Scorpion Venom as a Source of Antimicrobial Peptides: Overview of Biomolecule Separation, Analysis and Characterization Methods
Abstract
1. Introduction
2. Methods Used for Separation of Venom Complex Mixtures
2.1. Electrophoretic Separation Techniques
2.1.1. One-Dimension Gel Electrophoresis (1-DGE)
2.1.2. Two-Dimension Gel Electrophoresis (2-DGE)
2.2. Chromatographic Separation Techniques
2.2.1. Reverse-Phase High-Performance Liquid Chromatography (RP-HPLC)
2.2.2. Size-Exclusion Chromatography (SEC)
2.2.3. Ion Exchange Chromatography (IEX)
2.2.4. Affinity Chromatography
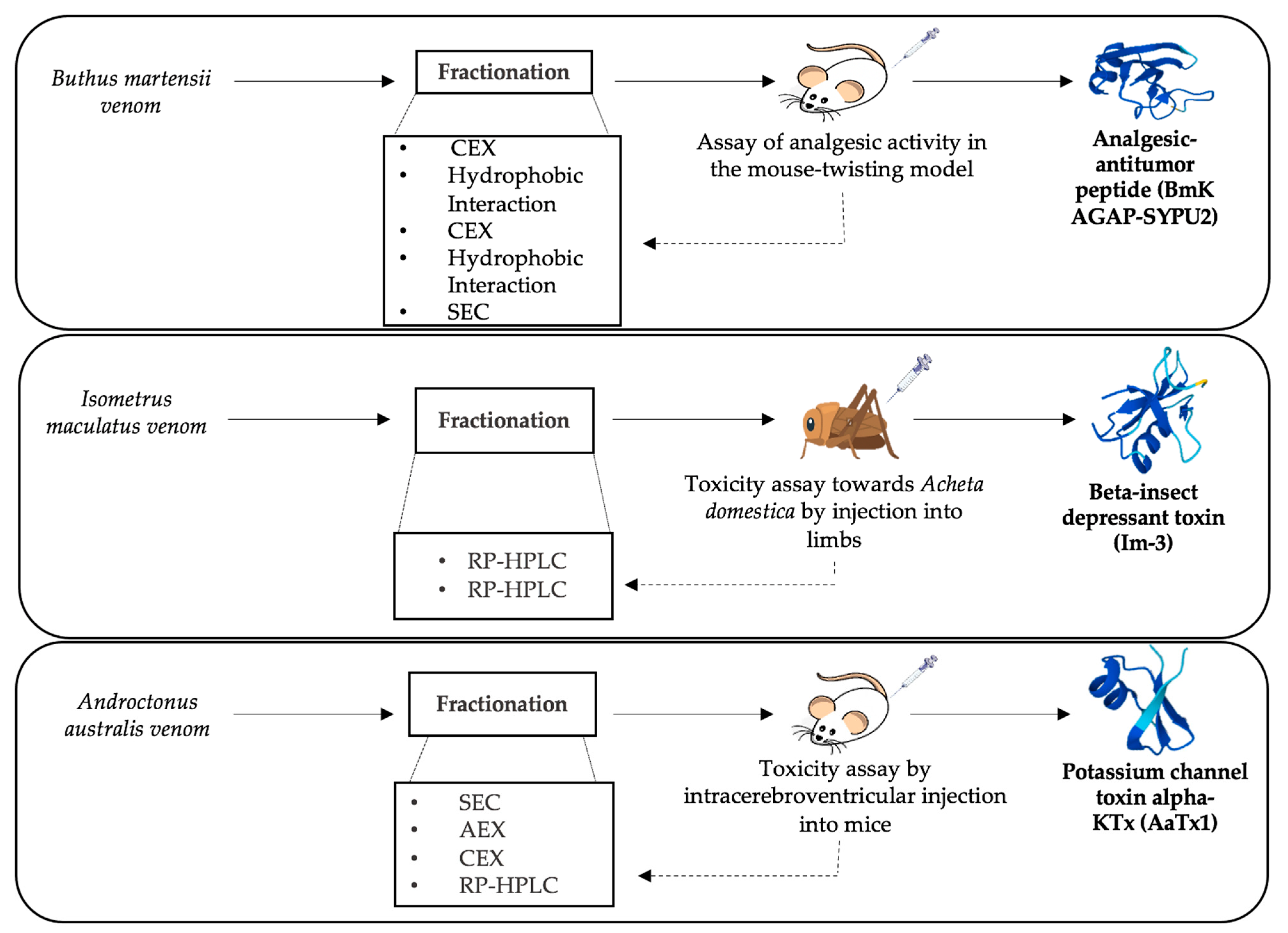
3. Implementation of Separation Methods for Scorpion Venoms
3.1. Bioassay-Guided Fractionation
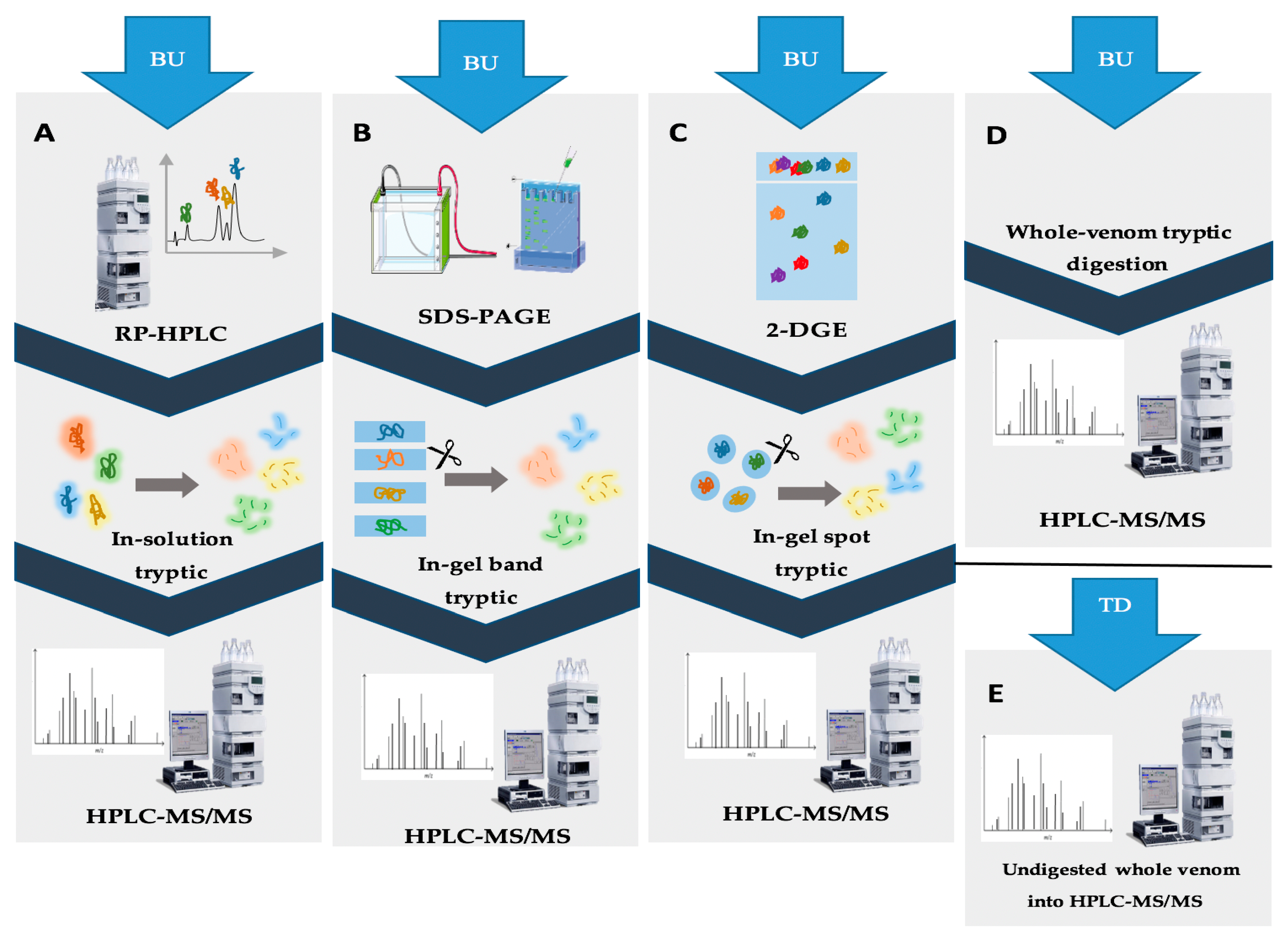
3.2. Whole Proteome Characterization
4. Scorpion Venom Antimicrobial Peptides (AMPs)
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chippaux, J.-P.; Goyffon, M. Epidemiology of scorpionism: A global appraisal. Acta Trop. 2008, 107, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Mendes, L.C.; Viana, G.M.M.; Nencioni, A.L.A.; Pimenta, D.C.; Beraldo-Neto, E. Scorpion Peptides and Ion Channels: An Insightful Review of Mechanisms and Drug Development. Toxins 2023, 15, 238. [Google Scholar] [CrossRef]
- Soudani, N.; Gharbi-Chihi, J.; Srairi-Abid, N.; Yazidi, C.M.-E.; Planells, R.; Margotat, A.; Torresani, J.; El Ayeb, M. Isolation and molecular characterization of LVP1 lipolysis activating peptide from scorpion Buthus occitanus tunetanus. Biochim. Et Biophys. Acta (BBA)—Proteins Proteom. 2005, 1747, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Prudencio, G.; Cid-Uribe, J.I.; Morales, J.A.; Possani, L.D.; Ortiz, E.; Romero-Gutiérrez, T. The Enzymatic Core of Scorpion Venoms. Toxins 2022, 14, 248. [Google Scholar] [CrossRef]
- Rincón-Cortés, C.A.; Bayona-Rojas, M.A.; Reyes-Montaño, E.A.; Vega-Castro, N.A. Antimicrobial Activity Developed by Scorpion Venoms and Its Peptide Component. Toxins 2022, 14, 740. [Google Scholar] [CrossRef]
- Verano-Braga, T.; Figueiredo-Rezende, F.; Melo, M.N.; Lautner, R.Q.; Gomes, E.R.; Mata-Machado, L.T.; Murari, A.; Rocha-Resende, C.; de Lima, M.E.; Guatimosim, S.; et al. Structure-function studies of Tityus serrulatus Hypotensin-I (TsHpt-I): A new agonist of B(2) kinin receptor. Toxicon 2010, 56, 1162–1171. [Google Scholar] [CrossRef]
- Queiroz, A.M.; Sampaio, V.S.; Mendonça, I.; Fé, N.F.; Sachett, J.; Ferreira, L.C.L.; Feitosa, E.; Wen, F.H.; Lacerda, M.; Monteiro, W. Severity of Scorpion Stings in the Western Brazilian Amazon: A Case-Control Study. PLoS ONE 2015, 10, e0128819. [Google Scholar] [CrossRef]
- Almaaytah, A.; Albalas, Q. Scorpion venom peptides with no disulfide bridges: A review. Peptides 2014, 51, 35–45. [Google Scholar] [CrossRef]
- Zhao, Z.; Ma, Y.; Dai, C.; Zhao, R.; Li, S.; Wu, Y.; Cao, Z.; Li, W. Imcroporin, a New Cationic Antimicrobial Peptide from the Venom of the Scorpion Isometrus maculates. Antimicrob. Agents Chemother. 2009, 53, 3472–3477. [Google Scholar] [CrossRef]
- Dai, C.; Ma, Y.; Zhao, Z.; Zhao, R.; Wang, Q.; Wu, Y.; Cao, Z.; Li, W. Mucroporin, the First Cationic Host Defense Peptide from the Venom of Lychas mucronatus. Antimicrob. Agents Chemother. 2008, 52, 3967–3972. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Aponte, C.A.; Silva-Sanchez, J.; Quintero-Hernández, V.; Rodríguez-Romero, A.; Balderas, C.; Possani, L.D.; Gurrola, G.B. Vejovine, a new antibiotic from the scorpion venom of Vaejovis mexicanus. Toxicon 2011, 57, 84–92. [Google Scholar] [CrossRef]
- Fang, W.; Vega-Rodríguez, J.; Ghosh, A.K.; Jacobs-Lorena, M.; Kang, A.; Leger, R.J.S. Development of Transgenic Fungi that Kill Human Malaria Parasites in Mosquitoes. Science 2011, 331, 1074–1077. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Xu, J.; Rodriguez, M.d.C.; Lanz-Mendoza, H.; Hernández-Rivas, R.; Du, W.; Zhu, S. Characterization of two linear cationic antimalarial peptides in the scorpion Mesobuthus eupeus. Biochimie 2010, 92, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Butte, P.V.; Mamelak, A.; Parrish-Novak, J.; Drazin, D.; Shweikeh, F.; Gangalum, P.R.; Chesnokova, A.; Ljubimova, J.Y.; Black, K. Near-infrared imaging of brain tumors using the Tumor Paint BLZ-100 to achieve near-complete resection of brain tumors. Neurosurg. Focus 2014, 36, E1. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Alewood, P.F. Modern Venom Profiling: Mining into Scorpion Venom Biodiversity. In Scorpion Venoms; Gopalakrishnakone, P., Possani, L.D., Schwartz, E.F., De La Vega, R.C.R., Eds.; Springer: Dordrecht, The Netherlands, 2015; pp. 547–561. [Google Scholar] [CrossRef]
- Rochat, C.; Rochat, H.; Miranda, F.; Lissitzky, S. Purification and Some Properties of the Neurotoxins of Androctonus australis Hector*. Biochemistry 1967, 6, 578–585. [Google Scholar] [CrossRef]
- de la Vega, R.C.R.; Corzo, G.; Possani, L.D. Scorpion Venoms as a Platform for Drug Development. In Drug Discovery; King, G.F., Ed.; Royal Society of Chemistry: Cambridge, UK, 2015; Chapter 7; pp. 204–220. [Google Scholar] [CrossRef]
- Cajado-Carvalho, D.; da Silva, C.C.F.; Kodama, R.T.; Mariano, D.O.C.; Pimenta, D.C.; Duzzi, B.; Kuniyoshi, A.K.; Portaro, F.V. Purification and Biochemical Characterization of TsMS 3 and TsMS 4: Neuropeptide-Degrading Metallopeptidases in the Tityus serrulatus Venom. Toxins 2019, 11, 194. [Google Scholar] [CrossRef]
- Paul, L.F.; Fletcher, M.D.; Weninger, K.; Anderson, T.E.; Martin, B.M. Vesicle-associated Membrane Protein (VAMP) Cleavage by a New Metalloprotease from the Brazilian Scorpion Tityus serrulatus. J. Biol. Chem. 2010, 285, 7405–7416. [Google Scholar] [CrossRef]
- Estrada-Gómez, S.; Cupitra, N.I.; Arango, W.M.; Muñoz, L.J.V. Intraspecific Variation of Centruroides edwardsii Venom from Two Regions of Colombia. Toxins 2014, 6, 2082. [Google Scholar] [CrossRef]
- Badhe, R.V.; Thomas, A.B.; Harer, S.L.; Deshpande, A.D.; Salvi, N.; Waghmare, A. Intraspecific variation in protein pattern of red scorpion (Mesobuthus tamulus, coconsis, pocock) venoms from Western and Southern India. J. Venom. Anim. Toxins Incl. Trop. Dis. 2006, 12, 612–619. [Google Scholar] [CrossRef]
- Omran, M.A.A.; McVean, A. Intraspecific Variation in Scorpion Leiurus Quinquestriatus Venom Collected from Egypt (Sinai and Aswan Deserts). J. Toxicol. Toxin Rev. 2000, 19, 247–264. [Google Scholar] [CrossRef]
- Khoobdel, M.; Zahraei-Salehi, T.; Nayeri-Fasaei, B.; Khosravi, M.; Omidian, Z.; Motedayen, M.H.; Akbari, A. Purification of the Immunogenic Fractions and Determination of Toxicity in Mesobuthus eupeus (Scorpionida: Buthidae) Venom. J. Arthropod Borne Dis. 2013, 7, 139–146. [Google Scholar] [PubMed]
- Borges, A.; Lomonte, B.; Angulo, Y.; de Patiño, H.A.; Pascale, J.M.; Otero, R.; Miranda, R.J.; De Sousa, L.; Graham, M.R.; Gómez, A.; et al. Venom diversity in the Neotropical scorpion genus Tityus: Implications for antivenom design emerging from molecular and immunochemical analyses across endemic areas of scorpionism. Acta Trop. 2020, 204, 105346. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Gómez, S.; Vargas-Muñoz, L.J.; Saldarriaga-Córdoba, M.M.; van der Meijden, A. MS/MS analysis of four scorpion venoms from Colombia: A descriptive approach. J. Venom. Anim. Toxins Incl. Trop. Dis. 2021, 27, 1–13. [Google Scholar] [CrossRef]
- Maleki, M.; Dounighi, N.M. Purification and characterization of a novel type of neurotoxic peptides from the venom of the Iranian scorpion Hemiscorpius lepturus. Iran. J. Basic Med. Sci. 2019, 23, 195–201. [Google Scholar] [CrossRef]
- Uawonggul, N.; Thammasirirak, S.; Chaveerach, A.; Arkaravichien, T.; Bunyatratchata, W.; Ruangjirachuporn, W.; Jearranaiprepame, P.; Nakamura, T.; Matsuda, M.; Kobayashi, M.; et al. Purification and characterization of Heteroscorpine-1 (HS-1) toxin from Heterometrus laoticus scorpion venom. Toxicon 2007, 49, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Joshi, K.; Patil, D. Proteomics. In Innovative Approaches in Drug Discovery; Academic Press: Boston, MA, USA, 2017; pp. 273–294. [Google Scholar] [CrossRef]
- El-Aziz, T.M.A.; Soares, A.G.; Stockand, J.D. Advances in venomics: Modern separation techniques and mass spectrometry. J. Chromatogr. B 2020, 1160, 122352. [Google Scholar] [CrossRef]
- Chiou, S.-H.; Wu, S.-H. Evaluation of commonly used electrophoretic methods for the analysis of proteins and peptides and their application to biotechnology. Anal. Chim. Acta 1999, 383, 47–60. [Google Scholar] [CrossRef]
- Ni, Y.-T.; Guo, M.-H.; Wang, J.-D.; Liu, Y.; Xin, Y.; Wu, D.-C. Identification of Scorpion Venom in Buthus martensii Karsch from Different Regions by Two-dimensional Gel Electrophoresis (2-DE). Her. Med. 2012, 31, 1200–1202. [Google Scholar]
- Incamnoi, P.; Patramanon, R.; Thammasirirak, S.; Chaveerach, A.; Uawonggul, N.; Sukprasert, S.; Rungsa, P.; Daduang, J.; Daduang, S. Heteromtoxin (HmTx), a novel heterodimeric phospholipase A2 from Heterometrus laoticus scorpion venom. Toxicon 2013, 61, 62–71. [Google Scholar] [CrossRef]
- Xu, X.; Duan, Z.; Di, Z.; He, Y.; Li, J.; Li, Z.; Xie, C.; Zeng, X.; Cao, Z.; Wu, Y.; et al. Proteomic analysis of the venom from the scorpion Mesobuthus martensii. J. Proteom. 2014, 106, 162–180. [Google Scholar] [CrossRef]
- Mouchbahani-Constance, S.; Sharif-Naeini, R. Proteomic and Transcriptomic Techniques to Decipher the Molecular Evolution of Venoms. Toxins 2021, 13, 154. [Google Scholar] [CrossRef]
- Borges, A.; García, C.C.; Lugo, E.; Alfonzo, M.J.; Jowers, M.J.; Den Camp, H.J.M.O. Diversity of long-chain toxins in Tityus zulianus and Tityus discrepans venoms (Scorpiones, Buthidae): Molecular, immunological, and mass spectral analyses. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2006, 142, 240–252. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, L.; Borges, A.; Vásquez-Suárez, A.; Camp, H.J.O.D.; Chadee-Burgos, R.I.; Romero-Bellorín, M.; Espinoza, J.; De Sousa-Insana, L.; Pino-García, O. Differences in venom toxicity and antigenicity between females and males Tityus nororientalis (Buthidae) scorpions. J. Venom Res. 2010, 1, 61–70. [Google Scholar] [PubMed]
- Nocks, B.C.; Learner, R.M.; Blackman, D.; Brown, T.E. The effects of a community-based long term care project on nursing home utilization. Gerontologist 1986, 26, 150–157. [Google Scholar] [CrossRef]
- Juichi, H.; Miyashita, M.; Nakagawa, Y.; Miyagawa, H. Isolation and characterization of the insecticidal, two-domain toxin LaIT3 from the Liocheles australasiae scorpion venom. Biosci. Biotechnol. Biochem. 2019, 83, 2183–2189. [Google Scholar] [CrossRef]
- Evans, E.R.J.; McIntyre, L.; Northfield, T.D.; Daly, N.L.; Wilson, D.T. Small Molecules in the Venom of the Scorpion Hormurus waigiensis. Biomedicines 2020, 8, 259. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Sadilek, M.; Lehmberg, E.; Herrmann, R.; Herrmann, R.; Moskowitz, H.; Lee, Y.M.; Thomas, B.A.; Shimizu, R.; Kuroda, M.; et al. Rapid purification and molecular modeling of AaIT peptides from venom of Androctonus australis. Arch. Insect Biochem. Physiol. 1998, 38, 53–65. [Google Scholar] [CrossRef]
- ElFessi-Magouri, R.; Peigneur, S.; Othman, H.; Srairi-Abid, N.; ElAyeb, M.; Tytgat, J.; Kharrat, R. Characterization of Kbot21 Reveals Novel Side Chain Interactions of Scorpion Toxins Inhibiting Voltage-Gated Potassium Channels. PLoS ONE 2015, 10, e0137611. [Google Scholar] [CrossRef]
- Ramirez-Dominguez, M.E.; Olamendi-Portugal, T.; Garcia, U.; Garcia, C.; Arechiga, H.; Possani, L.D. Cn11, the first example of a scorpion toxin that is a true blocker of Na+ currents in crayfish neurons. J. Exp. Biol. 2002, 205, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.-H. Quantification and Analysis of Proteins. In Diagnostic Molecular Biology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 187–214. [Google Scholar] [CrossRef]
- Al Asmari, A.; Khan, H.A.; Manthiri, R.A. Rapid profiling of crude scorpion venom using liquid chromatography and its relevance to species identification. Acta Chromatogr. 2012, 24, 501–509. [Google Scholar] [CrossRef]
- Rappuoli, R.; Montecucco, C. Guidebook to Protein Toxins and Their Use in Cell Biology; OUP/Sambrook and Tooze Publications: Nashville, TN, USA, 1997. [Google Scholar]
- Céard, B.; De Lima, M.-E.; Bougis, P.E.; Martin-Eauclaire, M.-F. Purification of the main β-toxin from Tityus serrulatus scorpion venom using high-performance liquid chromatography. Toxicon 1992, 30, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Zerrouk, H.; Bougis, P.E.; Céard, B.; Benslimane, A.; Martin-Eauclaire, M.F. Analysis by high-performance liquid chromatography of Androctonus mauretanicus mauretanicus (black scorpion) venom. Toxicon 1991, 29, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.F.; Rochat, H. Large scale purification of toxins from the venom of the scorpion Androctonus australis Hector. Toxicon 1986, 24, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Valdez-Velázquez, L.L.; Olamendi-Portugal, T.; Restano-Cassulini, R.; Zamudio, F.Z.; Possani, L.D. Mass fingerprinting and electrophysiological analysis of the venom from the scorpion Centruroides hirsutipalpus (Scorpiones: Buthidae). J. Venom. Anim. Toxins Incl. Trop. Dis. 2018, 24, 17. [Google Scholar] [CrossRef]
- El-Bitar, A.M.H.; Sarhan, M.; Abdel-Rahman, M.A.; Quintero-Hernandez, V.; Aoki-Utsubo, C.; Moustafa, M.A.; Possani, L.D.; Hotta, H. Smp76, a Scorpine-Like Peptide Isolated from the Venom of the Scorpion Scorpio maurus palmatus, with a Potent Antiviral Activity Against Hepatitis C Virus and Dengue Virus. Int. J. Pept. Res. Ther. 2020, 26, 811–821. [Google Scholar] [CrossRef]
- ElFessi, R.; Khamessi, O.; Srairi-Abid, N.; Sabatier, J.-M.; Tytgat, J.; Peigneur, S.; Kharrat, R. Purification and Characterization of Bot33: A Non-Toxic Peptide from the Venom of Buthus occitanus tunetanus Scorpion. Molecules 2022, 27, 7278. [Google Scholar] [CrossRef]
- Morey, S.S.; Kiran, K.M.; Gadag, J.R. Purification and properties of hyaluronidase from Palamneus gravimanus (Indian black scorpion) venom. Toxicon 2006, 47, 188–195. [Google Scholar] [CrossRef]
- Opiteck, G.J.; Jorgenson, J.W.; Anderegg, R.J. Two-Dimensional SEC/RPLC Coupled to Mass Spectrometry for the Analysis of Peptides. Anal. Chem. 1997, 69, 2283–2291. [Google Scholar] [CrossRef]
- Aissaoui-Zid, D.; Saada, M.-C.; Moslah, W.; Potier-Cartereau, M.; Lemettre, A.; Othman, H.; Gaysinski, M.; Abdelkafi-Koubaa, Z.; Souid, S.; Marrakchi, N.; et al. AaTs-1: A Tetrapeptide from Androctonus australis Scorpion Venom, Inhibiting U87 Glioblastoma Cells Proliferation by p53 and FPRL-1 Up-Regulations. Molecules 2021, 26, 7610. [Google Scholar] [CrossRef]
- Bayatzadeh, M.A.; Mirakabadi, A.Z.; Babaei, N.; Doulah, A.H.; Doosti, A. Characterization, molecular modeling and phylogenetic analysis of a long mammalian neurotoxin from the venom of the Iranian scorpion Androctonus crassicauda. Biologia 2020, 75, 1029–1041. [Google Scholar] [CrossRef]
- Khataminia, A.; Jalali, M.R.; Jalali, S.M.; Jafari, H. The Effect of Various Fractions of Hemiscorpius lepturus Scorpion (Scorpionida: Hemiscorpiidae) Venom on Hemostatic System in Peripheral Blood of Rats in Comparison to Whole Venom. Jundishapur J. Health Sci. 2020, 12, 102586. [Google Scholar] [CrossRef]
- Zerouti, K.; Khemili, D.; Laraba-Djebari, F.; Hammoudi-Triki, D. Nontoxic fraction of scorpion venom reduces bacterial growth and inflammatory response in a mouse model of infection. Toxin Rev. 2021, 40, 310–324. [Google Scholar] [CrossRef]
- Advantages and Disadvantages of HPLC. Available online: https://aspiringyouths.com/advantages-disadvantages/hplc/ (accessed on 13 August 2023).
- Lee, K.; Shin, S.Y.; Kim, K.; Lim, S.S.; Hahm, K.-S.; Kim, Y. Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs. Biochem. Biophys. Res. Commun. 2004, 323, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.-J.; Liu, Z.; Kong, F.-Z.; Fan, L.-Y.; Xiao, H.; Cao, C.-X. Comparison of antimicrobial peptide purification via free-flow electrophoresis and gel filtration chromatography. Electrophoresis 2017, 38, 3147–3154. [Google Scholar] [CrossRef] [PubMed]
- Erdeş, E.; Doğan, T.S.; Coşar, İ.; Danışman, T.; Kunt, K.B.; Şeker, T.; Yücel, M.; Özen, C. Characterization of Leiurus abdullahbayrami (Scorpiones: Buthidae) venom: Peptide profile, cytotoxicity and antimicrobial activity. J. Venom. Anim. Toxins Incl. Trop. Dis. 2014, 20, 48. [Google Scholar] [CrossRef] [PubMed]
- Conde, R.; Zamudio, F.Z.; Rodríguez, M.H.; Possani, L.D. Scorpine, an anti-malaria and anti-bacterial agent purified from scorpion venom. FEBS Lett. 2000, 471, 165–168. [Google Scholar] [CrossRef]
- Torres-Larios, A.; Gurrola, G.B.; Zamudio, F.Z.; Possani, L.D. Hadrurin, a new antimicrobial peptide from the venom of the scorpion Hadrurus aztecus: Scorpion antimicrobial peptide. Eur. J. Biochem. 2000, 267, 5023–5031. [Google Scholar] [CrossRef]
- Acikara, Ö.B. Ion-Exchange Chromatography and Its Applications; IntechOpen: London, UK, 2013. [Google Scholar] [CrossRef]
- Rack, M.; Richter, D.; Rubly, N. Purification and characterization of a β-toxin from the venom of the African scorpion Leiurus quinquestriatus. FEBS Lett. 1987, 214, 163–166. [Google Scholar] [CrossRef]
- Shirmardi, S.; Shamsaei, M.; Gandomkar, M.; Saniei, E.; Ghannadi, M.; Zare, A. Comparison of two purified toxic fractions from Mesobuthus eupeus scorpion venom. J. Venom. Anim. Toxins Incl. Trop. Dis. 2009, 16, 639–646. [Google Scholar] [CrossRef]
- Newton, K.A.; Clench, M.R.; Deshmukh, R.; Jeyaseelan, K.; Strong, P.N. Mass fingerprinting of toxic fractions from the venom of the Indian red scorpion, Mesobuthus tamulus: Biotope-specific variation in the expression of venom peptides. Rapid Commun. Mass Spectrom. 2007, 21, 3467–3476. [Google Scholar] [CrossRef]
- Gómez-Ramírez, I.; Riaño-Umbarila, L.; Olamendi-Portugal, T.; Restano-Cassulini, R.; Possani, L.; Becerril, B. Biochemical, electrophysiological and immunological characterization of the venom from Centruroides baergi, a new scorpion species of medical importance in Mexico. Toxicon 2020, 184, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Louati, H.; Krayem, N.; Fendri, A.; Aissa, I.; Sellami, M.; Bezzine, S.; Gargouri, Y. A thermoactive secreted phospholipase A2 purified from the venom glands of Scorpio maurus: Relation between the kinetic properties and the hemolytic activity. Toxicon 2013, 72, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Mirakabadi, Z.; Habibi, E. Identification and Purification of two mammalian nourotoxins from Iranian Scorpion (Buthotus schach) venom. Arch. Razi Inst. 2008, 63, 39–45. [Google Scholar]
- El-Aziz, T.M.A.; Xiao, Y.; Kline, J.; Gridley, H.; Heaston, A.; Linse, K.D.; Ward, M.J.; Rokyta, D.R.; Stockand, J.D.; Cummins, T.R.; et al. Identification and Characterization of Novel Proteins from Arizona Bark Scorpion Venom That Inhibit Nav1.8, a Voltage-Gated Sodium Channel Regulator of Pain Signaling. Toxins 2021, 13, 501. [Google Scholar] [CrossRef]
- Wang, Y.; Xing, B.; Li, T.; Wang, C.; Zhou, M.; Liu, Y.; Fan, L.; Hu, L.; Peng, X.; Xiang, Y.; et al. SVP-B5 peptide from Buthus martensii Karsch scorpion venom exerts hyperproliferative effects on irradiated hematopoietic cells. Exp. Ther. Med. 2017, 14, 5081–5086. [Google Scholar] [CrossRef]
- Li, H.; Xia, Y. Recombinant production of the insecticidal scorpion toxin BjαIT in Escherichia coli. Protein Expr. Purif. 2018, 142, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Bayatzadeh, M.A.; Mirakabadi, A.Z.; Babaei, N.; Doulah, A.; Doosti, A. Expression and purification of recombinant alpha-toxin AnCra1 from the scorpion Androctonus crassicauda and its functional characterization on mammalian sodium channels. Mol. Biol. Rep. 2021, 48, 6303–6312. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Xiao, F.; Peng, F.; Jiang, D.; Mao, X.; Liu, H.; Li, W.; Hu, D.; Wang, T. Expression, purification and functional characterization of a recombinant scorpion venom peptide BmTXKβ. Peptides 2003, 24, 187–192. [Google Scholar] [CrossRef]
- Aubrey, N.; Devaux, C.; di Luccio, E.; Goyffon, M.; Rochat, H.; Billiald, P. A Recombinant scFv/Streptavidin-Binding Peptide Fusion Protein for the Quantitative Determination of the Scorpion Venom Neurotoxin AahI. Biol. Chem. 2001, 382, 1621–1628. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Cui, Y.; Song, Y.-B.; Liu, Y.-F.; Zhang, R.; Wu, C.-F.; Zhang, J.-H. Purification, characterization and functional expression of a new peptide with an analgesic effect from Chinese scorpion Buthus martensii Karsch (BmK AGP-SYPU1). Biomed. Chromatogr. 2011, 25, 801–807. [Google Scholar] [CrossRef]
- Slagboom, J.; Otvos, R.A.; Cardoso, F.C.; Iyer, J.; Visser, J.C.; van Doodewaerd, B.R.; McCleary, R.J.; Niessen, W.M.; Somsen, G.W.; Lewis, R.J.; et al. Neurotoxicity fingerprinting of venoms using on-line microfluidic AChBP profiling. Toxicon 2018, 148, 213–222. [Google Scholar] [CrossRef]
- Heemskerk, A.A.M.; Busnel, J.-M.; Schoenmaker, B.; Derks, R.J.E.; Klychnikov, O.; Hensbergen, P.J.; Deelder, A.M.; Mayboroda, O.A. Ultra-Low Flow Electrospray Ionization-Mass Spectrometry for Improved Ionization Efficiency in Phosphoproteomics. Anal. Chem. 2012, 84, 4552–4559. [Google Scholar] [CrossRef]
- Martin-Eauclaire, M.-F.; Granjeaud, S.; Belghazi, M.; Bougis, P.E. Achieving automated scorpion venom mass fingerprinting (VMF) in the nanogram range. Toxicon 2013, 69, 211–218. [Google Scholar] [CrossRef]
- Daoudi, K.; Malosse, C.; Lafnoune, A.; Darkaoui, B.; Chakir, S.; Sabatier, J.; Chamot-Rooke, J.; Cadi, R.; Oukkache, N. Mass spectrometry-based top-down and bottom-up approaches for proteomic analysis of the Moroccan Buthus occitanus scorpion venom. FEBS Open Bio 2021, 11, 1867–1892. [Google Scholar] [CrossRef]
- Phillipson, D.W.; Milgram, K.E.; Yanovsky, A.I.; Rusnak, L.S.; Haggerty, D.A.; Farrell, W.P.; Greig, M.J.; Xiong, X.; Proefke, M.L. High-Throughput Bioassay-Guided Fractionation: A Technique for Rapidly Assigning Observed Activity to Individual Components of Combinatorial Libraries, Screened in HTS Bioassays. J. Comb. Chem. 2002, 4, 591–599. [Google Scholar] [CrossRef]
- Malviya, N.; Malviya, S. Bioassay guided fractionation-an emerging technique influence the isolation, identification and characterization of lead phytomolecules. Int. J. Hosp. Pharm. 2017, 2, 1–6. [Google Scholar] [CrossRef][Green Version]
- Rezaei, A.; Asgari, S.; Komijani, S.; Sadat, S.N.; Sabatier, J.-M.; Nasrabadi, D.; Bagheri, K.P.; Shahbazzadeh, D.; Eidgahi, M.R.A.; De Waard, M.; et al. Discovery of Leptulipin, a New Anticancer Protein from the Iranian Scorpion, Hemiscorpius lepturus. Molecules 2022, 27, 2056. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.; Ayed, A.S.; Abdel-Mottaleb, Y.; Omran, M.A.A.; Nabil, Z.I. Cardiac disorders and mode of action of the Egyptian scorpion venom Androctonus bicolor on isolated toad’s heart. J. Basic Appl. Zool. 2015, 72, 137–144. [Google Scholar] [CrossRef]
- Yazdkhasti, M.; Jalali, M.R.; Khadjeh, G.H.; Jafari, H.; Rezaie, A. Cardiotoxic Effects of Hemiscorpius lepturus Scorpion Venom Fractions in Rats. Iran. J. Toxicol. 2021, 15, 27–36. [Google Scholar] [CrossRef]
- Goudarzi, H.R.; Nazari, A.; Noofeli, M.; Samiani, M. Bioassay of derived components from venom of Iranian medically important scorpions to identify the bradykinin potentiating factors. Pharmacol. Toxicol. 2017, preprint. [Google Scholar] [CrossRef]
- Shao, J.-H.; Cui, Y.; Zhao, M.-Y.; Wu, C.-F.; Liu, Y.-F.; Zhang, J.-H. Purification, characterization, and bioactivity of a new analgesic-antitumor peptide from Chinese scorpion Buthus martensii Karsch. Peptides 2014, 53, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Kawachi, T.; Miyashita, M.; Nakagawa, Y.; Miyagawa, H. Isolation and Characterization of an Anti-Insect β-Toxin from the Venom of the Scorpion Isometrus maculatus. Biosci. Biotechnol. Biochem. 2013, 77, 205–207. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mlayah-Bellalouna, S.; Dufour, M.; Mabrouk, K.; Mejdoub, H.; Carlier, E.; Othman, H.; Belghazi, M.; Tarbe, M.; Goaillard, J.M.; Gigmes, D.; et al. AaTX1, from Androctonus australis scorpion venom: Purification, synthesis and characterization in dopaminergic neurons. Toxicon 2014, 92, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, M.; Mitani, N.; Kitanaka, A.; Yakio, M.; Chen, M.; Nishimoto, S.; Uchiyama, H.; Sue, M.; Hotta, H.; Nakagawa, Y.; et al. Identification of an antiviral component from the venom of the scorpion Liocheles australasiae using transcriptomic and mass spectrometric analyses. Toxicon 2021, 191, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Jridi, I.; Catacchio, I.; Majdoub, H.; Shahbazzadeh, D.; El Ayeb, M.; Frassanito, M.A.; Solimando, A.G.; Ribatti, D.; Vacca, A.; Borchani, L. The small subunit of Hemilipin2, a new heterodimeric phospholipase A2 from Hemiscorpius lepturus scorpion venom, mediates the antiangiogenic effect of the whole protein. Toxicon 2017, 126, 38–46. [Google Scholar] [CrossRef]
- Díaz-García, A.; Ruiz-Fuentes, J.L.; Yglesias-Rivera, A.; Rodríguez-Sánchez, H.; Garlobo, Y.R.; Martinez, O.F.; Castro, J.A.F. Enzymatic analysis of venom from Cuban scorpion Rhopalurus junceus. J. Venom Res. 2015, 6, 11–18. [Google Scholar]
- Abreu, C.B.; Bordon, K.C.F.; Cerni, F.A.; Oliveira, I.S.; Balenzuela, C.; Alexandre-Silva, G.M.; Zoccal, K.F.; Reis, M.B.; Wiezel, G.A.; Peigneur, S.; et al. Pioneering Study on Rhopalurus crassicauda Scorpion Venom: Isolation and Characterization of the Major Toxin and Hyaluronidase. Front. Immunol. 2020, 11, 2011. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Zhang, Y.; Gopalakrishnakone, P. A Novel Trypsin-like Serine Proteinase from the Venom of the Chinese Scorpion Buthus martensii Karsch. In Proceedings of the 4th Kuala Lumpur International Conference on Biomedical Engineering 2008, Kuala Lumpur, Malaysia, 25–28 June 2008; Abu Osman, N.A., Ibrahim, F., Wan Abas, W.A.B., Abdul Rahman, H.S., Ting, H.-N., Eds.; IFMBE Proceedings 21. Springer: Berlin/Heidelberg, Germany, 2008; pp. 829–832. [Google Scholar] [CrossRef]
- Louati, H.; Zouari, N.; Miled, N.; Gargouri, Y. A new chymotrypsin-like serine protease involved in dietary protein digestion in a primitive animal, Scorpio maurus: Purification and biochemical characterization. Lipids Health Dis. 2011, 10, 121. [Google Scholar] [CrossRef]
- Espino-Solis, G.P.; Estrada, G.; Olamendi-Portugal, T.; Villegas, E.; Zamudio, F.; Cestele, S.; Possani, L.D.; Corzo, G. Isolation and molecular cloning of beta-neurotoxins from the venom of the scorpion Centruroides suffusus suffusus. Toxicon 2011, 57, 739–746. [Google Scholar] [CrossRef]
- Corzo, G.; Escoubas, P.; Villegas, E.; Barnham, K.J.; He, W.; Norton, R.S.; Nakajima, T. Characterization of unique amphipathic antimicrobial peptides from venom of the scorpion Pandinus imperator. Biochem. J. 2001, 359, 35–45. [Google Scholar] [CrossRef]
- Vetter, I.; Davis, J.L.; Rash, L.D.; Anangi, R.; Mobli, M.; Alewood, P.F.; Lewis, R.J.; King, G.F. Venomics: A new paradigm for natural products-based drug discovery. Amino Acids 2011, 40, 15–28. [Google Scholar] [CrossRef]
- Wilson, D.; Daly, N. Venomics: A Mini-Review. High-Throughput 2018, 7, 19. [Google Scholar] [CrossRef]
- Schwartz, E.F.; Diego-Garcia, E.; de la Vega, R.C.R.; Possani, L.D. Transcriptome analysis of the venom gland of the Mexican scorpion Hadrurus gertschi (Arachnida: Scorpiones). BMC Genom. 2007, 8, 119. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, M.A.; Quintero-Hernandez, V.; Possani, L.D. Venom proteomic and venomous glands transcriptomic analysis of the Egyptian scorpion Scorpio maurus palmatus (Arachnida: Scorpionidae). Toxicon 2013, 74, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Yu, Y.; Wu, Y.; Hao, P.; Di, Z.; He, Y.; Chen, Z.; Yang, W.; Shen, Z.; He, X.; et al. The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat. Commun. 2013, 4, 2602. [Google Scholar] [CrossRef]
- Rendón-Anaya, M.; Delaye, L.; Possani, L.D.; Herrera-Estrella, A. Global Transcriptome Analysis of the Scorpion Centruroides noxius: New Toxin Families and Evolutionary Insights from an Ancestral Scorpion Species. PLoS ONE 2012, 7, e43331. [Google Scholar] [CrossRef]
- Luna-Ramírez, K.; Quintero-Hernández, V.; Juárez-González, V.R.; Possani, L.D. Whole Transcriptome of the Venom Gland from Urodacus yaschenkoi Scorpion. PLoS ONE 2015, 10, e0127883. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.; Harrison, P.L.; Strong, P.N. Snapshots of scorpion venomics. J. Arid. Environ. 2015, 112, 170–176. [Google Scholar] [CrossRef]
- Rokyta, D.R.; Ward, M.J. Venom-gland transcriptomics and venom proteomics of the black-back scorpion (Hadrurus spadix) reveal detectability challenges and an unexplored realm of animal toxin diversity. Toxicon 2017, 128, 23–37. [Google Scholar] [CrossRef]
- Ward, M.J.; Ellsworth, S.A.; Rokyta, D.R. Venom-gland transcriptomics and venom proteomics of the Hentz striped scorpion (Centruroides hentzi; Buthidae) reveal high toxin diversity in a harmless member of a lethal family. Toxicon 2018, 142, 14–29. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, U.C.; Nishiyama, M.Y.J.; Dos Santos, M.B.V.; Santos-Da-Silva, A.D.P.; Chalkidis, H.D.M.; Souza-Imberg, A.; Candido, D.M.; Yamanouye, N.; Dorce, V.A.C.; Junqueira-De-Azevedo, I.D.L.M. Proteomic endorsed transcriptomic profiles of venom glands from Tityus obscurus and T. serrulatus scorpions. PLoS ONE 2018, 13, e0193739. [Google Scholar] [CrossRef]
- Díaz, C.; Chang-Castillo, A.; Lomonte, B.; Bonilla, F.; Víquez, C.; Alfaro-Chinchilla, A.; Triana, F.; Sasa, M. Venomics of the scorpion Tityus ocelote (Scorpiones, Buthidae): Understanding venom evolution in the subgenus Archaeotityus. Int. J. Pept. Res. Ther. 2022, 29, 2. [Google Scholar] [CrossRef]
- Ghezellou, P.; Jakob, K.; Atashi, J.; Ghassempour, A.; Spengler, B. Mass-Spectrometry-Based Lipidome and Proteome Profiling of Hottentotta saulcyi (Scorpiones: Buthidae) Venom. Toxins 2022, 14, 370. [Google Scholar] [CrossRef]
- Slagboom, J.; Kaal, C.; Arrahman, A.; Vonk, F.J.; Somsen, G.W.; Calvete, J.J.; Wüster, W.; Kool, J. Analytical strategies in venomics. Microchem. J. 2022, 175, 107187. [Google Scholar] [CrossRef]
- Solovyeva, E.M.; Lobas, A.A.; Kopylov, A.T.; Ilina, I.Y.; Levitsky, L.I.; Moshkovskii, S.A.; Gorshkov, M.V. FractionOptimizer: A method for optimal peptide fractionation in bottom-up proteomics. Anal. Bioanal. Chem. 2018, 410, 3827–3833. [Google Scholar] [CrossRef] [PubMed]
- Cid-Uribe, J.I.; Veytia-Bucheli, J.I.; Romero-Gutierrez, T.; Ortiz, E.; Possani, L.D. Scorpion venomics: A 2019 overview. Expert Rev. Proteom. 2020, 17, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Matallana-Surget, S.; Leroy, B.; Wattiez, R. Shotgun proteomics: Concept, key points and data mining. Expert Rev. Proteom. 2010, 7, 5–7. [Google Scholar] [CrossRef]
- Lomonte, B.; Calvete, J.J. Strategies in ‘snake venomics’ aiming at an integrative view of compositional, functional, and immunological characteristics of venoms. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 26. [Google Scholar] [CrossRef]
- Kim, B.; Araujo, R.; Howard, M.; Magni, R.; Liotta, L.A.; Luchini, A. Affinity enrichment for mass spectrometry: Improving the yield of low abundance biomarkers. Expert Rev. Proteom. 2018, 15, 353–366. [Google Scholar] [CrossRef]
- Magalhães, A.C.M.; de Santana, C.J.C.; Melani, R.D.; Domont, G.B.; Castro, M.S.; Fontes, W.; Roepstorff, P.; Júnior, O.R.P. Exploring the biological activities and proteome of Brazilian scorpion Rhopalurus agamemnon venom. J. Proteom. 2021, 237, 104119. [Google Scholar] [CrossRef]
- Bringans, S.; Eriksen, S.; Kendrick, T.; Gopalakrishnakone, P.; Livk, A.; Lock, R.; Lipscombe, R. Proteomic analysis of the venom of Heterometrus longimanus (Asian black scorpion). Proteomics 2008, 8, 1081–1096. [Google Scholar] [CrossRef] [PubMed]
- Rogowska-Wrzesinska, A.; Le Bihan, M.-C.; Thaysen-Andersen, M.; Roepstorff, P. 2D gels still have a niche in proteomics. J. Proteom. 2013, 88, 4–13. [Google Scholar] [CrossRef]
- Uribe, J.I.C.; Vargas, J.M.J.; Batista, C.V.F.; Zuñiga, F.Z.; Possani, L.D. Comparative proteomic analysis of female and male venoms from the Mexican scorpion Centruroides limpidus: Novel components found. Toxicon 2017, 125, 91–98. [Google Scholar] [CrossRef]
- Romero-Gutiérrez, M.; Santibáñez-López, C.; Jiménez-Vargas, J.; Batista, C.; Ortiz, E.; Possani, L. Transcriptomic and Proteomic Analyses Reveal the Diversity of Venom Components from the Vaejovid Scorpion Serradigitus gertschi. Toxins 2018, 10, 359. [Google Scholar] [CrossRef]
- Verano-Braga, T.; Dutra, A.A.A.; León, I.R.; Melo-Braga, M.N.; Roepstorff, P.; Pimenta, A.M.C.; Kjeldsen, F. Moving Pieces in a Venomic Puzzle: Unveiling Post-translationally Modified Toxins from Tityus serrulatus. J. Proteome Res. 2013, 12, 3460–3470. [Google Scholar] [CrossRef] [PubMed]
- Melani, R.D.; Nogueira, F.C.S.; Domont, G.B. It is time for top-down venomics. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017, 23, 44. [Google Scholar] [CrossRef]
- Melani, R.D.; Nogueira, F.C.S.; Domont, G.B. Female-biased population divergence in the venom of the Hentz striped scorpion (Centruroides hentzi). Toxicon 2018, 152, 137–149. [Google Scholar] [CrossRef]
- Pandeswari, P.B.; Sabareesh, V. Middle-down approach: A choice to sequence and characterize proteins/proteomes by mass spectrometry. RSC Adv. 2019, 9, 313–344. [Google Scholar] [CrossRef]
- Harrison, P.L.; Abdel-Rahman, M.A.; Strong, P.N.; Tawfik, M.M.; Miller, K. Characterisation of three alpha-helical antimicrobial peptides from the venom of Scorpio maurus palmatus. Toxicon 2016, 117, 30–36. [Google Scholar] [CrossRef]
- de Melo, E.T.; Estrela, A.B.; Santos, E.C.G.; Machado, P.R.L.; Farias, K.J.S.; Torres, T.M.; Carvalho, E.; Lima, J.P.M.S.; Silva-Júnior, A.A.; Barbosa, E.G.; et al. Structural characterization of a novel peptide with antimicrobial activity from the venom gland of the scorpion Tityus stigmurus: Stigmurin. Peptides 2015, 68, 3–10. [Google Scholar] [CrossRef]
- Guo, X.; Ma, C.; Du, Q.; Wei, R.; Wang, L.; Zhou, M.; Chen, T.; Shaw, C. Two peptides, TsAP-1 and TsAP-2, from the venom of the Brazilian yellow scorpion, Tityus serrulatus: Evaluation of their antimicrobial and anticancer activities. Biochimie 2013, 95, 1784–1794. [Google Scholar] [CrossRef]
- Zeng, X.-C.; Corzo, G.; Hahin, R. Scorpion Venom Peptides without Disulfide Bridges. Int. Union Biochem. Mol. Biol. Life 2005, 57, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Yang, F.; Li, F.; Li, Z.; Lang, Y.; Shen, B.; Wu, Y.; Li, W.; Harrison, P.L.; Strong, P.N.; et al. Therapeutic Potential of a Scorpion Venom-Derived Antimicrobial Peptide and Its Homologs Against Antibiotic-Resistant Gram-Positive Bacteria. Front. Microbiol. 2018, 9, 1159. [Google Scholar] [CrossRef] [PubMed]
- Cesa-Luna, C.; Muñoz-Rojas, J.; Saab-Rincon, G.; Baez, A.; Morales-García, Y.E.; Juárez-González, V.R.; Quintero-Hernández, V. Structural characterization of scorpion peptides and their bactericidal activity against clinical isolates of multidrug-resistant bacteria. PLoS ONE 2019, 14, e0222438. [Google Scholar] [CrossRef]
- Harrison, P.L.; Abdel-Rahman, M.A.; Miller, K.; Strong, P.N. Antimicrobial peptides from scorpion venoms. Toxicon 2014, 88, 115–137. [Google Scholar] [CrossRef]
- Cao, L.; Li, Z.; Zhang, R.; Wu, Y.; Li, W.; Cao, Z. StCT2, a new antibacterial peptide characterized from the venom of the scorpion Scorpiops tibetanus. Peptides 2012, 36, 213–220. [Google Scholar] [CrossRef]
- Dai, L.; Corzo, G.; Naoki, H.; Andriantsiferana, M.; Nakajima, T. Purification; structure–function analysis, and molecular characterization of novel linear peptides from scorpion Opisthacanthus madagascariensis. Biochem. Biophys. Res. Commun. 2002, 293, 1514–1522. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Nomura, K.; Ferrat, G.; Nakajima, T.; Darbon, H.; Iwashita, T.; Corzo, G. Induction of Morphological Changes in Model Lipid Membranes and the Mechanism of Membrane Disruption by a Large Scorpion-Derived Pore-Forming Peptide. Biophys. J. 2005, 89, 4067–4080. [Google Scholar] [CrossRef] [PubMed]
- Belokoneva, O.S.; Satake, H.; Mal’Tseva, E.L.; Pal’Mina, N.P.; Villegas, E.; Nakajima, T.; Corzo, G. Pore formation of phospholipid membranes by the action of two hemolytic arachnid peptides of different size. Biochim. Biophys. Acta (BBA) Biomembr. 2004, 1664, 182–188. [Google Scholar] [CrossRef] [PubMed][Green Version]
- López-Giraldo, E.; Carrillo, E.; Titaux-Delgado, G.; Cano-Sánchez, P.; Colorado, A.; Possani, L.D.; del Río-Portilla, F. Structural and functional studies of scorpine: A channel blocker and cytolytic peptide. Toxicon 2023, 222, 106985. [Google Scholar] [CrossRef] [PubMed]
- Luna-Ramirez, K.; Tonk, M.; Rahnamaeian, M.; Vilcinskas, A. Bioactivity of Natural and Engineered Antimicrobial Peptides from Venom of the Scorpions Urodacus yaschenkoi and U. manicatus. Toxins 2017, 9, 22. [Google Scholar] [CrossRef]
| Separation Techniques | Advantages | Disadvantages | Example of Purified AMP | References |
|---|---|---|---|---|
| RP-HPLC |
|
| Vejovine [11] | [58] |
| Cytotoxic linear peptide (IsCT) [59] | ||||
| Scorpine-like peptide (Smp76) [50] | ||||
| SEC |
|
| First step in Heteroscorpine-1 purification [27] | [60,61,62] |
| First step in Scorpine purification | ||||
| First step in Hadurin purification [63] |
| Type of IEX | Column Name | Scorpion Species | Purified Molecule | References |
|---|---|---|---|---|
| Strong AEX | Quaternary ammonium (Q) column | Scorpio maurus | Phospholipase A2 (Sm-PLVG) | [69] |
| Weak AEX | Diethylaminoethyl (DEAE) column | Buthotus schach | BS311 and BS313 | [70] |
| Strong CEX | Sulphopropyl (SP) column | Centruroides sculpturatus | Proteins inhibiting Nav1.8 | [71] |
| Weak CEX | Carboxymethyl (CM) column | Mesobuthus martensii | Scorpion venom peptide (SVP-B5) | [72] |
| Purified Molecule | Scorpion Species | Molecular Mass (Da) | Separation Process | Column Used | References |
|---|---|---|---|---|---|
| Phospholipase A2 | Liocheles australasiae | 13,079.8 | RP-HPLC RP-HPLC LC/MS | C4 C18 C18 | [91] |
| Hemiscorpius lepturus | 14,000 | SEC RP-HPLC RP-HPLC | Sephadex G-50 Semi preparative C8 Analytical C8 | [92] | |
| Scorpio maurus | 17,000 | SEC AEX Hydrophobic Interaction HPLC | Sephadex G-100 Q-Sepharose Phenyl-Sepharose Nucleogel GFC 300-8 | [69] | |
| Heterometrus laoticus | 14,018.4 | SEC CEX RP-HPLC | Sephadex G-50 CM-650 M C4 | [32] | |
| Hyaluronidase | Rhopalurus junceus | 45,000–60,000 | SEC | Superdex 75 | [93] |
| Tityus serrulatus | 49,312 | SEC RP-HPLC | Sephadex G-50 Analytical C8 | [94] | |
| Palamneus gravimanus | 52,000 | SEC IEX SEC | Sephadex G-75 DEAE-cellulose | [52] | |
| Metalloproteinase | Tityus serrulatus | 22,000 24,000 | AEX SEC | DEAE Diol-300 | [18] |
| 25,500 | SEC RP-HPLC | Sephadex G-50 C18 | [19] | ||
| Serine proteinase | Mesobuthus martensii | 33,000 | SEC CEX RP-HPLC | Superdex G-75 UNO-Q C8 | [95] |
| Scorpio maurus | 25,000 | CEX SECFP AEXSEC | DEAE-SephadexSephadex G-100SP-SepharoseSephadex G-50 | [96] | |
| Neurotoxins | Mesobuthus martensii | 7246.40 | CEX Hydrophobic Interaction CEX Hydrophobic Interaction SEC | SP-Sepharose Phenyl Sepharose 4 SP-Sepharose Phenyl Sepharose 4 Superdex Peptide HR 10/30 | [88] |
| Centruroides suffusus suffusus | 7524.9 7537.6 7588.6 13,596 | RP-HPLC CEX RP-HPLC | C18 TSK-gel sulfopropyl C18 | [97] | |
| Isometrus maculatus | 6894 | RP-HPLC RP-HPLC | C4 C18 | [89] | |
| Androctonus australis | 3849.5 | SEC SEC Exchange FPLC RP-HPLC | Sephadex G-50 Resource S C18 | [90] | |
| Hemiscorpius lepturus | 4874 5107 | SEC AEX CEX RP-HPLC | Sephadex G-50 DEAE-Sepharose CM-Sepharose C8 | [26] | |
| AMPs | Heterometrus laoticus (Heteroscorpine-1) | 8293 | SEC CEX | Sephadex G-50 CM-Sepharose | [27] |
| Pandinus imperator (Scorpine) | 8350 | SEC CEX RP-HPLC | Sephadex G-50 CM-Cellulose C18 | [62] | |
| Hoffmannihadrurus aztecus (Hadrurin) | 4436 | SEC HPLC HPLC | Sephadex G-50 C18 C18 | [63] | |
| Pandinus imperator (Pandinin-1) | 4799 | RP-HPLC CEX RP-HPLC | C18 TSK-gel sulphopropyl C4 | [98] |
| Workflow | Scorpion Species | No. of Proteins | Main Protein Distribution/Most Abundant Venom Components | References |
|---|---|---|---|---|
| Workflow 1 Shotgun strategy | Tityus obscurus | ND | Metalloproteinase (47.48%) NaScTxs (13.80%) KScTxs (11.45%) Conserved venom components (10.26%) AMPs (3.51%) Other proteinases (5.74%) Other components (7.76%) | [109] |
| Tityus serrulatus | ND | Metalloproteinase (36.55%) NaScTxs (14.19%) KScTxs (15.60%) Conserved venom components (14.99%) Hypotensin (4.91%) Other component (15.98%) | ||
| Rhopalurus agamemnon | 230 | NaScTxs (16.95%) KScTxs (2.17%) AMPs (1.73%) Housekeeping proteins (40.43%) Metalloproteinase (6.12%) Amylase (2.825%) Others (29.775%) | [118] | |
| Combination of workflow 2 + 3 + 4 | Mesobuthus martensii | 227 | NaScTxs (9.69%) KScTxs (5.32%) AMPs (0.44%) Regulation proteins (11.2%) Structure proteins (7.04%) Metabolism proteins (7.04%) Other components (59.27%) | [33] |
| Combination of workflow 1 + 3 + TD | Buthus occitanus | 102 | NaScTxs (77%) KScTxs (14%) ClScTxs (3%) CaScTxs (1%) Toxin Acra (1%) Other components (4%) | [81] |
| Combination of workflow 2 + 3 + TD | Tityus serrulatus | 147 | KScTxs (12.19%) NaScTxs (10.81%) Enzymes (32%) AMPs (2%) Other components (43%) | [123] |
| Scorpion Species | AMP | Mechanism of Action | Structure | Reference |
|---|---|---|---|---|
| Heterometrus laoticus | Heteroscorpine-1 (HS-1) | Formation of blebs on the membrane | 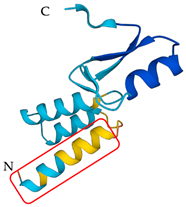 | [27] |
| Hoffmannihadrurus aztecus | Hadrurin | Lysis of zwitterionic phospholipids using hydrophobic interactions and acidic liposomes using electrostatic forces |  | [63] |
| Pandinus imperator | Pandinin-1 | Membrane disruption and pore formation |  | [138] |
| Pandinus imperator | Pandinin-2 | Liaison with degradation of lipid membrane and pore formation |  | [139] |
| Pandinus imperator | Scorpine | Membrane disruption by hydrophobic liaisons and cell penetration | 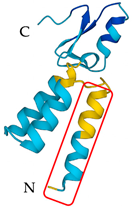 | [140] |
| Scorpiops tibetanus | Amphipathic peptide CT2 | Immediate disruption of bacterial membrane causing rapid killing | 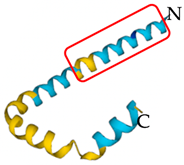 | [134] |
| Vaejovis mexicanus | Vejovine | Membrane disruption by direct interaction through the N-terminal region | 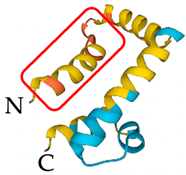 | [11] |
| Scorpion AMPs | Molecular Mass | Net Charge | Length | Amino Acid Sequence | Reference |
|---|---|---|---|---|---|
| Amphipathic peptide CT2 | 7930 Da | +2 | 69 | N- MKTQFAVLIISMILMQMLVQTEAGFWGKLWEGVKSAIGKRSLRNQDQFDNMFDSDLSDADLKLLDDLFD -C | [136,137] |
| Cytotoxic linear peptide IsCT2 | 1463.92 Da | +2 | 71 | N- MKTQFAILLVALVLFQMFAQSEAIFGAIWNGIKSLFGRRALNNDLDLDGLDELFDGEISQADVDFLKELMR -C | |
| Hadrurin | 4436 Da | +5 | 41 | N- GILDTIKSIASKVWNSKTVQDLKRKGINWVANKLGVSPQAA -C | |
| Imcroporin | 1760 Da | +2 | 74 | N- MKFQYLLAVFLIVLVVTDHCQAFFSLLPSLIGGLVSAIKGRRRRQLEARFEPKQRNFRKRELDFEKLFANMPDY -C | |
| Meucin-24 | 2753.95 Da | +4 | 88 | N- MMKQQFFLFLVIVMISSVIEAGRGREFMSNLKEKL SGVKEKMKNSWNRLTSMSEYACPVIEKWCEDHCQAKNAIGRCENTECKCLSK -C | |
| Meucin-25 | 3095.56 Da | +4 | 56 | N- MFRIEYSLVQLLLRNVTIPLLLIIQMHIMSSVKLIQIRIWIQYVTVLQMFSMKTKQ -C | |
| Mucroporin | 2031.58 Da | +2 | 74 | N- MKVKFLLAVFLIVLVVTDHCHALFGLIPSLIGGLVSAFKGRRKRQMEARFEPQNRNYRKRELDLEKLFANMPDY -C | |
| Pandinin-1 | 4799.2 Da | +1 | 44 | N- GKVWDWIKSAAKKIWSSEPVSQLKGQVLNAAKNYVAEKIGATPT -C | |
| Pandinin-2 | 2612.6 Da | +1 | 24 | N- FWGALAKGALKLIPSLFSSFSKKD -C | |
| Scorpine | 8350 Da | +3 | 94 | N- MNSKLTALIFLGLIAIAYCGWINEEKIQKKIDERMGNTVLGGMAKAIVHKMAKNEFQCMANMDMLGNCEKHCQTSGEKGYCHGTKCKCGTPLSY -C | |
| Vejovine | 4873 Da | +4 | 82 | N- MNAKTLFVVFLIGMLVTEQVEAGIWSSIKNLASKAWNSDIGQSLRNKAAGAINKFVADKIGVTPSQAASMTLDEIVDAMYYD -C | |
| Uy17 | 1369.43 Da | +2 | 13 | N- ILSAIWSGIKGLL -C | [141] |
| Uy192 | 1459.98 Da | +2 | 13 | N- FLSTIWNGIKGLL -C | |
| Uy234 | 1986.19 Da | +3 | 18 | N- FPFLLSLIPSAISAIKRL -C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasr, S.; Borges, A.; Sahyoun, C.; Nasr, R.; Roufayel, R.; Legros, C.; Sabatier, J.-M.; Fajloun, Z. Scorpion Venom as a Source of Antimicrobial Peptides: Overview of Biomolecule Separation, Analysis and Characterization Methods. Antibiotics 2023, 12, 1380. https://doi.org/10.3390/antibiotics12091380
Nasr S, Borges A, Sahyoun C, Nasr R, Roufayel R, Legros C, Sabatier J-M, Fajloun Z. Scorpion Venom as a Source of Antimicrobial Peptides: Overview of Biomolecule Separation, Analysis and Characterization Methods. Antibiotics. 2023; 12(9):1380. https://doi.org/10.3390/antibiotics12091380
Chicago/Turabian StyleNasr, Sara, Adolfo Borges, Christina Sahyoun, Riad Nasr, Rabih Roufayel, Christian Legros, Jean-Marc Sabatier, and Ziad Fajloun. 2023. "Scorpion Venom as a Source of Antimicrobial Peptides: Overview of Biomolecule Separation, Analysis and Characterization Methods" Antibiotics 12, no. 9: 1380. https://doi.org/10.3390/antibiotics12091380
APA StyleNasr, S., Borges, A., Sahyoun, C., Nasr, R., Roufayel, R., Legros, C., Sabatier, J.-M., & Fajloun, Z. (2023). Scorpion Venom as a Source of Antimicrobial Peptides: Overview of Biomolecule Separation, Analysis and Characterization Methods. Antibiotics, 12(9), 1380. https://doi.org/10.3390/antibiotics12091380








