Bidirectional Interaction between Tetracyclines and Gut Microbiome
Abstract
1. Introduction
2. General Information about Tetracyclines Used in Humans
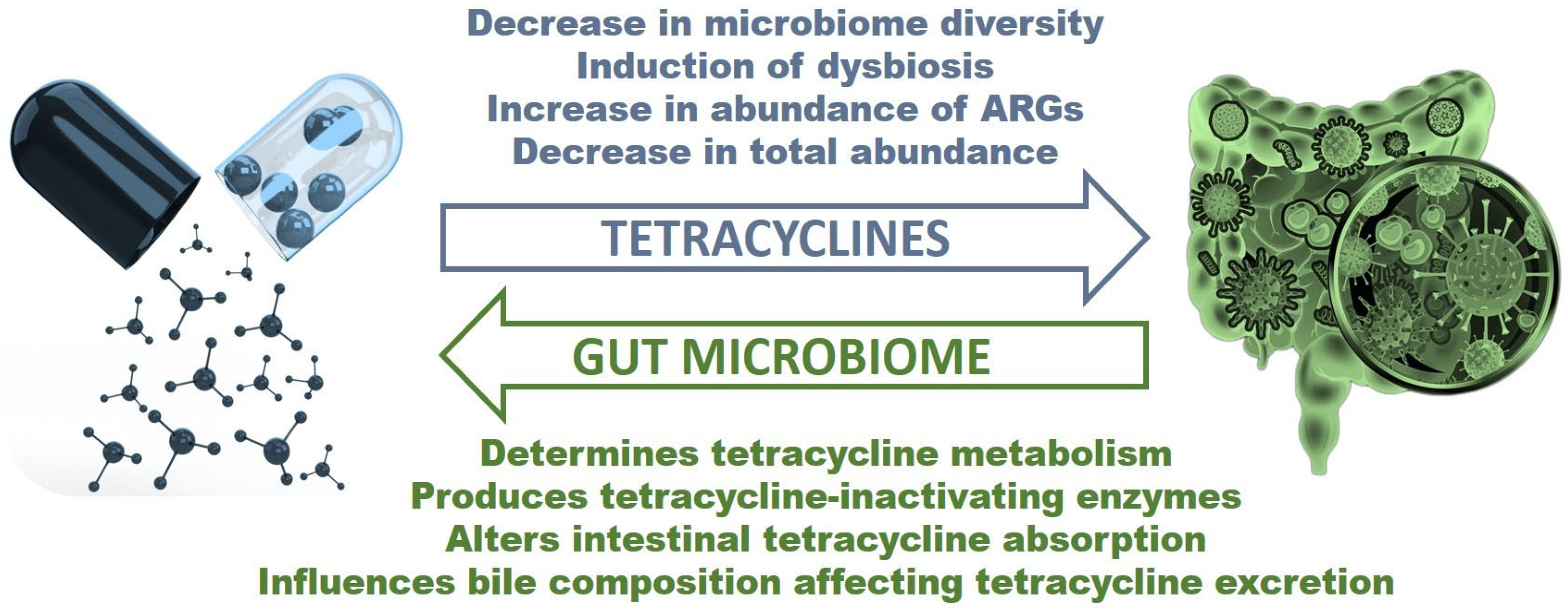
3. Tetracycline Effects on the Gut Microbiome
| Antibiotic | Experimental Model | Bacteria Decreased | Bacteria Increased | Time for Gut to Recover | Long Term Impacts |
|---|---|---|---|---|---|
| Oxytetracycline [63] | Wistar rat | Lactobacillaceae, Aerococcaceae, Helicobacteraceae, and Pasteurellaceae | Bifidobacteriaceae, Enterococcaceae, and Actinomycetacaea | Composition started recovering after treatment but still displayed abnormalities two weeks after treatment | 4-epi-oxytetracycline was found in blood and tissue samples two weeks after treatment, ARGs increased, and metabolism significantly changed in treatment groups |
| Oxytetracycline [64] | Nile Tilapia | Actinobacteria, Lamia, Aeromonas, Pseudomonas, Reyranella, Nocardioides, Mycobacterium, Smaragdicoccus, Pedomicrobium, Chlamydiae, Verrucomicrobia, Gematta, Planctopirus | Plesiomonas, Aquicella, Hyphomicrobium, Actinobacteria, Bacteroidetes, Chlroflexi, Firmicutes, Acidobacteria, Cetobacterium, Macellibacteroides | Not Applicable | Disruption of microbiome could act as a pressure in resistance development in the recovered community |
| Oxytetracycline [65] | Zebrafish | Cetobacterium, Aeromonas, Shewanella, Plesiomonas, Enterobacterales | Mesorhizobium, Rhodobacteraceae, Rhizobiaceae, Pseudomonas, Variovorax, Shewanella, Bacteroides, | Up to 1 month after treatment | Post-exposure changes in gut flora were observed |
| Minocycline (oral) [49] | Human | Lactobacillus salivarius, Bifidobacterium adolescentis, Bifidobacterium pseudolongum, and Bifidobacterium breve | Bacteroidetes | Not Applicable | Not Applicable |
| Minocycline (oral) [68] | Human | Lactobacillus spp. | Enterobacteriaceae and Enterococcus spp. | Almost entirely recovered 3 weeks after treatment | Enterococcus spp. remained high Several families failed to recover |
| Minocycline (oral) [71] | Rat | Not Applicable | Lachnospiraceae, Clostridiales Family XIII | Not Applicable | Antidepressant effects observed depending on traits and sex |
| Minocycline (oral) [72] | Rat | Lactobacillus, Blautia | Lachnospiraceae, Porphyromonadaceae | Not Applicable | Prevented and reversed impairments in spatial recognition memory caused by diet |
4. How Can the Gut Microbiome Alter Tetracycline Treatment?
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef] [PubMed]
- Browne, A.J.; Chipeta, M.G.; Haines-Woodhouse, G.; Kumaran, E.P.A.; Hamadani, B.H.K.; Zaraa, S.; Henry, N.J.; Deshpande, A.; Reiner, R.C., Jr.; Day, N.P.J.; et al. Global antibiotic consumption and usage in humans, 2000–2018: A spatial modelling study. Lancet Planet Health 2021, 5, e893–e904. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Measuring Outpatient Antibiotic Prescribing; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2022. [Google Scholar]
- Wang, D.; Liu, C.; Zhang, X.; Liu, C. Does diagnostic uncertainty increase antibiotic prescribing in primary care? NPJ Prim. Care Respir. Med. 2021, 31, 17. [Google Scholar] [CrossRef] [PubMed]
- Fletcher-Lartey, S.; Yee, M.; Gaarslev, C.; Khan, R. Why do general practitioners prescribe antibiotics for upper respiratory tract infections to meet patient expectations: A mixed methods study. BMJ Open 2016, 6, e012244. [Google Scholar] [CrossRef] [PubMed]
- Demeke, C.A.; Adinew, G.M.; Abebe, T.B.; Gelaye, A.T.; Gemeda, S.G.; Yimenu, D.K. Comparative analysis of the effectiveness of narrow-spectrum versus broad-spectrum antibiotics for the treatment of childhood pneumonia. SAGE Open Med. 2021, 9, 20503121211044379. [Google Scholar] [CrossRef]
- Gandra, S.; Kotwani, A. Need to improve availability of “access” group antibiotics and reduce the use of “watch” group antibiotics in India for optimum use of antibiotics to contain antimicrobial resistance. J. Pharm. Policy Pract. 2019, 12, 20. [Google Scholar] [CrossRef]
- World Health Organization. 2021 AWaRe Classification; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Klein, E.Y.; Milkowska-Shibata, M.; Tseng, K.K.; Sharland, M.; Gandra, S.; Pulcini, C.; Laxminarayan, R. Assessment of WHO antibiotic consumption and access targets in 76 countries, 2000–2015: An analysis of pharmaceutical sales data. Lancet Infect. Dis. 2021, 21, 107–115. [Google Scholar] [CrossRef]
- Brenon, J.R.; Shulder, S.E.; Munsiff, S.S.; Burgoyne, C.M.; Nagel, A.K.; Pillinger, K.E. Rate of broad-spectrum antibiotic overuse in patients receiving outpatient parenteral antibiotic therapy (OPAT). Antimicrob. Steward. Healthc. Epidemiol. 2021, 1, e36. [Google Scholar] [CrossRef]
- Alsaleh, N.A.; Al-Omar, H.A.; Mayet, A.Y.; Mullen, A.B. Exploring Physicians’ Views, Perceptions and Experiences about Broad-Spectrum Antimicrobial Prescribing in a Tertiary Care Hospital Riyadh, Saudi Arabia: A Qualitative Approach. Antibiotics 2021, 10, 366. [Google Scholar] [CrossRef]
- Frank, R.G.; Zeckhauser, R.J. Custom-made versus ready-to-wear treatments: Behavioral propensities in physicians’ choices. J. Health Econ. 2007, 26, 1101–1127. [Google Scholar] [CrossRef]
- Nguyen, F.; Starosta, A.L.; Arenz, S.; Sohmen, D.; Donhofer, A.; Wilson, D.N. Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 2014, 395, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Lambert, T. Antibiotics that affect the ribosome. Rev. Sci. Tech. 2012, 31, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed]
- Eloe-Fadrosh, E.A.; Rasko, D.A. The human microbiome: From symbiosis to pathogenesis. Annu. Rev. Med. 2013, 64, 145–163. [Google Scholar] [CrossRef] [PubMed]
- Ogunrinola, G.A.; Oyewale, J.O.; Oshamika, O.O.; Olasehinde, G.I. The Human Microbiome and Its Impacts on Health. Int. J. Microbiol. 2020, 2020, 8045646. [Google Scholar] [CrossRef]
- Weinstein, N.; Garten, B.; Vainer, J.; Minaya, D.; Czaja, K. Managing the Microbiome: How the Gut Influences Development and Disease. Nutrients 2020, 13, 74. [Google Scholar] [CrossRef]
- Baran, A.; Kwiatkowska, A.; Potocki, L. Antibiotics and Bacterial Resistance-A Short Story of an Endless Arms Race. Int. J. Mol. Sci. 2023, 24, 5777. [Google Scholar] [CrossRef]
- Yoo, D.H.; Kim, I.S.; Van Le, T.K.; Jung, I.H.; Yoo, H.H.; Kim, D.H. Gut microbiota-mediated drug interactions between lovastatin and antibiotics. Drug Metab. Dispos. 2014, 42, 1508–1513. [Google Scholar] [CrossRef]
- Markley, J.L.; Wencewicz, T.A. Tetracycline-Inactivating Enzymes. Front. Microbiol. 2018, 9, 1058. [Google Scholar] [CrossRef]
- Miyake, Y.; Tsuruda, K.; Okuda, K.; Widowati; Iwamoto, Y.; Suginaka, H. In vitro activity of tetracyclines, macrolides, quinolones, clindamycin and metronidazole against periodontopathic bacteria. J. Periodontal Res. 1995, 30, 290–293. [Google Scholar] [CrossRef]
- Petersen, P.J.; Jacobus, N.V.; Weiss, W.J.; Sum, P.E.; Testa, R.T. In vitro and in vivo antibacterial activities of a novel glycylcycline, the 9-t-butylglycylamido derivative of minocycline (GAR-936). Antimicrob. Agents Chemother. 1999, 43, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Duggar, B.M. Aureomycin; a product of the continuing search for new antibiotics. Ann. N. Y. Acad. Sci. 1948, 51, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Finlay, A.C.; Hobby, G.L.; P’An, S.Y.; Regna, P.P.; Routien, J.B.; Seeley, D.B.; Shull, G.M.; Sobin, B.A.; Solomons, I.A.; Vinson, J.W.; et al. Terramycin, a new antibiotic. Science 1950, 111, 85. [Google Scholar] [CrossRef] [PubMed]
- McCormick, J.R.D.; Sjolander, N.O.; Hirsch, U.; Jensen, E.R.; Doerschuk, A.P. A New Family of Antibiotics: The Demethyltetracyclines. J. Am. Chem. Soc. 1957, 79, 4561–4563. [Google Scholar] [CrossRef]
- Blackwood, R.K.; English, A.R. Structure–Activity Relationships in the Tetracycline Series. Adv. Appl. Microbiol. 1970, 13, 237–266. [Google Scholar] [CrossRef]
- Nelson, M.L.; Levy, S.B. The history of the tetracyclines. Ann. N. Y. Acad. Sci. 2011, 1241, 17–32. [Google Scholar] [CrossRef]
- Tariq, S.; Rizvi, S.F.A.; Anwar, U. Tetracycline: Classification, Structure Activity Relationship and Mechanism of Action as a Theranostic Agent for Infectious Lesions-A Mini Review. Biomed. J. Sci. Tech. Res. 2018, 7, 5787–5796. [Google Scholar] [CrossRef]
- Greer, N.D. Tigecycline (Tygacil): The first in the glycylcycline class of antibiotics. Proc. (Bayl. Univ. Med. Cent.) 2006, 19, 155–161. [Google Scholar] [CrossRef]
- Food and Drug Administration. All Approvals and Tentative Approvals; Food and Drug Administration: Silver Spring, MD, USA, 2023. [Google Scholar]
- Agwuh, K.N.; MacGowan, A. Pharmacokinetics and pharmacodynamics of the tetracyclines including glycylcyclines. J. Antimicrob. Chemother. 2006, 58, 256–265. [Google Scholar] [CrossRef]
- MacGowan, A.P. Tigecycline pharmacokinetic/pharmacodynamic update. J. Antimicrob. Chemother. 2008, 62 (Suppl. 1), i11–i16. [Google Scholar] [CrossRef]
- Deeks, E.D. Sarecycline: First Global Approval. Drugs 2019, 79, 325–329. [Google Scholar] [CrossRef]
- Sun, H.; Ting, L.; Machineni, S.; Praestgaard, J.; Kuemmell, A.; Stein, D.S.; Sunkara, G.; Kovacs, S.J.; Villano, S.; Tanaka, S.K. Randomized, Open-Label Study of the Pharmacokinetics and Safety of Oral and Intravenous Administration of Omadacycline to Healthy Subjects. Antimicrob. Agents Chemother. 2016, 60, 7431–7435. [Google Scholar] [CrossRef] [PubMed]
- Rodvold, K.A.; Pai, M.P. Pharmacokinetics and Pharmacodynamics of Oral and Intravenous Omadacycline. Clin. Infect. Dis. 2019, 69, S16–S22. [Google Scholar] [CrossRef] [PubMed]
- Zhanel, G.G.; Cheung, D.; Adam, H.; Zelenitsky, S.; Golden, A.; Schweizer, F.; Gorityala, B.; Lagace-Wiens, P.R.; Walkty, A.; Gin, A.S.; et al. Review of Eravacycline, a Novel Fluorocycline Antibacterial Agent. Drugs 2016, 76, 567–588. [Google Scholar] [CrossRef] [PubMed]
- Jasiecka-Mikolajczyk, A.; Ziolkowski, H.; Jaroszewski, J.J. Pharmacokinetics of tigecycline in turkeys following different routes of administration. J. Vet. Pharmacol. Ther. 2018, 41, e22–e29. [Google Scholar] [CrossRef]
- Qamar, M.U.; Aatika; Chughtai, M.I.; Ejaz, H.; Mazhari, B.B.Z.; Maqbool, U.; Alanazi, A.; Alruwaili, Y.; Junaid, K. Antibiotic-Resistant Bacteria, Antimicrobial Resistance Genes, and Antibiotic Residue in Food from Animal Sources: One Health Food Safety Concern. Microorganisms 2023, 11, 161. [Google Scholar] [CrossRef]
- Litichevskiy, L.; Thaiss, C.A. The Oscillating Gut Microbiome and Its Effects on Host Circadian Biology. Annu. Rev. Nutr. 2022, 42, 145–164. [Google Scholar] [CrossRef]
- Tzanis, E.; Manley, A.; Villano, S.; Tanaka, S.K.; Bai, S.; Loh, E. Effect of Food on the Bioavailability of Omadacycline in Healthy Participants. J. Clin. Pharmacol. 2017, 57, 321–327. [Google Scholar] [CrossRef]
- Neuvonen, P.J. Interactions with the absorption of tetracyclines. Drugs 1976, 11, 45–54. [Google Scholar] [CrossRef]
- Grada, A.; Del Rosso, J.Q.; Graber, E.; Bunick, C.G.; Stein Gold, L.; Moore, A.Y.; Baldwin, H.; Obagi, Z.; Damiani, G.; Carrothers, T.; et al. Sarecycline treatment for acne vulgaris: Rationale for weight-based dosing and limited impact of food intake on clinical efficacy. Dermatol. Ther. 2022, 35, e15275. [Google Scholar] [CrossRef]
- Lakota, E.A.; Van Wart, S.A.; Trang, M.; Tzanis, E.; Bhavnani, S.M.; Safir, M.C.; Friedrich, L.; Steenbergen, J.N.; Ambrose, P.G.; Rubino, C.M. Population Pharmacokinetic Analyses for Omadacycline Using Phase 1 and 3 Data. Antimicrob. Agents Chemother. 2020, 64, e02263-19. [Google Scholar] [CrossRef] [PubMed]
- Perret, L.J.; Tait, C.P. Non-antibiotic properties of tetracyclines and their clinical application in dermatology. Australas. J. Dermatol. 2014, 55, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Di Caprio, R.; Lembo, S.; Di Costanzo, L.; Balato, A.; Monfrecola, G. Anti-inflammatory properties of low and high doxycycline doses: An in vitro study. Mediat. Inflamm. 2015, 2015, 329418. [Google Scholar] [CrossRef] [PubMed]
- Kircik, L.H. Doxycycline and minocycline for the management of acne: A review of efficacy and safety with emphasis on clinical implications. J. Drugs Dermatol. 2010, 9, 1407–1411. [Google Scholar] [PubMed]
- Moore, A.Y.; Charles, J.E.M.; Moore, S. Sarecycline: A narrow spectrum tetracycline for the treatment of moderate-to-severe acne vulgaris. Future Microbiol. 2019, 14, 1235–1242. [Google Scholar] [CrossRef]
- Thompson, K.G.; Rainer, B.M.; Antonescu, C.; Florea, L.; Mongodin, E.F.; Kang, S.; Chien, A.L. Minocycline and Its Impact on Microbial Dysbiosis in the Skin and Gastrointestinal Tract of Acne Patients. Ann. Dermatol. 2020, 32, 21–30. [Google Scholar] [CrossRef]
- Hoofnagle, J.H. LiverTox: Clinical and Research Information on Drug-Induced Liver Injury; National Institute of Diabetes and Digestive and Kidney Diseases: Bethesda, MD, USA, 2012. [Google Scholar]
- Brauncajs, M.; Bielec, F.; Macieja, A.; Pastuszak-Lewandoska, D. In Vitro Activity of Eravacycline against Carbapenemase-Producing Gram-Negative Bacilli Clinical Isolates in Central Poland. Biomedicines 2023, 11, 1784. [Google Scholar] [CrossRef]
- Patangia, D.V.; Anthony Ryan, C.; Dempsey, E.; Paul Ross, R.; Stanton, C. Impact of antibiotics on the human microbiome and consequences for host health. Microbiologyopen 2022, 11, e1260. [Google Scholar] [CrossRef]
- Gawey, B.; Czaja, K. Broad-Spectrum Antibiotic Abuse and its Connection to Obesity. J. Nutr. Health Food Sci. 2017, 5, 1–21. [Google Scholar] [CrossRef]
- Hurkacz, M.; Dobrek, L.; Wiela-Hojenska, A. Antibiotics and the Nervous System-Which Face of Antibiotic Therapy Is Real, Dr. Jekyll (Neurotoxicity) or Mr. Hyde (Neuroprotection)? Molecules 2021, 26, 7456. [Google Scholar] [CrossRef]
- Hawrelak, J.A.; Myers, S.P. The causes of intestinal dysbiosis: A review. Altern. Med. Rev. 2004, 9, 180–197. [Google Scholar]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current Understanding of Dysbiosis in Disease in Human and Animal Models. Inflamm. Bowel Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef]
- Heimdahl, A.; Nord, C.E. Influence of doxycycline on the normal human flora and colonization of the oral cavity and colon. Scand. J. Infect. Dis. 1983, 15, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Elvers, K.T.; Wilson, V.J.; Hammond, A.; Duncan, L.; Huntley, A.L.; Hay, A.D.; van der Werf, E.T. Antibiotic-induced changes in the human gut microbiota for the most commonly prescribed antibiotics in primary care in the UK: A systematic review. BMJ Open 2020, 10, e035677. [Google Scholar] [CrossRef] [PubMed]
- Matto, J.; Maukonen, J.; Alakomi, H.L.; Suihko, M.L.; Saarela, M. Influence of oral doxycycline therapy on the diversity and antibiotic susceptibility of human intestinal bifidobacterial population. J. Appl. Microbiol. 2008, 105, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Wu, Y.; Chen, G.; Wang, S.; Hu, F.; Zheng, H. The Pass-on Effect of Tetracycline-Induced Honey Bee (Apis mellifera) Gut Community Dysbiosis. Front. Microbiol. 2021, 12, 781746. [Google Scholar] [CrossRef]
- Raymann, K.; Shaffer, Z.; Moran, N.A. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 2017, 15, e2001861. [Google Scholar] [CrossRef]
- Rashid, M.U.; Panagiotidis, G.; Backstrom, T.; Weintraub, A.; Nord, C.E. Ecological impact of doxycycline at low dose on normal oropharyngeal and intestinal microflora. Int. J. Antimicrob. Agents 2013, 41, 352–357. [Google Scholar] [CrossRef]
- Han, H.; Xiao, H.; Zhang, K.; Lu, Z. Impact of 4-epi-oxytetracycline on the gut microbiota and blood metabolomics of Wistar rats. Sci. Rep. 2016, 6, 23141. [Google Scholar] [CrossRef]
- Payne, C.J.; Turnbull, J.F.; MacKenzie, S.; Crumlish, M. Investigating the Effect of an Oxytetracycline Treatment on the Gut Microbiome and Antimicrobial Resistance Gene Dynamics in Nile Tilapia (Oreochromis niloticus). Antibiotics 2021, 10, 1213. [Google Scholar] [CrossRef]
- Almeida, A.R.; Domingues, I.; Henriques, I. Zebrafish and water microbiome recovery after oxytetracycline exposure. Environ. Pollut. 2021, 272, 116371. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Breiman, A.; le Pendu, J.; Uyttendaele, M. Anti-viral Effect of Bifidobacterium adolescentis against Noroviruses. Front. Microbiol. 2016, 7, 864. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Messaoudi, S.; Manai, M.; Kergourlay, G.; Prevost, H.; Connil, N.; Chobert, J.M.; Dousset, X. Lactobacillus salivarius: Bacteriocin and probiotic activity. Food Microbiol. 2013, 36, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Moura, I.B.; Grada, A.; Spittal, W.; Clark, E.; Ewin, D.; Altringham, J.; Fumero, E.; Wilcox, M.H.; Buckley, A.M. Profiling the Effects of Systemic Antibiotics for Acne, Including the Narrow-Spectrum Antibiotic Sarecycline, on the Human Gut Microbiota. Front. Microbiol. 2022, 13, 901911. [Google Scholar] [CrossRef]
- Krawczyk, B.; Wityk, P.; Galecka, M.; Michalik, M. The Many Faces of Enterococcus spp.-Commensal, Probiotic and Opportunistic Pathogen. Microorganisms 2021, 9, 1900. [Google Scholar] [CrossRef]
- Asadi, A.; Abdi, M.; Kouhsari, E.; Panahi, P.; Sholeh, M.; Sadeghifard, N.; Amiriani, T.; Ahmadi, A.; Maleki, A.; Gholami, M. Minocycline, focus on mechanisms of resistance, antibacterial activity, and clinical effectiveness: Back to the future. J. Glob. Antimicrob. Resist. 2020, 22, 161–174. [Google Scholar] [CrossRef]
- Schmidtner, A.K.; Slattery, D.A.; Glasner, J.; Hiergeist, A.; Gryksa, K.; Malik, V.A.; Hellmann-Regen, J.; Heuser, I.; Baghai, T.C.; Gessner, A.; et al. Minocycline alters behavior, microglia and the gut microbiome in a trait-anxiety-dependent manner. Transl. Psychiatry 2019, 9, 223. [Google Scholar] [CrossRef]
- Leigh, S.J.; Kaakoush, N.O.; Westbrook, R.F.; Morris, M.J. Minocycline-induced microbiome alterations predict cafeteria diet-induced spatial recognition memory impairments in rats. Transl. Psychiatry 2020, 10, 92. [Google Scholar] [CrossRef]
- Moura, I.B.; Buckley, A.M.; Ewin, D.; Shearman, S.; Clark, E.; Wilcox, M.H.; Chilton, C.H. Omadacycline Gut Microbiome Exposure Does Not Induce Clostridium difficile Proliferation or Toxin Production in a Model That Simulates the Proximal, Medial, and Distal Human Colon. Antimicrob. Agents Chemother. 2019, 63, e01581-18. [Google Scholar] [CrossRef]
- Mutuyemungu, E.; Singh, M.; Liu, S.; Rose, D.J. Intestinal gas production by the gut microbiota: A review. J. Funct. Foods 2023, 100, 105367. [Google Scholar] [CrossRef]
- Baldelli, V.; Scaldaferri, F.; Putignani, L.; Del Chierico, F. The Role of Enterobacteriaceae in Gut Microbiota Dysbiosis in Inflammatory Bowel Diseases. Microorganisms 2021, 9, 6979. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Gan, X.P.; Li, F.F.; Zhang, D.Y.; Chen, L.; Cao, Y.N.; Qiu, H.H.; Cheng, D.C.; Zu, J.F.; Liu, W.Y.; et al. Effect of exposure to antibiotics on the gut microbiome and biochemical indexes of pregnant women. BMJ Open Diabetes Res. Care 2021, 9, e002321. [Google Scholar] [CrossRef] [PubMed]
- Griffin, M.O.; Fricovsky, E.; Ceballos, G.; Villarreal, F. Tetracyclines: A pleitropic family of compounds with promising therapeutic properties. Review of the literature. Am. J. Physiol. Cell Physiol. 2010, 299, C539–C548. [Google Scholar] [CrossRef] [PubMed]
- Maier, L.; Goemans, C.V.; Wirbel, J.; Kuhn, M.; Eberl, C.; Pruteanu, M.; Muller, P.; Garcia-Santamarina, S.; Cacace, E.; Zhang, B.; et al. Unravelling the collateral damage of antibiotics on gut bacteria. Nature 2021, 599, 120–124. [Google Scholar] [CrossRef]
- Mihai, M.M.; Ion, A.; Giurcaneanu, C.; Nitipir, C.; Popa, A.M.; Chifiriuc, M.C.; Popa, M.I.; Ricar, J.; Popa, L.G.; Sarbu, I.; et al. The Impact of Long-Term Antibiotic Therapy of Cutaneous Adverse Reactions to EGFR Inhibitors in Colorectal Cancer Patients. J. Clin. Med. 2021, 10, 3219. [Google Scholar] [CrossRef]
- Swallow, M.A.; Fan, R.; Cohen, J.M.; Bunick, C.G. Antibiotic Resistance Risk with Oral Tetracycline Treatment of Acne Vulgaris. Antibiotics 2022, 11, 1032. [Google Scholar] [CrossRef]
- Bunick, C.G.; Keri, J.; Tanaka, S.K.; Furey, N.; Damiani, G.; Johnson, J.L.; Grada, A. Antibacterial Mechanisms and Efficacy of Sarecycline in Animal Models of Infection and Inflammation. Antibiotics 2021, 10, 439. [Google Scholar] [CrossRef]
- Ternak, G.; Nemeth, M.; Rozanovic, M.; Bogar, L. Antibiotic Consumption Patterns in European Countries Might Be Associated with the Prevalence of Type 1 and 2 Diabetes. Front. Endocrinol. 2022, 13, 870465. [Google Scholar] [CrossRef]
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; Relman, D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008, 6, e280. [Google Scholar] [CrossRef]
- Garrido-Mesa, J.; Algieri, F.; Rodriguez-Nogales, A.; Vezza, T.; Utrilla, M.P.; Garcia, F.; Chueca, N.; Rodriguez-Cabezas, M.E.; Garrido-Mesa, N.; Galvez, J. Immunomodulatory tetracyclines ameliorate DNBS-colitis: Impact on microRNA expression and microbiota composition. Biochem. Pharmacol. 2018, 155, 524–536. [Google Scholar] [CrossRef]
- Delara, M.; McMillan, D.E.; Nickel, N.C.; Jong, G.W.; Seitz, D.P.; Mignone, J. Early life exposure to antibiotics and the risk of mood and anxiety disorders in children and adolescents: A population-based cohort study. J. Psychiatr. Res. 2021, 137, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, B.; Liu, S.; Zhang, Y.; Chen, C.; Jin, Y.; Shen, Z.; Yuan, T.; Yu, X. Antibiotic exposure and risk of overweight/obesity in school children: A multicenter, case-control study from China. Ecotoxicol. Environ. Saf. 2022, 240, 113702. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.C.; Cirillo, P.M.; Krigbaum, N.Y.; Singal, A.G.; Jones, D.P.; Zaki, T.; Cohn, B.A. In-utero exposure to antibiotics and risk of colorectal cancer in a prospective cohort of 18,000 adult offspring. Int. J. Epidemiol. 2023, dyad004. [Google Scholar] [CrossRef] [PubMed]
- Dias, D.; Hipolito, D.; Figueiredo, A.; Fonseca, C.; Caetano, T.; Mendo, S. Unravelling the Diversity and Abundance of the Red Fox (Vulpes vulpes) Faecal Resistome and the Phenotypic Antibiotic Susceptibility of Indicator Bacteria. Animals 2022, 12, 2572. [Google Scholar] [CrossRef]
- Shekhawat, S.S.; Kulshreshtha, N.M.; Saini, P.; Upadhyay, A.; Gupta, A.B.; Jenifer, M.H.; Subramanian, V.; Kumari, A.; Pareek, N.; Vivekanand, V. Antibiotic resistance genes and bacterial diversity: A comparative molecular study of treated sewage from different origins and their impact on irrigated soils. Chemosphere 2022, 307, 136175. [Google Scholar] [CrossRef]
- Werner, K.A.; Schneider, D.; Poehlein, A.; Diederich, N.; Feyen, L.; Axtmann, K.; Hubner, T.; Bruggemann, N.; Prost, K.; Daniel, R.; et al. Metagenomic Insights Into the Changes of Antibiotic Resistance and Pathogenicity Factor Pools Upon Thermophilic Composting of Human Excreta. Front. Microbiol. 2022, 13, 826071. [Google Scholar] [CrossRef]
- Yarahmadi, N.; Halimi, S.; Moradi, P.; Zamanian, M.H.; Rezaei, A.; Vaziri, S.; Akya, A.; Alvandi, A.; Yazdani, S.; Ghadimi, D.; et al. Prevalence of Antibiotic-Resistant Lactobacilli in Sepsis Patients with Long-Term Antibiotic Therapy. Curr. Microbiol. 2022, 79, 318. [Google Scholar] [CrossRef]
- Gehring, M.; Wieczorek, D.; Kapp, A.; Wedi, B. Potent Anti-Inflammatory Effects of Tetracyclines on Human Eosinophils. Front. Allergy 2021, 2, 754501. [Google Scholar] [CrossRef]
- Itoh, K.; Shigemi, H.; Chihara, K.; Sada, K.; Yamauchi, T.; Iwasaki, H. Caspofungin suppresses zymosan-induced cytokine and chemokine release in THP-1 cells: Possible involvement of the spleen tyrosine kinase pathway. Transl. Res. 2021, 227, 53–63. [Google Scholar] [CrossRef]
- Behr, C.; Kamp, H.; Fabian, E.; Krennrich, G.; Mellert, W.; Peter, E.; Strauss, V.; Walk, T.; Rietjens, I.; van Ravenzwaay, B. Gut microbiome-related metabolic changes in plasma of antibiotic-treated rats. Arch. Toxicol. 2017, 91, 3439–3454. [Google Scholar] [CrossRef]
- Zhang, X.; Han, Y.; Huang, W.; Jin, M.; Gao, Z. The influence of the gut microbiota on the bioavailability of oral drugs. Acta Pharm. Sin. B 2021, 11, 1789–1812. [Google Scholar] [CrossRef] [PubMed]
- Serio, A.W.; Keepers, T.R.; Wright, K.; Anastasiou, D. Pathogens susceptible to tetracycline are also susceptible to omadacycline: Tetracycline as a one-sided surrogate to predict omadacycline susceptible pathogens. Diagn. Microbiol. Infect. Dis. 2022, 104, 115785. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Li, Y.; Yu, R.; Ma, M.; Yang, M.; Si, H. Identification of Novel tet(X3) Variants Resistant To Tigecycline in Acinetobacter Species. Microbiol. Spectr. 2022, 10, e0133322. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhou, W.; Wu, J.; Liu, X.; Lin, J.; Ji, X.; Lin, H.; Wang, J.; Jiang, H.; Zhou, Q.; et al. Large-Scale Studies on Antimicrobial Resistance and Molecular Characterization of Escherichia coli from Food Animals in Developed Areas of Eastern China. Microbiol. Spectr. 2022, 10, e0201522. [Google Scholar] [CrossRef]
- Pazra, D.F.; Latif, H.; Basri, C.; Wibawan, I.W.T.; Rahayu, P. Distribution analysis of tetracycline resistance genes in Escherichia coli isolated from floor surface and effluent of pig slaughterhouses in Banten Province, Indonesia. Vet. World 2023, 16, 509–517. [Google Scholar] [CrossRef]
- Xu, L.; Zhou, Y.; Niu, S.; Liu, Z.; Zou, Y.; Yang, Y.; Feng, H.; Liu, D.; Niu, X.; Deng, X.; et al. A novel inhibitor of monooxygenase reversed the activity of tetracyclines against tet(X3)/tet(X4)-positive bacteria. EBioMedicine 2022, 78, 103943. [Google Scholar] [CrossRef]
- Zhang, R.M.; Sun, J.; Sun, R.Y.; Wang, M.G.; Cui, C.Y.; Fang, L.X.; Liao, M.N.; Lu, X.Q.; Liu, Y.X.; Liao, X.P.; et al. Source Tracking and Global Distribution of the Tigecycline Non-Susceptible tet(X). Microbiol. Spectr. 2021, 9, e0116421. [Google Scholar] [CrossRef]
- Kusunur, A.B.; Mogilipuri, S.S.; Moturu, D.; Benala, M.; Vaiyapuri, M.; Panda, S.K.; George, J.C.; Badireddy, M.R. Tetracycline resistance potential of heterotrophic bacteria isolated from freshwater fin-fish aquaculture system. J. Appl. Microbiol. 2023, 134, lxad060. [Google Scholar] [CrossRef]
- Zakerifar, M.; Kaboosi, H.; Goli, H.R.; Rahmani, Z.; Peyravii Ghadikolaii, F. Antibiotic resistance genes and molecular typing of Streptococcus agalactiae isolated from pregnant women. BMC Pregnancy Childbirth 2023, 23, 43. [Google Scholar] [CrossRef]
- Butiuc-Keul, A.; Carpa, R.; Podar, D.; Szekeres, E.; Muntean, V.; Iordache, D.; Farkas, A. Antibiotic Resistance in Pseudomonas spp. Through the Urban Water Cycle. Curr. Microbiol. 2021, 78, 1227–1237. [Google Scholar] [CrossRef]
- Ramos, M.S.; Furlan, J.P.R.; Gallo, I.F.L.; Dos Santos, L.D.R.; de Campos, T.A.; Savazzi, E.A.; Stehling, E.G. High Level of Resistance to Antimicrobials and Heavy Metals in Multidrug-Resistant Pseudomonas sp. Isolated from Water Sources. Curr. Microbiol. 2020, 77, 2694–2701. [Google Scholar] [CrossRef] [PubMed]
- Khlaif, M.M.; Hussein, N.H. Sequencing analysis of tigecycline resistance among tigecycline non-susceptible in three species of G-ve bacteria isolated from clinical specimens in Baghdad. Mol. Biol. Rep. 2022, 49, 11811–11820. [Google Scholar] [CrossRef] [PubMed]
- Sivaraman, G.K.; Rajan, V.; Vijayan, A.; Elangovan, R.; Prendiville, A.; Bachmann, T.T. Antibiotic Resistance Profiles and Molecular Characteristics of Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli and Klebsiella pneumoniae Isolated From Shrimp Aquaculture Farms in Kerala, India. Front. Microbiol. 2021, 12, 622891. [Google Scholar] [CrossRef] [PubMed]
- McMahan, R.H.; Hulsebus, H.J.; Najarro, K.M.; Giesy, L.E.; Frank, D.N.; Kovacs, E.J. Changes in gut microbiome correlate with intestinal barrier dysfunction and inflammation following a 3-day ethanol exposure in aged mice. Alcohol. 2023, 107, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Liu, J.; Ma, Y.; Wei, Y.; Liu, J.; Wang, H. Impairment of Intestinal Barrier Function Induced by Early Weaning via Autophagy and Apoptosis Associated With Gut Microbiome and Metabolites. Front. Immunol. 2021, 12, 804870. [Google Scholar] [CrossRef]
- Usuda, H.; Okamoto, T.; Wada, K. Leaky Gut: Effect of Dietary Fiber and Fats on Microbiome and Intestinal Barrier. Int. J. Mol. Sci. 2021, 22, 7613. [Google Scholar] [CrossRef]
- Adir, J. Enterohepatic circulation of tetracycline in rats. J. Pharm. Sci. 1975, 64, 1847–1850. [Google Scholar] [CrossRef]
- Sayin, S.I.; Wahlstrom, A.; Felin, J.; Jantti, S.; Marschall, H.U.; Bamberg, K.; Angelin, B.; Hyotylainen, T.; Oresic, M.; Backhed, F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Wang, Z.; Zhang, M.; Wang, S.; Xiang, Z.; Pan, H.; Li, M. The Relationship Between Gut Microbiome and Bile Acids in Primates With Diverse Diets. Front. Microbiol. 2022, 13, 899102. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B.; Bajaj, J.S. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 2014, 30, 332–338. [Google Scholar] [CrossRef]
- Krajmalnik-Brown, R.; Ilhan, Z.E.; Kang, D.W.; DiBaise, J.K. Effects of gut microbes on nutrient absorption and energy regulation. Nutr. Clin. Pract. 2012, 27, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Tang, Y.; Li, Y.; Xu, Z.; Zhang, D.; Liu, J.; Wang, X.; Xia, W.; Xu, S. Perinatal High-Salt Diet Induces Gut Microbiota Dysbiosis, Bile Acid Homeostasis Disbalance, and NAFLD in Weanling Mice Offspring. Nutrients 2021, 13, 2135. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Moorthy, B.; Haribabu, B.; Jala, V.R. Cytochrome P450 1A1 is essential for the microbial metabolite, Urolithin A-mediated protection against colitis. Front. Immunol. 2022, 13, 1004603. [Google Scholar] [CrossRef] [PubMed]
- Girvan, H.M.; Munro, A.W. Applications of microbial cytochrome P450 enzymes in biotechnology and synthetic biology. Curr. Opin. Chem. Biol. 2016, 31, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Mosa, A.; Gerber, A.; Neunzig, J.; Bernhardt, R. Products of gut-microbial tryptophan metabolism inhibit the steroid hormone-synthesizing cytochrome P450 11A1. Endocrine 2016, 53, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Van Bogaert, I.N.; Groeneboer, S.; Saerens, K.; Soetaert, W. The role of cytochrome P450 monooxygenases in microbial fatty acid metabolism. FEBS J. 2011, 278, 206–221. [Google Scholar] [CrossRef]
- Wang, S.; Wen, Q.; Qin, Y.; Xia, Q.; Shen, C.; Song, S. Gut microbiota and host cytochrome P450 characteristics in the pseudo germ-free model: Co-contributors to a diverse metabolic landscape. Gut Pathog. 2023, 15, 15. [Google Scholar] [CrossRef]
- Garcia, W.L.; Miller, C.J.; Lomas, G.X.; Gaither, K.A.; Tyrrell, K.J.; Smith, J.N.; Brandvold, K.R.; Wright, A.T. Profiling How the Gut Microbiome Modulates Host Xenobiotic Metabolism in Response to Benzo[a]pyrene and 1-Nitropyrene Exposure. Chem. Res. Toxicol. 2022, 35, 585–596. [Google Scholar] [CrossRef]
- Patel, D.; Sharma, D.; Mandal, P. Gut Microbiota: Target for Modulation of Gut-Liver-Adipose Tissue Axis in Ethanol-Induced Liver Disease. Mediat. Inflamm. 2022, 2022, 4230599. [Google Scholar] [CrossRef]
- Montassier, E.; Valdes-Mas, R.; Batard, E.; Zmora, N.; Dori-Bachash, M.; Suez, J.; Elinav, E. Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner. Nat. Microbiol. 2021, 6, 1043–1054. [Google Scholar] [CrossRef]
- Mutalub, Y.B.; Abdulwahab, M.; Mohammed, A.; Yahkub, A.M.; AL-Mhanna, S.B.; Yusof, W.; Tang, S.P.; Rasool, A.H.G.; Mokhtar, S.S. Gut Microbiota Modulation as a Novel Therapeutic Strategy in Cardiometabolic Diseases. Foods 2022, 11, 2575. [Google Scholar] [CrossRef] [PubMed]
- Penumutchu, S.; Korry, B.J.; Hewlett, K.; Belenky, P. Fiber supplementation protects from antibiotic-induced gut microbiome dysbiosis by modulating gut redox potential. Nat. Commun. 2023, 14, 5161. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, R.A.; Cabral, V.; Xavier, K.B. Microbiome-diet interactions drive antibiotic efficacy. Nat. Microbiol. 2021, 6, 824–825. [Google Scholar] [CrossRef] [PubMed]
| Active Substance [Reference] | Administration Routes | Bioavailability from GI (%) | Metabolism | Biological Half-Fife (h) | Excretion |
|---|---|---|---|---|---|
| Tetracycline [29,32] | Oral, topical | 75–88 | Minimally metabolized | 6–11 | Renal, feces |
| Oxytetracycline [29,32] | Oral, ophthalmic | 58 | Not metabolized | 6–9.2 | Renal, feces |
| Chlortetracycline [29,32] | Oral, topical | 25–30 | Not metabolized | 5.6–9 | Renal, biliary |
| Demeclocycline [29,32] | Oral | 60–80 | Hepatic | 10–17 | Renal |
| Lymecycline [29] | Oral | 100 | - | 10 | Renal |
| Rolitetracycline [32] | Intravenous | - | Not metabolized | 5.8 | Renal |
| Doxycycline [29,32] | Oral, intravenous | 80–100 | Not metabolized | 15–25 | Feces, renal |
| Minocycline [29,32] | Oral | 100 | Hepatic | 11–18 | Renal, feces |
| Tigecycline [29,33] | Intravenous | - * | Not metabolized | 42.4 | Biliary, renal |
| Sarecycline [34] | Oral | - | Minimally metabolized | 21–22 | Rena, feces |
| Omadacycline [35,36] | Oral, intravenous | 34.5 | Not metabolized | 16.8 | Feces, renal |
| Eravacycline [37] | Intravenous | 28 | Minimally metabolized | 48 | Biliary, renal |
| Antibiotic | Experimental Model | Bacteria Decreased | Bacteria Increased | Time for Gut to Recover | Long Term Impacts |
|---|---|---|---|---|---|
| Doxycycline [57] | Human | Enterobacteriaceae, Enterococcus spp., Escherichia coli, Streptococcus spp., and Fusobacterium spp. | Not Applicable | 9 days after treatment | Not Applicable |
| Doxycycline [59] | Human | Bifidobacterium | Not Applicable | Not Applicable | Increase in tetracycline resistance |
| Doxycycline [62] | Human | Escherichia coli, Enterococcaceae | Not Applicable | 4 weeks after 16-week treatment | Increased doxycycline resistance |
| Tetracycline [60] | Honeybee | Lactobacillus, Frischella, Commensalibacter, Bartonella, Gilliamella, Snodgrassella | Not Applicable | Did not recover | Gut microbiota did not recover in treated bees. This could harm the colony as contact with hive mates is a major contributor to bee microbiota |
| Tetracycline [61] | Honeybee | Bifidobacterium, Firm-4, Firm-5, Snodgrassella alvi, Alpha 2.1, Frischella perrara, Lactobacillus kunkeei, Bartonella apis | Serratia, Halomonadaceae, Gilliamella apicola | 32% of treated bees recovered 3 days after treatment | Tetracycline-treated bees have increased mortality |
| Antibiotic | Experimental Model | Bacteria Decreased | Bacteria Increased | Time for Gut to Recover | Long Term Impacts |
|---|---|---|---|---|---|
| Omadacycline [73] | Human | Bacteroides fragilis, Bifidobacteria, Lactobacilli, and Enterococcus spp. | Lactose-fermenting Enterobacteriaceae | Within 3 weeks | Not Applicable |
| Antibiotic | Mechanism of Dysbiosis | References |
|---|---|---|
| Doxycycline | Decrease in bacterial diversity | [57,59,62] |
| Tetracycline | Reduction of absolute bacterial abundance Increase in some opportunistic bacteria | [60,61] |
| Antibiotic | Mechanism of Dysbiosis | References |
|---|---|---|
| Oxytetracycline | Increase in opportunistic bacteria Decrease in microbial diversity and evenness | [63,64,65] |
| Minocycline | Decrease in bacterial diversity Failure to recover to pre-treatment levels in some bacteria Reduction in microbial richness Increase in opportunistic bacteria | [49,68,71,72] |
| Antibiotic | Mechanism of Dysbiosis | References |
|---|---|---|
| Omadacycline | Decrease in total bacterial abundance Decrease in bacterial diversity Reduction of some species below limit of detection Increase in lactose-fermenting Enterobacteriaceae | [73] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaroszewski, J.; Mamun, N.; Czaja, K. Bidirectional Interaction between Tetracyclines and Gut Microbiome. Antibiotics 2023, 12, 1438. https://doi.org/10.3390/antibiotics12091438
Jaroszewski J, Mamun N, Czaja K. Bidirectional Interaction between Tetracyclines and Gut Microbiome. Antibiotics. 2023; 12(9):1438. https://doi.org/10.3390/antibiotics12091438
Chicago/Turabian StyleJaroszewski, Jerzy, Niles Mamun, and Krzysztof Czaja. 2023. "Bidirectional Interaction between Tetracyclines and Gut Microbiome" Antibiotics 12, no. 9: 1438. https://doi.org/10.3390/antibiotics12091438
APA StyleJaroszewski, J., Mamun, N., & Czaja, K. (2023). Bidirectional Interaction between Tetracyclines and Gut Microbiome. Antibiotics, 12(9), 1438. https://doi.org/10.3390/antibiotics12091438






