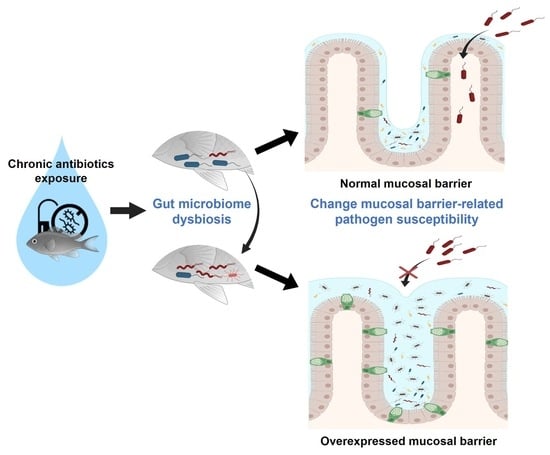
Effects of Antibiotic Residues on Fish Gut Microbiome Dysbiosis and Mucosal Barrier-Related Pathogen Susceptibility in Zebrafish Experimental Model
Abstract
1. Introduction
2. Results
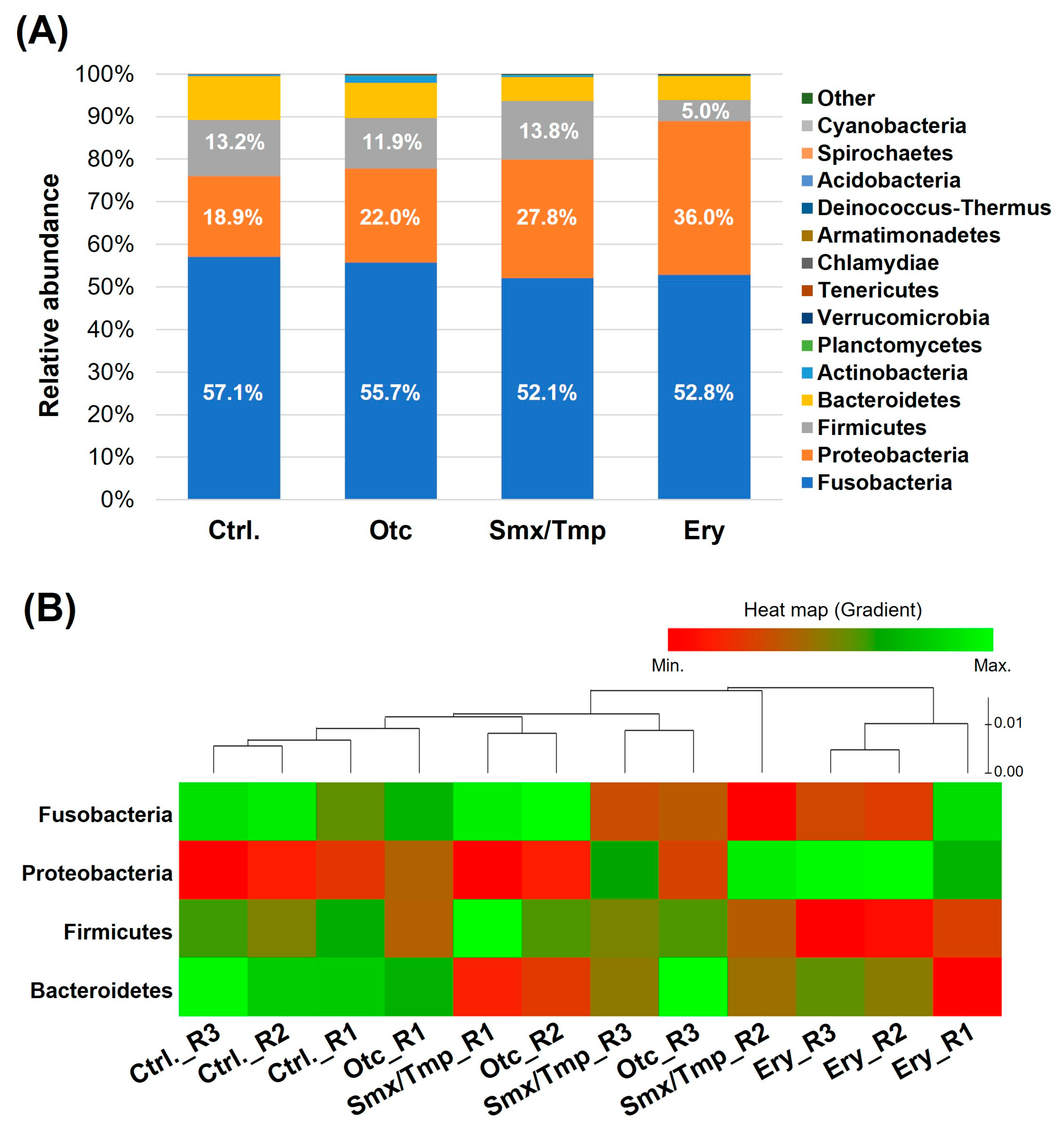
2.1. Zebrafish Gut Microbiome Dysbiosis Induced by Chronic Antibiotics Exposure under Laboratory Conditions
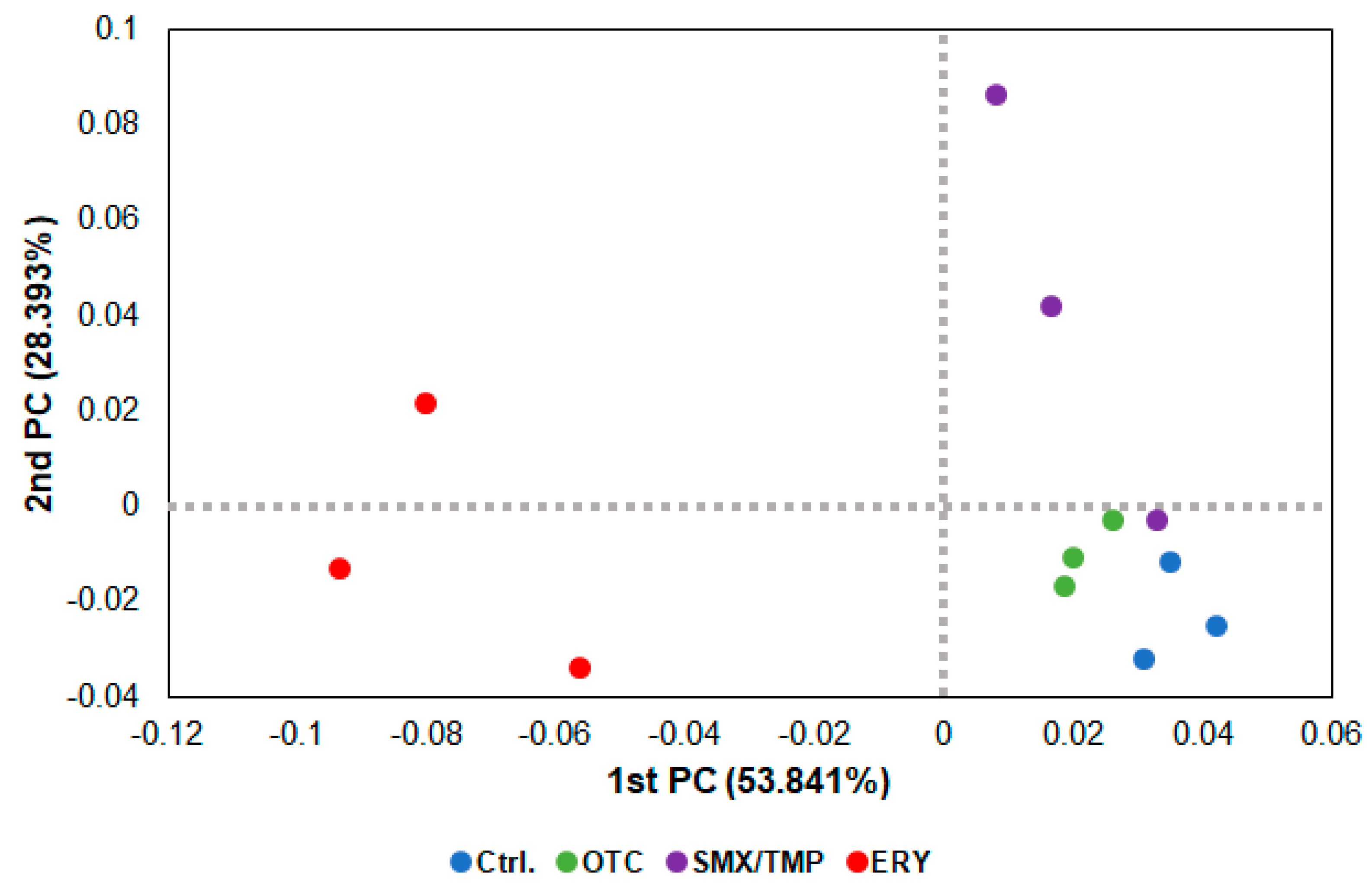
2.2. Chronic Antibiotics Exposure Did Not Affect the Zebrafish Gut Microbiome Diversity
2.3. Alteration of Pathogen Susceptibility Due to the Zebrafish Gut Microbiome Dysbiosis
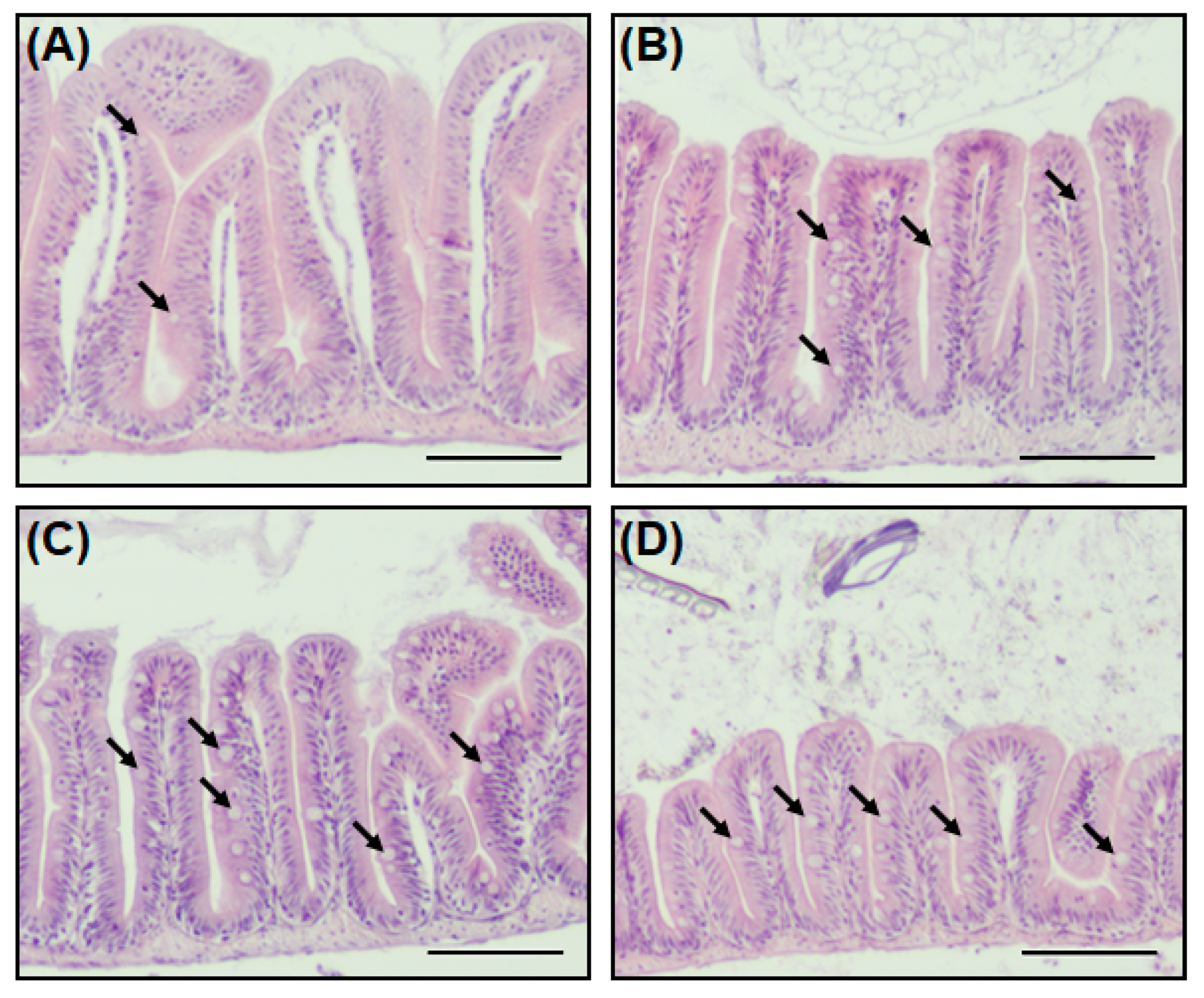
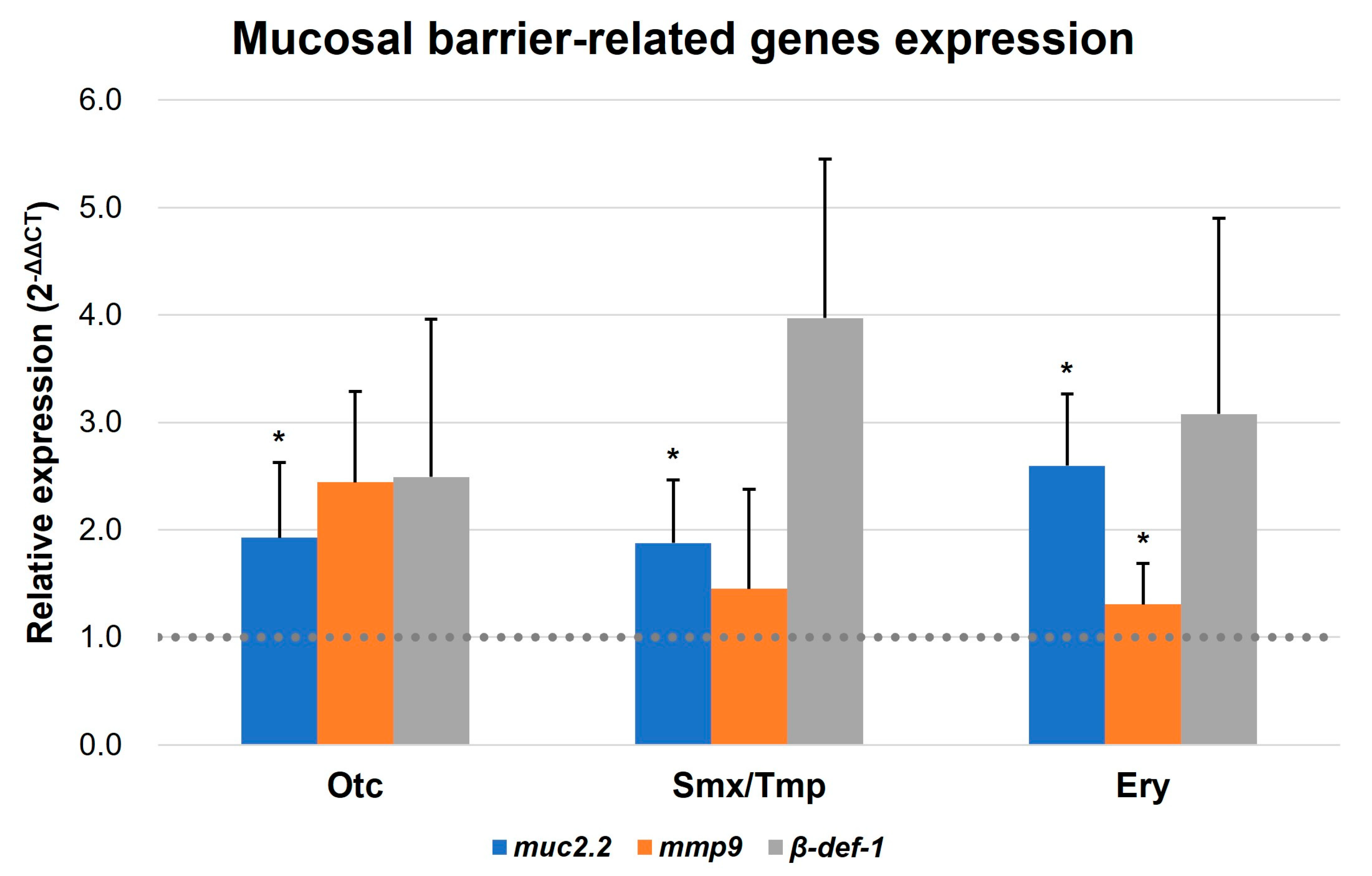
2.4. A New Hypothesis of Low Pathogen Susceptibility: An Increase in the Intestinal Mucosal Barrier
2.5. Alteration of Immune and Stress-Related Gene Expression by Overexpressed Intestinal Mucosal Barrier
3. Discussion
4. Materials and Methods
4.1. Experimental Animals
4.2. Chronic Antibiotics Exposure Assay
4.3. Zebrafish Gut Microbiome Analysis
4.4. Lethal Dose 50 Assay on Edwardsiella piscicida
4.5. Histological Analysis
4.6. Quantitative Reverse Transcription PCR
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barko, P.C.; McMichael, M.A.; Swanson, K.S.; Williams, D.A. The Gastrointestinal Microbiome: A Review. J. Vet. Intern. Med. 2018, 32, 9–25. [Google Scholar] [CrossRef]
- Laukens, D.; Brinkman, B.M.; Raes, J.; De Vos, M.; Vandenabeele, P. Heterogeneity of the gut microbiome in mice: Guidelines for optimizing experimental design. FEMS Microbiol. Rev. 2016, 40, 117–132. [Google Scholar] [CrossRef]
- Tarnecki, A.M.; Burgos, F.A.; Ray, C.L.; Arias, C.R. Fish intestinal microbiome: Diversity and symbiosis unravelled by metagenomics. J. Appl. Microbiol. 2017, 123, 2–17. [Google Scholar] [CrossRef]
- Wiens, J.J. Explaining large-scale patterns of vertebrate diversity. Biol. Lett. 2015, 11, 20150506. [Google Scholar] [CrossRef]
- Kim, P.S.; Shin, N.-R.; Lee, J.-B.; Kim, M.-S.; Whon, T.W.; Hyun, D.-W.; Yun, J.-H.; Jung, M.-J.; Kim, J.Y.; Bae, J.-W. Host habitat is the major determinant of the gut microbiome of fish. Microbiome 2021, 9, 166. [Google Scholar] [CrossRef]
- Action Sustainability Inc. World Fisheries and Aquacultur; Food and Agriculture Organization: Rome, Italy, 2020; Volume 2020, pp. 1–244. [Google Scholar]
- Ashley, P.J. Fish welfare: Current issues in aquaculture. Appl. Anim. Behav. Sci. 2007, 104, 199–235. [Google Scholar] [CrossRef]
- Lafferty, K.D.; Harvell, C.D.; Conrad, J.M.; Friedman, C.S.; Kent, M.L.; Kuris, A.M.; Powell, E.N.; Rondeau, D.; Saksida, S.M. Infectious diseases affect marine fisheries and aquaculture economics. Ann. Rev. Mar. Sci. 2015, 7, 471–496. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Sánchez, T.; Mora-Sánchez, B.; Balcázar, J.L. Biological approaches for disease control in aquaculture: Advantages, limitations and challenges. Trends Microbiol. 2018, 26, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Sun, R.; Pan, C.; Sun, Y.; Mai, B.; Li, Q.X. Antibiotics and food safety in aquaculture. J. Agric. Food Chem. 2020, 68, 11908–11919. [Google Scholar] [CrossRef]
- Maki, T.; Hasegawa, H.; Kitami, H.; Fumoto, K.; Munekage, Y.; Ueda, K. Bacterial degradation of antibiotic residues in marine fish farm sediments of Uranouchi Bay and phylogenetic analysis of antibiotic-degrading bacteria using 16S rDNA sequences. Fish. Sci. 2006, 72, 811–820. [Google Scholar] [CrossRef]
- Björklund, H.; Bondestam, J.; Bylund, G. Residues of oxytetracycline in wild fish and sediments from fish farms. Aquaculture 1990, 86, 359–367. [Google Scholar] [CrossRef]
- Li, W.; Shi, Y.; Gao, L.; Liu, J.; Cai, Y. Occurrence of antibiotics in water, sediments, aquatic plants, and animals from Baiyangdian Lake in North China. Chemosphere 2012, 89, 1307–1315. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Song, G.; Lim, W. A review of the toxicity in fish exposed to antibiotics. Comp. Biochem. Physiol. Part C Toxicol. 2020, 237, 108840. [Google Scholar] [CrossRef]
- Kim, B.; Ji, K.; Kho, Y.; Kim, P.-G.; Park, K.; Kim, K.; Kim, Y.; Kim, K.-T.; Choi, K. Effects of chronic exposure to cefadroxil and cefradine on Daphnia magna and Oryzias latipes. Chemosphere 2017, 185, 844–851. [Google Scholar] [CrossRef]
- Li, Z.; Limbu, S.M.; Fang, Q.; Du, Z.-Y.; Zhang, M. Influence of Long-Term Feeding Antibiotics on the Gut Health of Zebrafish. Zebrafish 2018, 15, 340–348. [Google Scholar] [CrossRef]
- da Silva Morais, A.; Oliveira, J.M.; Reis, R.L. Small animal models. In Osteochondral Tissue Engineering; Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2018; pp. 423–439. [Google Scholar]
- Flores, E.M.; Nguyen, A.T.; Odem, M.A.; Eisenhoffer, G.T.; Krachler, A.M. The zebrafish as a model for gastrointestinal tract–microbe interactions. Cell. Microbiol. 2020, 22, e13152. [Google Scholar] [CrossRef] [PubMed]
- Torraca, V.; Mostowy, S. Zebrafish infection: From pathogenesis to cell biology. Trends Cell Biol. 2018, 28, 143–156. [Google Scholar] [CrossRef]
- Liu, X.; Steele, J.C.; Meng, X.-Z. Usage, residue, and human health risk of antibiotics in Chinese aquaculture: A review. Environ. Pollut. 2017, 223, 161–169. [Google Scholar] [CrossRef]
- Armstrong, S.; Hargrave, B.; Haya, K. Antibiotic Use in Finfish Aquaculture: Modes of Action, Environmental Fate, and Microbial Resistance; Springer: Berlin/Heidelberg, Germany, 2005; pp. 341–357. [Google Scholar]
- Buján, N.; Toranzo, A.E.; Magariños, B. Edwardsiella piscicida: A significant bacterial pathogen of cultured fish. Dis. Aquat. 2018, 131, 59–71. [Google Scholar] [CrossRef]
- Specian, R.D.; Oliver, M.G. Functional biology of intestinal goblet cells. Am. J. Physiol. Cell Physiol. 1991, 260, C183–C193. [Google Scholar] [CrossRef]
- McDermott, A.J.; Huffnagle, G.B. The microbiome and regulation of mucosal immunity. Immunology 2014, 142, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Ulloa, P.E.; Solís, C.J.; De la Paz, J.F.; Alaurent, T.G.; Caruffo, M.; Hernández, A.J.; Dantagnan, P.; Feijóo, C.G. Lactoferrin decreases the intestinal inflammation triggered by a soybean meal-based diet in zebrafish. J. Immunol. Res. 2016, 2016, 1639720. [Google Scholar] [CrossRef]
- Jevtov, I.; Samuelsson, T.; Yao, G.; Amsterdam, A.; Ribbeck, K. Zebrafish as a model to study live mucus physiology. Sci. Rep. 2014, 4, 6653. [Google Scholar] [CrossRef]
- Pujada, A.; Walter, L.; Patel, A.; Bui, T.A.; Zhang, Z.; Zhang, Y.; Denning, T.L.; Garg, P. Matrix metalloproteinase MMP9 maintains epithelial barrier function and preserves mucosal lining in colitis associated cancer. Oncotarget 2017, 8, 94650. [Google Scholar] [CrossRef]
- García-Valtanen, P.; Martinez-Lopez, A.; Ortega-Villaizan, M.; Perez, L.; Coll, J.; Estepa, A. In addition to its antiviral and immunomodulatory properties, the zebrafish β-defensin 2 (zfBD2) is a potent viral DNA vaccine molecular adjuvant. Antivir. Res. 2014, 101, 136–147. [Google Scholar] [CrossRef]
- Loes, A.N.; Hinman, M.N.; Farnsworth, D.R.; Miller, A.C.; Guillemin, K.; Harms, M.J. Identification and characterization of zebrafish Tlr4 coreceptor Md-2. J. Immunol. 2021, 206, 1046–1057. [Google Scholar] [CrossRef]
- Xu, J.; Qian, Q.; Xia, M.; Wang, X.; Wang, H. Trichlorocarban induces developmental and immune toxicity to zebrafish (Danio rerio) by targeting TLR4/MyD88/NF-κB signaling pathway. Environ. Pollut. 2021, 273, 116479. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, D.; Chattopadhyay, A. Induction of oxidative stress and related transcriptional effects of sodium fluoride in female zebrafish liver. Bull. Environ. Contam. Toxicol. 2014, 93, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Malek, R.L.; Sajadi, H.; Abraham, J.; Grundy, M.A.; Gerhard, G.S. The effects of temperature reduction on gene expression and oxidative stress in skeletal muscle from adult zebrafish. Comp. Biochem. Physiol. Part C Toxicol. 2004, 138, 363–373. [Google Scholar] [CrossRef]
- Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Rev. Infect. Dis. 1980, 2, 129–139. [Google Scholar] [CrossRef]
- Terreni, M.; Taccani, M.; Pregnolato, M. New antibiotics for multidrug-resistant bacterial strains: Latest research developments and future perspectives. Molecules 2021, 26, 2671. [Google Scholar] [CrossRef] [PubMed]
- Browne, A.J.; Chipeta, M.G.; Haines-Woodhouse, G.; Kumaran, E.P.; Hamadani, B.H.K.; Zaraa, S.; Henry, N.J.; Deshpande, A.; Reiner, R.C.; Day, N.P. Global antibiotic consumption and usage in humans, 2000–2018: A spatial modelling study. Lancet Planet. Health 2021, 5, e893–e904. [Google Scholar] [CrossRef]
- FDA. 2019 Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals; FDA: Silver Spring, MD, USA, 2014. [Google Scholar]
- Berkner, S.; Konradi, S.; Schönfeld, J. Antibiotic resistance and the environment—There and back again: Science & Society series on Science and Drugs. EMBO Rep. 2014, 15, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Larsson, D.J. Antibiotics in the environment. Ups. J. Med. Sci. 2014, 119, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Wammer, K.H.; Slattery, M.T.; Stemig, A.M.; Ditty, J.L. Tetracycline photolysis in natural waters: Loss of antibacterial activity. Chemosphere 2011, 85, 1505–1510. [Google Scholar] [CrossRef]
- de Bruijn, I.; Liu, Y.; Wiegertjes, G.F.; Raaijmakers, J.M. Exploring fish microbial communities to mitigate emerging diseases in aquaculture. FEMS Microbiol. Ecol. 2018, 94, fix161. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Hao, Q.; Xia, R.; Olsen, R.E.; Ringø, E.; Yang, Y.; Zhang, Z.; Ran, C.; Zhou, Z. Nuclease-treated stabilized fermentation product of Cetobacterium somerae improves growth, non-specific immunity, and liver health of zebrafish (Danio rerio). Front. Nutr. 2022, 9, 918327. [Google Scholar] [CrossRef]
- Xie, M.; Xie, Y.; Li, Y.; Zhou, W.; Zhang, Z.; Yang, Y.; Olsen, R.E.; Ringø, E.; Ran, C.; Zhou, Z. Stabilized fermentation product of Cetobacterium somerae improves gut and liver health and antiviral immunity of zebrafish. Fish Shellfish Immunol. 2022, 120, 56–66. [Google Scholar] [CrossRef]
- Huang, H.; Zhou, P.; Chen, P.; Xia, L.; Hu, S.; Yi, G.; Lu, J.; Yang, S.; Xie, J.; Peng, J. Alteration of the gut microbiome and immune factors of grass carp infected with Aeromonas veronii and screening of an antagonistic bacterial strain (Streptomyces flavotricini). Microb. Pathog. 2020, 143, 104092. [Google Scholar] [CrossRef]
- Arias-Jayo, N.; Abecia, L.; Alonso-Saez, L.; Ramirez-Garcia, A.; Rodriguez, A.; Pardo, M.A. High-Fat Diet Consumption Induces Microbiota Dysbiosis and Intestinal Inflammation in Zebrafish. Microb. Ecol. 2018, 76, 1089–1101. [Google Scholar] [CrossRef]
- Zheng, M.; Lu, J.; Lin, G.; Su, H.; Sun, J.; Luan, T. Dysbiosis of gut microbiota by dietary exposure of three graphene-family materials in zebrafish (Danio rerio). Environ. Pollut. 2019, 254, 112969. [Google Scholar] [CrossRef]
- Johansson, M.E.; Hansson, G.C. Goblet cells need some stress. J. Clin. Investig. 2022, 132, e162030. [Google Scholar] [CrossRef]
- Uribe, C.; Folch, H.; Enríquez, R.; Moran, G. Innate and adaptive immunity in teleost fish: A review. Vet. Med. 2011, 56, 486–503. [Google Scholar] [CrossRef]
- Sun, B.-Y.; He, W.; Yang, H.-X.; Tian, D.-Y.; Jian, P.-Y.; Wu, K.; Yang, C.-G.; Song, X.-H. Increased susceptibility to Aeromonas hydrophila infection in grass carp with antibiotic-induced intestinal dysbiosis. Aquaculture 2022, 552, 737969. [Google Scholar] [CrossRef]
- He, S.; Wang, Q.; Li, S.; Ran, C.; Guo, X.; Zhang, Z.; Zhou, Z. Antibiotic growth promoter olaquindox increases pathogen susceptibility in fish by inducing gut microbiota dysbiosis. Sci. China Life Sci. 2017, 60, 1260–1270. [Google Scholar] [CrossRef]
- Barreto-Curiel, F.; Ramirez-Puebla, S.T.; Ringø, E.; Escobar-Zepeda, A.; Godoy-Lozano, E.; Vazquez-Duhalt, R.; Sanchez-Flores, A.; Viana, M.T. Effects of extruded aquafeed on growth performance and gut microbiome of juvenile Totoaba macdonaldi. Anim. Feed Sci. Technol. 2018, 245, 91–103. [Google Scholar] [CrossRef]
- Comizzoli, P.; Power, M.L.; Bornbusch, S.L.; Muletz-Wolz, C.R. Interactions between reproductive biology and microbiomes in wild animal species. Anim. Microbiome 2021, 3, 87. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Myers, E.W.; Miller, W. Optimal alignments in linear space. Bioinformatics 1988, 4, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.J.; Eddy, S.R. nhmmer: DNA homology search with profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Chao, A.; Lee, S.-M. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 1992, 87, 210–217. [Google Scholar] [CrossRef]
- Chao, A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 1987, 43, 783–791. [Google Scholar] [CrossRef]
- Magurran, A.E. Measuring biological diversity. Curr. Biol. 2021, 31, R1174–R1177. [Google Scholar] [CrossRef]
- Lin, J. Divergence measures based on the Shannon entropy. IEEE Trans. Inf. Theory 1991, 37, 145–151. [Google Scholar] [CrossRef]
- Fournie, J.W.; Krol, R.M.; Hawkins, W.E. Fixation of Fish Tissues; Elsevier: Amsterdam, The Netherlands, 2000; pp. 569–578. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Group | Average Survival Rate (%) | LD50 | p Value (a) | ||||
|---|---|---|---|---|---|---|---|
| 107 | 106 | 105 | 104 | 103 | |||
| Ctrl. | 0 | 0 | 0 | 13.33 | 66.67 | 9.79 × 103 CFU | - |
| Otc | 0 | 0 | 3.33 | 53.33 | 76.67 | 3.25 × 104 CFU | 0.0736 |
| Smx/Tmp | 0 | 0 | 6.67 | 43.33 | 73.33 | 2.59 × 104 CFU | 0.0287 |
| Ery | 0 | 0 | 0 | 36.67 | 76.67 | 2.03 × 104 CFU | 0.0473 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.H.; Park, J.W.; Kim, H.S.; Lee, S.; Yerke, A.M.; Jaiswal, Y.S.; Williams, L.L.; Hwang, S.; Moon, K.H. Effects of Antibiotic Residues on Fish Gut Microbiome Dysbiosis and Mucosal Barrier-Related Pathogen Susceptibility in Zebrafish Experimental Model. Antibiotics 2024, 13, 82. https://doi.org/10.3390/antibiotics13010082
Yang JH, Park JW, Kim HS, Lee S, Yerke AM, Jaiswal YS, Williams LL, Hwang S, Moon KH. Effects of Antibiotic Residues on Fish Gut Microbiome Dysbiosis and Mucosal Barrier-Related Pathogen Susceptibility in Zebrafish Experimental Model. Antibiotics. 2024; 13(1):82. https://doi.org/10.3390/antibiotics13010082
Chicago/Turabian StyleYang, Jun Hyeok, Jeong Woo Park, Ho Sung Kim, Seungki Lee, Aaron M. Yerke, Yogini S. Jaiswal, Leonard L. Williams, Sungmin Hwang, and Ki Hwan Moon. 2024. "Effects of Antibiotic Residues on Fish Gut Microbiome Dysbiosis and Mucosal Barrier-Related Pathogen Susceptibility in Zebrafish Experimental Model" Antibiotics 13, no. 1: 82. https://doi.org/10.3390/antibiotics13010082
APA StyleYang, J. H., Park, J. W., Kim, H. S., Lee, S., Yerke, A. M., Jaiswal, Y. S., Williams, L. L., Hwang, S., & Moon, K. H. (2024). Effects of Antibiotic Residues on Fish Gut Microbiome Dysbiosis and Mucosal Barrier-Related Pathogen Susceptibility in Zebrafish Experimental Model. Antibiotics, 13(1), 82. https://doi.org/10.3390/antibiotics13010082