant to an antibiotic and cases where they are not.
5.3. Epidemiological Insights
5.4. Real-World Applications
5.5. Case Studies
6. Challenges and Future Directions
6.1. Technical and Ethical Challenges
6.2. Future Directions
- Standardization of data collection. The establishment of standardized protocols for data collection and reporting is critical. This will enhance the quality and comparability of datasets, which are vital for the effectiveness of ML models in epidemiological studies [113].
- Investment in infrastructure. Governments and organizations should invest in the necessary infrastructure, including high-performance computing resources and secure data storage solutions, to support the integration of ML and epidemiological methods [114].
- Interdisciplinary collaboration. It is essential to foster collaboration among data scientists, epidemiologists, healthcare professionals, and policymakers. Such interdisciplinary partnerships can drive the creation of effective and practical solutions for tackling antimicrobial resistance (AMR) and other public health challenges [40].
- Ethical and regulatory frameworks. Developing comprehensive ethical and regulatory frameworks is crucial. These frameworks should address privacy concerns, data security, and the responsible use of AI and ML technologies, thereby ensuring public trust and the successful deployment of these approaches in real-world settings [115].
6.3. The Role of Interdisciplinary Collaboration in Advancing This Field
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom and Wellcome Trust: London, UK, 2016. Available online: https://wellcomecollection.org/works/thvwsuba (accessed on 8 April 2021).
- Walsh, T.R.; Gales, A.C.; Laxminarayan, R.; Dodd, P.C. Antimicrobial resistance: Addressing a global threat to humanity. PLoS Med. 2023, 20, e1004264. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization; Food and Agriculture Organization of the United Nations; United Nations Environment Programme; World Organisation for Animal Health. A One Health Priority Research Agenda for Antimicrobial Resistance; WHO: Geneva, Switzerland, 2023; Available online: https://www.who.int/publications/i/item/9789240075924 (accessed on 2 August 2024).
- Anderson, M.; Panteli, D.; Mossialos, E. Strengthening the EU Response to Prevention and Control of Antimicrobial Resistance (AMR): Policy Priorities for Effective Implementation; WHO Regional Office for Europe: Geneva, Switzerland, 2024. [Google Scholar]
- Laxminarayan, R.; Impalli, I.; Rangarajan, R.; Cohn, J.; Ramjeet, K.; Trainor, B.W.; Strathdee, S.; Sumpradit, N.; Berman, D.; Wertheim, H.; et al. Expanding antibiotic, vaccine, and diagnostics development and access to tackle antimicrobial resistance. Lancet 2024, 403, 2534–2550. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Naas, T.; Poirel, L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerging Infectious Diseases 2011, 17, 1791–1798. [Google Scholar] [CrossRef] [PubMed]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatile beta-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef]
- Poirel, L.; Pitout, J.D.; Nordmann, P. Carbapenemases: Molecular diversity and clinical consequences. Future Microbiol. 2007, 2, 501–512. [Google Scholar] [CrossRef]
- Piddock, L.J.V. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 2006, 19, 382–402. [Google Scholar] [CrossRef]
- Fernández, L.; Hancock, R.E. Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2012, 25, 661–681. [Google Scholar] [CrossRef]
- Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef]
- Michaelis, C.; Grohmann, E. Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms. Antibiotics 2023, 12, 328. [Google Scholar] [CrossRef]
- Mó, I.; da Silva, G.J. Tackling Carbapenem Resistance and the Imperative for One Health Strategies—Insights from the Portuguese Perspective. Antibiotics 2024, 13, 557. [Google Scholar] [CrossRef] [PubMed]
- Logan, L.K.; Weinstein, R.A. The epidemiology of carbapenem-resistant Enterobacteriaceae: The impact and evolution of a global menace. J. Infect. Dis. 2017, 215 (Suppl. S1), S28–S36. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report: 2022; World Health Organization: Geneva, Switzerland, 2022; Available online: https://www.who.int/publications/i/item/9789240062702 (accessed on 2 August 2024).
- van Duin, D.; Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017, 8, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Barmpouni, M.; Gordon, J.P.; Miller, R.L.; Dennis, J.W.; Grammelis, V.; Rousakis, A.; Souliotis, K.; Poulakou, G.; Daikos, G.L.; Al-Taie, A. Clinical and Economic Value of Reducing Antimicrobial Resistance in the Management of Hospital-Acquired Infections with Limited Treatment Options in Greece. Infect. Dis. Ther. 2023, 12, 1891–1905. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Song, X.; Li, M.; Yu, Z.; Cheng, W.; Yu, Z.; Zhang, W.; Zhang, Y.; Shen, A.; Sun, H.; et al. Global spread of carbapenem-resistant Enterobacteriaceae: Epidemiological features, resistance mechanisms, detection and therapy. Microbiol. Res. 2023, 266, 127249. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States, 2019; U.S. Department of Health and Human Services: Washington, DC, USA, 2019. Available online: https://www.cdc.gov/antimicrobial-resistance/data-research/threats/index.html (accessed on 19 August 2024).
- Zhang, S.; Di, L.; Qi, Y.; Qian, X.; Wang, S. Treatment of infections caused by carbapenem-resistant Acinetobacter baumannii. Front. Cell. Infect. Microbiol. 2024, 14, 1395260. [Google Scholar] [CrossRef]
- Tenover, F.C.; Nicolau, D.P.; Gill, C.M. Carbapenemase-producing Pseudomonas aeruginosa—An emerging challenge. Emerg. Microbes Infect. 2022, 11, 811–814. [Google Scholar] [CrossRef]
- Cai, B.; Echols, R.; Magee, G.; Arjona Ferreira, J.C.; Morgan, G.; Ariyasu, M.; Sawada, T.; Nagata, T.D. Prevalence of carbapenem-resistant gram-negative infections in the United States predominated by Acinetobacter baumannii and Pseudomonas aeruginosa. Open Forum Infect. Dis. 2017, 4, ofx176. [Google Scholar] [CrossRef]
- Dossouvi, K.M.; Ametepe, A.S. Carbapenem Resistance in Animal-Environment-Food from Africa: A Systematic Review, Recommendations and Perspectives. Infect. Drug Resist. 2024, 17, 1699–1728. [Google Scholar] [CrossRef]
- Tumbarello, M.; Trecarichi, E.M.; De Rosa, F.G.; Giannella, M.; Giacobbe, D.R.; Bassetti, M.; Losito, A.R.; Bartoletti, M.; Del Bono, V.; Corcione, S.; et al. Infections caused by KPC-producing Klebsiella pneumoniae: Differences in therapy and mortality in a multicentre study. J. Antimicrob. Chemother. 2015, 70, 2133–2143. [Google Scholar] [CrossRef]
- Falagas, M.E.; Tansarli, G.S.; Karageorgopoulos, D.E.; Vardakas, K.Z. Deaths attributable to carbapenem-resistant Enterobacteriaceae infections. Emerg. Infect. Dis. 2014, 20, 1170–1175. [Google Scholar] [CrossRef] [PubMed]
- Bonomo, R.A.; Burd, E.M.; Conly, J.; Limbago, B.M.; Poirel, L.; A Segre, J.; Westblade, L.F. Carbapenemase-producing organisms: A global scourge. Clin. Infect. Dis. 2018, 66, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.J.; Corey, B.W.; Sannio, F.; Hall, L.R.; MacDonald, U.; Jones, B.T.; Mills, E.G.; Harless, C.; Stam, J.; Maybank, R.; et al. Anatomy of an extensively drug-resistant Klebsiella pneumoniae outbreak in Tuscany, Italy. Proc. Natl. Acad. Sci. USA 2021, 118, e2110227118. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.K.; Page, A.; Lohsen, S.; Haider, A.A.; Waggoner, J.; Smith, G.; Babiker, A.; Jacob, J.T.; Howard-Anderson, J.; Satola, S.W. Rates of resistance and heteroresistance to newer β-lactam/β-lactamase inhibitors for carbapenem-resistant Enterobacterales. JAC-Antimicrob. Resist. 2024, 6, dlae048. [Google Scholar] [CrossRef]
- Epidemiology is a science of high importance. Nat. Commun. 2018, 9, 1703. [CrossRef]
- Wiemken, T.L.; Kelley, R.R. Machine learning in epidemiology and health outcomes research. Annu. Rev. Public Health 2020, 41, 21–36. [Google Scholar] [CrossRef]
- Saqib, M.; Iftikhar, M.; Neha, F.; Karishma, F.; Mumtaz, H. Artificial intelligence in critical illness and its impact on patient care: A comprehensive review. Front. Med. 2023, 10, 1176192. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Antimicrobial Resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report 2022; ECDC: Stockholm, Switzerland, 2023; Available online: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2022 (accessed on 16 August 2024).
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modeling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Ayobami, O.; Willrich, N.; Suwono, B.; Eckmanns, T.; Markwart, R. The epidemiology of carbapenem-non-susceptible Acinetobacter species in Europe: Analysis of EARS-Net data from 2013 to 2017. Antimicrob. Resist. Infect. Control 2020, 9, 89. [Google Scholar] [CrossRef]
- Musa, K.; Okoliegbe, I.; Abdalaziz, T.; Aboushady, A.T.; Stelling, J.; Gould, I.M. Laboratory surveillance, quality management, and its role in addressing antimicrobial resistance in Africa: A narrative review. Antibiotics 2023, 12, 1313. [Google Scholar] [CrossRef]
- Holmes, A.H.; Moore, L.S.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J.V. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Wall, S. Prevention of antibiotic resistance—An epidemiological scoping review to identify research categories and knowledge gaps. Glob. Health Action 2019, 12, 1756191. [Google Scholar] [CrossRef] [PubMed]
- Leek, J.T.; Peng, R.D. Statistics: P values are just the tip of the iceberg. Nature 2015, 520, 612. [Google Scholar] [CrossRef] [PubMed]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.P. Machine Learning: A Probabilistic Perspective; MIT Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Sutton, R.S.; Barto, A.G. Reinforcement Learning: An Introduction; MIT Press: Cambridge, MA, USA, 2018. [Google Scholar]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning: With Applications in R; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Breiman, L.; Friedman, J.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees, 1st ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 1984. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine learning in medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef]
- Obermeyer, Z.; Emanuel, E.J. Predicting the future: Big data, machine learning, and clinical medicine. N. Engl. J. Med. 2016, 375, 1216–1219. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Sakagianni, A.; Feretzakis, G.; Kalles, D.; Koufopoulou, C.; Kaldis, V. Setting up an Easy-to-Use Machine Learning Pipeline for Medical Decision Support: A Case Study for COVID-19 Diagnosis Based on Deep Learning with CT Scans. Stud. Health Technol. Inform. 2020, 272, 13–16. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.N.; Weber, G.; Mendis, M.; Gainer, V.; Chueh, H.C.; Churchill, S.; Kohane, I. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J. Am. Med. Inform. Assoc. 2010, 17, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.A.; Southworth, F.; Sarzynski, A. The geography of metropolitan carbon footprints. Policy Soc. 2009, 27, 285–304. [Google Scholar] [CrossRef]
- Marmot, M. Social determinants of health inequalities. Lancet 2005, 365, 1099–1104. [Google Scholar] [CrossRef] [PubMed]
- Rahm, E.; Do, H.H. Data cleaning: Problems and current approaches. IEEE Data Eng. Bull. 2000, 23, 3–13. [Google Scholar]
- Jain, A.K.; Duin, R.P.W.; Mao, J. Statistical pattern recognition: A review. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 4–37. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar] [CrossRef][Green Version]
- Ali, T.; Ahmed, S.; Aslam, M. Artificial Intelligence for Antimicrobial Resistance Prediction: Challenges and Opportunities towards Practical Implementation. Antibiotics 2023, 12, 523. [Google Scholar] [CrossRef]
- Bianconi, I.; Aschbacher, R.; Pagani, E. Current Uses and Future Perspectives of Genomic Technologies in Clinical Microbiology. Antibiotics 2023, 12, 1580. [Google Scholar] [CrossRef]
- Robertson, A.J.; Mallett, A.J.; Stark, Z.; Sullivan, C. It is in our DNA: Bringing electronic health records and genomic data together for precision medicine. JMIR Bioinform. Biotechnol. 2024, 5, e55632. [Google Scholar] [CrossRef]
- Armstrong, G.L.; MacCannell, D.R.; Taylor, J.; Carleton, H.A.; Neuhaus, E.B.; Bradbury, R.S.; Posey, J.E.; Gwinn, M. Pathogen Genomics in Public Health. N. Engl. J. Med. 2019, 381, 2569–2580. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Ding, S.; Chen, J.; Chen, X.; Xu, Y.; Xu, Z.; Huang, M. Prediction of carbapenem-resistant gram-negative bacterial bloodstream infection in intensive care unit based on machine learning. BMC Med. Inform. Decis. Mak. 2024, 24, 123. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Gao, J.; Liu, X.F.; Rao, G.; Luo, J.; Han, P.; Hu, W.; Zhang, Z.; Zhao, Q.; Han, L.; et al. Direct prediction of carbapenem resistance in Pseudomonas aeruginosa by whole genome sequencing and metagenomic sequencing. J. Clin. Microbiol. 2023, 61, e0061723. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cao, Y.; Wang, M.; Wang, L.; Wu, Y.; Fang, Y.; Zhao, Y.; Fan, Y.; Liu, X.; Liang, H.; et al. Development and validation of machine learning models to predict MDRO colonization or infection on ICU admission by using electronic health record data. Antimicrob. Resist. Infect. Control 2024, 13, 74. [Google Scholar] [CrossRef] [PubMed]
- Feretzakis, G.; Loupelis, E.; Sakagianni, A.; Kalles, D.; Martsoukou, M.; Lada, M.; Skarmoutsou, N.; Christopoulos, C.; Valakis, K.; Velentza, A.; et al. Using Machine Learning Techniques to Aid Empirical Antibiotic Therapy Decisions in the Intensive Care Unit of a General Hospital in Greece. Antibiotics 2020, 9, 50. [Google Scholar] [CrossRef] [PubMed]
- Feretzakis, G.; Sakagianni, A.; Loupelis, E.; Kalles, D.; Skarmoutsou, N.; Martsoukou, M.; Christopoulos, C.; Lada, M.; Petropoulou, S.; Velentza, A.; et al. Machine Learning for Antibiotic Resistance Prediction: A Prototype Using Off-the-Shelf Techniques and Entry-Level Data to Guide Empiric Antimicrobial Therapy. Healthc. Inform. Res. 2021, 27, 214–221. [Google Scholar] [CrossRef]
- Tang, R.; Luo, R.; Tang, S.; Song, H.; Chen, X. Machine learning in predicting antimicrobial resistance: A systematic review and meta-analysis. Int. J. Antimicrob. Agents 2022, 60, 106684. [Google Scholar] [CrossRef]
- Kim, J.I.; Maguire, F.; Tsang, K.K.; Gouliouris, T.; Peacock, S.J.; McAllister, T.A.; Beiko, R.G. Machine learning for antimicrobial resistance prediction: Current practice, limitations, and clinical perspective. Clin. Microbiol. Rev. 2022, 35, e0017921. [Google Scholar] [CrossRef]
- Amin, D.; Garzόn-Orjuela, N.; Garcia Pereira, A.; Parveen, S.; Vornhagen, H.; Vellinga, A. Artificial Intelligence to Improve Antibiotic Prescribing: A Systematic Review. Antibiotics 2023, 12, 1293. [Google Scholar] [CrossRef]
- Khaledi, A.; Weimann, A.; Schniederjans, M.; Asgari, E.; Kuo, T.; Oliver, A.; Cabot, G.; Kola, A.; Gastmeier, P.; Hogardt, M.; et al. Predicting antimicrobial resistance in Pseudomonas aeruginosa with machine learning-enabled molecular diagnostics. EMBO Mol. Med. 2020, 12, e10264. [Google Scholar] [CrossRef]
- Ravkin, H.D.; Ravkin, R.M.; Rubin, E.; Nesher, L. Machine-learning-based risk assessment tool to rule out empirical use of ESBL-targeted therapy in endemic areas. J. Hosp. Infect. 2024, 149, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Sophonsri, A.; Lou, M.; Ny, P.; Minejima, E.; Nieberg, P.; Wong-Beringer, A. Machine learning to identify risk factors associated with the development of ventilated hospital-acquired pneumonia and mortality: Implications for antibiotic therapy selection. Front. Med. 2023, 10, 1268488. [Google Scholar] [CrossRef] [PubMed]
- Powers, D.M. Evaluation: From precision, recall and F-measure to ROC, informedness, markedness and correlation. J. Mach. Learn. Technol. 2011, 2, 37–63. [Google Scholar] [CrossRef]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Davis, J.; Goadrich, M. The relationship between precision-recall and ROC curves. In Proceedings of the 23rd International Conference on Machine Learning, Pittsburgh, PA, USA, 25–29 June 2006; pp. 233–240. [Google Scholar] [CrossRef]
- Goutte, C.; Gaussier, E. A probabilistic interpretation of precision, recall and F-score, with implication for evaluation. In Advances in Information Retrieval; Losada, D.E., Fernández-Luna, J.M., Eds.; ECIR 2005; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2005; Volume 3408, pp. 345–359. [Google Scholar] [CrossRef]
- Bradley, A.P. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997, 30, 1145–1159. [Google Scholar] [CrossRef]
- Liang, Q.; Zhao, Q.; Xu, X.; Zhou, Y.; Huang, M. Early prediction of carbapenem-resistant gram-negative bacterial carriage in intensive care units using machine learning. J. Glob. Antimicrob. Resist. 2022, 29, 225–231. [Google Scholar] [CrossRef]
- McGuire, R.J.; Yu, S.C.; Payne, P.R.O.; Lai, A.M.; Vazquez-Guillamet, M.C.; Kollef, M.H.; Michelson, A.P. A Pragmatic Machine Learning Model To Predict Carbapenem Resistance. Antimicrob. Agents Chemother. 2021, 65, e0006321. [Google Scholar] [CrossRef]
- Olawade, D.B.; Wada, O.J.; David-Olawade, A.C.; Kunonga, E.; Abaire, O.; Ling, J. Using artificial intelligence to improve public health: A narrative review. Front. Public Health 2023, 11, 1196397. [Google Scholar] [CrossRef]
- Branda, F.; Scarpa, F. Implications of artificial intelligence in addressing antimicrobial resistance: Innovations, global challenges, and healthcare’s future. Antibiotics 2024, 13, 502. [Google Scholar] [CrossRef]
- Raghupathi, W.; Raghupathi, V. Big data analytics in healthcare: Promise and potential. Health Inf. Sci. Syst. 2014, 2, 3. [Google Scholar] [CrossRef]
- Lepper, H.C.; Woolhouse, M.E.J.; van Bunnik, B.A.D. The role of the environment in dynamics of antibiotic resistance in humans and animals: A modelling study. Antibiotics 2022, 11, 1361. [Google Scholar] [CrossRef]
- Li, L.G.; Yin, X.; Zhang, T. Tracking antibiotic resistance gene pollution from different sources using machine-learning classification. Microbiome 2018, 6, 93. [Google Scholar] [CrossRef]
- Zhang, T.; Rabhi, F.; Chen, X.; Paik, H.Y.; MacIntyre, C.R. A machine learning-based universal outbreak risk prediction tool. Comput. Biol. Med. 2024, 169, 107876. [Google Scholar] [CrossRef]
- Cho, G.; Park, J.R.; Choi, Y.; Ahn, H.; Lee, H. Detection of COVID-19 epidemic outbreak using machine learning. Front. Public Health 2023, 11, 1252357. [Google Scholar] [CrossRef]
- Zeng, D.; Cao, Z.; Neill, D.B. Artificial intelligence–enabled public health surveillance—From local detection to global epidemic monitoring and control. Artif. Intell. Med. 2021, 437–453. [Google Scholar] [CrossRef]
- MacIntyre, C.R.; Chen, X.; Kunasekaran, M.; Quigley, A.; Lim, S.; Stone, H.; Paik, H.Y.; Yao, L.; Heslop, D.; Wei, W.; et al. Artificial intelligence in public health: The potential of epidemic early warning systems. J. Int. Med. Res. 2023, 51, 3000605231159335. [Google Scholar] [CrossRef]
- Giannella, M.; Freire, M.; Rinaldi, M.; Abdala, E.; Rubin, A.; Mularoni, A.; Gruttadauria, S.; Grossi, P.; Shbaklo, N.; Tandoi, F.; et al. Development of a risk prediction model for carbapenem-resistant Enterobacteriaceae infection after liver transplantation: A multinational cohort study. Clin. Infect. Dis. 2021, 73, e955–e966. [Google Scholar] [CrossRef]
- Freire, M.P.; Rinaldi, M.; Terrabuio, D.R.B.; Furtado, M.; Pasquini, Z.; Bartoletti, M.; de Oliveira, T.A.; Nunes, N.N.; Lemos, G.T.; Maccaro, A.; et al. Prediction models for carbapenem-resistant Enterobacterales carriage at liver transplantation: A multicenter retrospective study. Transpl. Infect. Dis. 2022, 24, e13920. [Google Scholar] [CrossRef]
- Çağlayan, Ç.; Barnes, S.L.; Pineles, L.L.; Harris, A.D.; Klein, E.Y. A data-driven framework for identifying intensive care unit admissions colonized with multidrug-resistant organisms. Front. Public Health 2022, 10, 853757. [Google Scholar] [CrossRef]
- Li, Y.; Wang, B.; Wen, L.; Li, H.; He, F.; Wu, J.; Gao, S.; Hou, D. Machine learning and radiomics for the prediction of multidrug resistance in cavitary pulmonary tuberculosis: A multicentre study. Eur. Radiol. 2023, 33, 391–400. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, F.; Li, L.; Pang, Y. Clinical utilization of artificial intelligence in predicting therapeutic efficacy in pulmonary tuberculosis. J. Infect. Public Health 2024, 17, 632–641. [Google Scholar] [CrossRef]
- Burdick, H.; Pino, E.; Gabel-Comeau, D.; Gu, C.; Roberts, J.; Le, S.; Slote, J.; Saber, N.; Pellegrini, E.; Green-Saxena, A.; et al. Validation of a machine learning algorithm for early severe sepsis prediction: A retrospective study predicting severe sepsis up to 48 h in advance using a diverse dataset from 461 US hospitals. BMC Med. Inform. Decis. Mak. 2020, 20, 276. [Google Scholar] [CrossRef]
- Mao, Q.; Jay, M.; Hoffman, J.L.; Calvert, J.; Barton, C.; Shimabukuro, D.; Shieh, L.; Chettipally, U.; Fletcher, G.; Kerem, Y.; et al. Multicenter validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward, and ICU. BMJ Open 2018, 8, e017833. [Google Scholar] [CrossRef]
- Moran, E.; Robinson, E.; Green, C.; Keeling, M.; Collyer, B. Towards personalized guidelines: Using machine-learning algorithms to guide antimicrobial selection. J. Antimicrob. Chemother. 2020, 75, 2677–2680. [Google Scholar] [CrossRef]
- Beam, A.L.; Kohane, I.S. Big data and machine learning in health care. JAMA 2018, 319, 1317–1318. [Google Scholar] [CrossRef]
- Sullivan, T.; Ichikawa, O.; Dudley, J.; Li, L.; Aberg, J. The rapid prediction of carbapenem resistance in patients with Klebsiella pneumoniae bacteremia using electronic medical record data. Open Forum Infect. Dis. 2018, 5, ofy091. [Google Scholar] [CrossRef]
- Arzilli, G.; De Vita, E.; Pasquale, M.; Carloni, L.M.; Pellegrini, M.; Di Giacomo, M.; Esposito, E.; Porretta, A.D.; Rizzo, C. Innovative techniques for infection control and surveillance in hospital settings and long-term care facilities: A scoping review. Antibiotics 2024, 13, 77. [Google Scholar] [CrossRef]
- Elbehiry, A.; Marzouk, E.; Abalkhail, A.; El-Garawany, Y.; Anagreyyah, S.; Alnafea, Y.; Almuzaini, A.M.; Alwarhi, W.; Rawway, M.; Draz, A. The development of technology to prevent, diagnose, and manage antimicrobial resistance in healthcare-associated infections. Vaccines 2022, 10, 2100. [Google Scholar] [CrossRef]
- OECD. Embracing a One Health Framework to Fight Antimicrobial Resistance; OECD Health Policy Studies: Paris, France, 2023. [Google Scholar] [CrossRef]
- AlQudah, A.A.; Al-Emran, M.; Shaalan, K. Medical data integration using HL7 standards for patient’s early identification. PLoS ONE 2021, 16, e0262067. [Google Scholar] [CrossRef]
- Kahn, M.G.; Brown, J.S.; Chun, A.T.; Davidson, B.N.; Meeker, D.; Ryan, P.B.; Schilling, L.M.; Weiskopf, N.G.; Williams, A.E.; Zozus, M.N. Transparent reporting of data quality in distributed data networks. Egems (Gener. Evid. Methods Improv. Patient Outcomes) 2015, 3, 1052. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. Why should I trust you? Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD ‘16), San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar] [CrossRef]
- Mittelstadt, B.D.; Allo, P.; Taddeo, M.; Wachter, S.; Floridi, L. The ethics of algorithms: Mapping the debate. Big Data Soc. 2016, 3. [Google Scholar] [CrossRef]
- Mehrabi, N.; Morstatter, F.; Saxena, N.A.; Lerman, K.; Galstyan, A.G. A survey on bias and fairness in machine learning. ACM Comput. Surv. (CSUR) 2019, 54, 1–35. [Google Scholar] [CrossRef]
- Sakagianni, A.; Koufopoulou, C.; Feretzakis, G.; Kalles, D.; Verykios, V.S.; Myrianthefs, P.; Fildisis, G. Using machine learning to predict antimicrobial resistance—A literature review. Antibiotics 2023, 12, 452. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Dean, J.; Ghemawat, S. MapReduce: Simplified data processing on large clusters. Commun. ACM 2019, 51, 107–113. [Google Scholar] [CrossRef]
- Schadt, E.E.; Linderman, M.D.; Sorenson, J.; Lee, L.; Nolan, G.P. Computational solutions to large-scale data management and analysis. Nat. Rev. Genet. 2010, 11, 647–657. [Google Scholar] [CrossRef]
- Osama, M.; Ateya, A.A.; Sayed, M.S.; Hammad, M.; Pławiak, P.; El-Latif, A.A.A.; Elsayed, R.A. Internet of medical things and healthcare 4.0: Trends, requirements, challenges, and research directions. Sensors 2023, 23, 7435. [Google Scholar] [CrossRef]
- Gliklich, R.E.; Leavy, M.B.; Dreyer, N.A. (Eds.) Tools and Technologies for Registry Interoperability, Registries for Evaluating Patient Outcomes: A User’s Guide, 3rd ed.; Addendum 2; Agency for Healthcare Research and Quality (US): Rockville, MD, USA, 2019. Available online: https://www.ncbi.nlm.nih.gov/books/NBK551879/ (accessed on 19 August 2024).
- OECD. Health at a Glance 2019: OECD Indicators; OECD Publishing: Paris, France, 2019. [Google Scholar] [CrossRef]
- Floridi, L.; Cowls, J.; Beltrametti, M.; Chatila, R.; Chazerand, P.; Dignum, V.; Luetge, C.; Madelin, R.; Pagallo, U.; Rossi, F.; et al. AI4People—An ethical framework for a good AI society: Opportunities, risks, principles, and recommendations. Minds Mach. 2018, 28, 689–707. [Google Scholar] [CrossRef]
- Krause-Jüttler, G.; Weitz, J.; Bork, U. Interdisciplinary Collaborations in Digital Health Research: Mixed Methods Case Study. JMIR Hum. Factors 2022, 9, e36579. [Google Scholar] [CrossRef]

| No. | Author | Geographical Setting | Publication Year | Medical Setting | Data Source | ML Algorithms | Performance Evaluation | Bacterial Species |
|---|---|---|---|---|---|---|---|---|
| 1 | Timothy Sullivan [99] | United States (Single Center) | 2018 | Hospital setting | EHR data, Klebsiella pneumoniae bacteremia cases | Multiple logistic regression | AUROC: 0.731, Sensitivity: 73%, Specificity: 59%, PPV: 16%, NPV: 95% | Klebsiella pneumoniae (Carbapenem-resistant) |
| 2 | Ariane Khaledi [71] | Germany, Spain | 2020 | Clinical settings, multicenter | Whole genome sequencing (WGS), transcriptomic data, gene presence/absence, expression profiles | Machine Learning (unspecified classifiers) | Sensitivity: 0.8–0.9, Predictive values: >0.9 | Pseudomonas aeruginosa (Carbapenem-resistant) |
| 3 | Ed Moran [97] | United Kingdom (Single Center) | 2020 | Hospital setting | Blood and urine cultures, demographics, microbiology and prescribing data | XGBoost | AUROC: 0.70, Point-scoring tools: AUROC 0.61 to 0.67, estimated reduction in broad-spectrum antibiotic use by 40% | Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa |
| 4 | Ryan J. McGuire [80] | United States (Single Center) | 2021 | Tertiary-care academic medical center | Demographics, medications, vital signs, procedures, lab results, cultures | Extreme gradient boosting (XGBoost) | AUROC: 0.846, Sensitivity: 30%, PPV: 30%, NPV: 99% | Carbapenem-resistant bacteria |
| 5 | Maddalena Giannella [90] | Multinational | 2021 | Liver transplantation units (multicenter) | Demographics, clinical data, mechanical ventilation, acute renal injury, surgical reintervention | Multivariable logistic regression, Fine-Gray subdistribution hazard model | AUROC: 74.6 (derivation), AUROC: 73.9 (bootstrapped validation), Brier Index: 16.6 | Carbapenem-resistant Enterobacteriaceae (CRE) |
| 6 | Qiqiang Liang [79] | China (Single Center) | 2022 | Intensive care unit (ICU) | Demographics, screening records, clinical data, vitals | Random forest, XGBoost, decision tree, logistic regression | AUROC: 0.91 (random forest), 0.89 (XGBoost, decision tree), 0.78 (logistic regression) | Carbapenem-resistant Gram-negative bacteria (CRGNB) |
| 7 | Maristela Pinheiro Freire [91] | Brazil, Italy | 2022 | Liver transplantation units (multicenter) | Antibiotic use, hepato-renal syndrome, CLIF-SOFA scores, cirrhosis complications | Machine learning (unspecified) | Sensitivity: 66%, Specificity: 83%, NPV: 97% | Carbapenem-resistant Enterobacterales (CRE) |
| 8 | Çaǧlar Çaǧlayan [92] | United States (Single Center) | 2022 | Intensive care unit (ICU) | EHR, MDRO screening program, sociodemographic and clinical factors | Logistic regression (LR), random forest (RF), XGBoost | Sensitivity: VRE 80%, CRE 73%, MRSA 76%, MDRO 82%; Specificity: VRE 66%, CRE 77%, MRSA 59%, MDRO 83% | MRSA, VRE, Carbapenem-resistant Enterobacteriaceae (CRE) |
| 9 | Qiqiang Liang [63] | China (Single Center) | 2024 | Intensive care unit (ICU) | Demographics, mechanical ventilation, invasive catheterization, carbapenem use history | Random forest, XGBoost, SVM | AUROC: random forest 0.86, XGBoost (infection): 0.86, SVM: 0.88, RF (CRGNB): 0.87 | Carbapenem-resistant Gram-negative bacteria (CRGNB) |
| 10 | Yun Li [65] | China/USA | 2024 | Intensive care unit (ICU) | Electronic health record data (PLAGH-ICU, MIMIC-IV) | Machine learning models | AUROC: 0.786 (PLAGH-ICU), 0.744 (MIMIC-IV) | Multidrug-resistant organisms (MDRO), including carbapenem-resistant species |
| 11 | Bing Liu [64] | China (Single Center) | 2024 | Multiple hospital settings | Whole-genome sequencing (WGS) data, metagenomic sequencing (MGS), genomic features | Machine learning (unspecified algorithms) | AUROC: 0.906 (IPM), 0.925 (MEM), PPV: 0.897 (IPM), 0.889 (MEM) | Pseudomonas aeruginosa (Carbapenem-resistant) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakagianni, A.; Koufopoulou, C.; Koufopoulos, P.; Feretzakis, G.; Kalles, D.; Paxinou, E.; Myrianthefs, P.; Verykios, V.S. The Synergy of Machine Learning and Epidemiology in Addressing Carbapenem Resistance: A Comprehensive Review. Antibiotics 2024, 13, 996. https://doi.org/10.3390/antibiotics13100996
Sakagianni A, Koufopoulou C, Koufopoulos P, Feretzakis G, Kalles D, Paxinou E, Myrianthefs P, Verykios VS. The Synergy of Machine Learning and Epidemiology in Addressing Carbapenem Resistance: A Comprehensive Review. Antibiotics. 2024; 13(10):996. https://doi.org/10.3390/antibiotics13100996
Chicago/Turabian StyleSakagianni, Aikaterini, Christina Koufopoulou, Petros Koufopoulos, Georgios Feretzakis, Dimitris Kalles, Evgenia Paxinou, Pavlos Myrianthefs, and Vassilios S. Verykios. 2024. "The Synergy of Machine Learning and Epidemiology in Addressing Carbapenem Resistance: A Comprehensive Review" Antibiotics 13, no. 10: 996. https://doi.org/10.3390/antibiotics13100996
APA StyleSakagianni, A., Koufopoulou, C., Koufopoulos, P., Feretzakis, G., Kalles, D., Paxinou, E., Myrianthefs, P., & Verykios, V. S. (2024). The Synergy of Machine Learning and Epidemiology in Addressing Carbapenem Resistance: A Comprehensive Review. Antibiotics, 13(10), 996. https://doi.org/10.3390/antibiotics13100996










