Ciprofloxacin and Tetracycline Resistance Cause Collateral Sensitivity to Aminoglycosides in Salmonella Typhimurium
Abstract
:1. Introduction
2. Results
2.1. Cross-Resistance and Collateral Sensitivity of Antibiotic-Induced Resistant S. Typhimurium
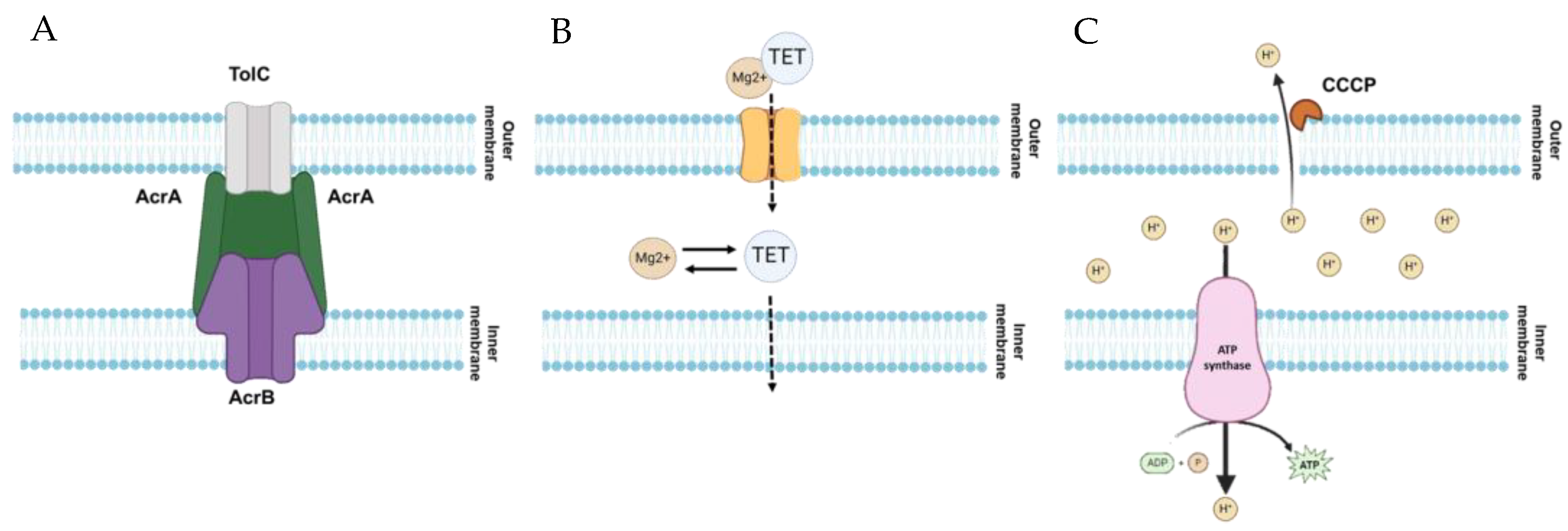
2.2. Role of Antibiotic Resistance Mechanisms in Evolving cross-Resistance and Collateral Sensitivity
3. Discussion
4. Materials and Methods
4.1. Strain and Culture Conditions
4.2. Preparation of Antibiotic Stock Solutions
4.3. Induction of Antibiotic-Resistant Salmonella
4.4. Antibiotic Susceptibility Assay
4.5. RT-qPCR Assay
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eng, S.-K.; Pusparajah, P.; Ab Mutalib, N.-S.; Ser, H.-L.; Chan, K.-G.; Lee, L.-H. Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 2015, 8, 284–293. [Google Scholar] [CrossRef]
- Wang, X.; Biswas, S.; Paudyal, N.; Pan, H.; Li, X.; Fang, W.; Yue, M. Antibiotic resistance in Salmonella Typhimurium isolates recovered from the food chain through national antimicrobial resistance monitoring system between 1996 and 2016. Front. Microbiol. 2019, 10, 985. [Google Scholar] [CrossRef]
- Seif, Y.; Kavvas, E.; Lachance, J.C.; Yurkovich, J.T.; Nuccio, S.P.; Fang, X.; Catoiu, E.; Raffatellu, M.; Palsson, B.O.; Monk, J.M. Genome-scale metabolic reconstructions of multiple Salmonella strains reveal serovar-specific metabolic traits. Nat. Commun. 2018, 9, 3771. [Google Scholar] [CrossRef]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.M.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef]
- Dawan, J.; Ahn, J. Assessment of cooperative antibiotic resistance of Salmonella Typhimurium within heterogeneous population. Microb. Pathog. 2021, 157, 104973. [Google Scholar] [CrossRef]
- Roope, L.S.J.; Smith, R.D.; Pouwels, K.B.; Buchanan, J.; Abel, L.; Eibich, P.; Butler, C.C.; Tan, P.S.; Walker, A.S.; Robotham, J.V.; et al. The challenge of antimicrobial resistance: What economics can contribute. Science 2019, 364, eaau4679. [Google Scholar] [CrossRef]
- Murugaiyan, J.; Kumar, P.A.; Rao, G.S.; Iskandar, K.; Hawser, S.; Hays, J.P.; Mohsen, Y.; Adukkadukkam, S.; Awuah, W.A.; Jose, R.A.M.; et al. Progress in alternative strategies to combat antimicrobial resistance: Focus on antibiotics. Antibiotics 2022, 11, 200. [Google Scholar] [CrossRef]
- Sincak, M.; Šoltisová, K.; Luptakova, A.; Sedlakova-Kadukova, J. Overproduction of efflux pumps as a mechanism of metal and antibiotic cross-resistance in the natural environment. Sustainability 2023, 15, 8767. [Google Scholar] [CrossRef]
- Kavya, I.K.; Kochhar, N.; Ghosh, A.; Shrivastava, S.; Singh Rawat, V.; Mondal Ghorai, S.; Kaur Sodhi, K.; James, A.; Kumar, M. Perspectives on systematic generation of antibiotic resistance with special emphasis on modern antibiotics. Total Environ. Res. Themes 2023, 8, 100068. [Google Scholar]
- Dawan, J.; Ahn, J. Assessment of cross-resistance potential to serial antibiotic treatments in antibiotic-resistant Salmonella Typhimurium. Microb. Pathog. 2020, 148, 104478. [Google Scholar] [CrossRef]
- Trampari, E.; Prischi, F.; Vargiu, A.V.; Abi-Assaf, J.; Bavro, V.N.; Webber, M.A. Functionally distinct mutations within AcrB underpin antibiotic resistance in different lifestyles. npj Antimicrob. Resist. 2023, 1, 2. [Google Scholar] [CrossRef]
- Akshay, S.D.; Nayak, S.; Deekshit, V.K.; Rohit, A.; Maiti, B. Differential expression of outer membrane proteins and quinolone resistance determining region mutations can lead to ciprofloxacin resistance in Salmonella Typhi. Arch. Microbiol. 2023, 205, 136. [Google Scholar] [CrossRef] [PubMed]
- Aulin, L.B.S.; Liakopoulos, A.; van der Graaf, P.H.; Rozen, D.E.; van Hasselt, J.G.C. Design principles of collateral sensitivity-based dosing strategies. Nat. Commun. 2021, 12, 5691. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Amado, S.; Laborda, P.; Martínez, J.L. Tackling antibiotic resistance by inducing transient and robust collateral sensitivity. Nat. Commun. 2023, 14, 1723. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Xi, W.; Yang, D.; Zhao, L.; Yu, W.; He, Y.; Ni, W.; Gao, Z. Collateral sensitivity between tetracyclines and aminoglycosides constrains resistance evolution in carbapenem-resistant Klebsiella pneumoniae. Drug Resist. Uptat. 2023, 68, 100961. [Google Scholar] [CrossRef] [PubMed]
- Pál, C.; Papp, B.; Lázár, V. Collateral sensitivity of antibiotic-resistant microbes. Trends Microbiol. 2015, 23, 401–407. [Google Scholar] [CrossRef]
- Podnecky, N.L.; Fredheim, E.G.A.; Kloos, J.; Sørum, V.; Primicerio, R.; Roberts, A.P.; Rozen, D.E.; Samuelsen, Ø.; Johnsen, P.J. Conserved collateral antibiotic susceptibility networks in diverse clinical strains of Escherichia coli. Nat. Commun. 2018, 9, 3673. [Google Scholar] [CrossRef]
- Imamovic, L.; Sommer, M.O.A. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med. 2013, 5, 204ra132. [Google Scholar] [CrossRef]
- Lázár, V.; Pal Singh, G.; Spohn, R.; Nagy, I.; Horváth, B.; Hrtyan, M.; Busa-Fekete, R.; Bogos, B.; Méhi, O.; Csörgő, B.; et al. Bacterial evolution of antibiotic hypersensitivity. Mol. Syst. Biol. 2013, 9, 700. [Google Scholar] [CrossRef]
- Barbosa, C.; Trebosc, V.; Kemmer, C.; Rosenstiel, P.; Beardmore, R.; Schulenburg, H.; Jansen, G. Alternative evolutionary paths to bacterial antibiotic resistance cause distinct collateral effects. Mol. Biol. Evol. 2017, 34, 2229–2244. [Google Scholar] [CrossRef]
- Barbosa, C.; Beardmore, R.; Schulenburg, H.; Jansen, G. Antibiotic combination efficacy (ACE) networks for a Pseudomonas aeruginosa model. PLoS Biol. 2018, 16, e2004356. [Google Scholar] [CrossRef] [PubMed]
- Maltas, J.; Wood, K.B. Pervasive and diverse collateral sensitivity profiles inform optimal strategies to limit antibiotic resistance. PLoS Biol. 2019, 17, e3000515. [Google Scholar] [CrossRef] [PubMed]
- Liakopoulos, A.; Aulin, L.B.S.; Buffoni, M.; Fragkiskou, E.; Hasselt, J.G.C.v.; Rozen, D.E. Allele-specific collateral and fitness effects determine the dynamics of fluoroquinolone resistance evolution. Proc. Natl. Acad. Sci. USA 2022, 119, e2121768119. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lieberman, T.D.; Kishony, R. Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance. Proc. Natl. Acad. Sci. USA 2014, 111, 14494–14499. [Google Scholar] [CrossRef]
- Brepoels, P.; Appermans, K.; Pérez-Romero, C.A.; Lories, B.; Marchal, K.; Steenackers, H.P. Antibiotic cycling affects resistance evolution independently of collateral sensitivity. Mol. Biol. Evol. 2022, 39, msac257. [Google Scholar] [CrossRef] [PubMed]
- Szybalski, W.; Bryson, V. Genetic studies on microbial cross resistance to toxic agents. I. Cross resistance of Escherichia coli to fifteen antibiotics. J. Bacteriol. 1952, 64, 489–499. [Google Scholar] [CrossRef]
- Liu, D.Y.; Phillips, L.; Wilson, D.M.; Fulton, K.M.; Twine, S.M.; Wong, A.; Linington, R.G. Collateral sensitivity profiling in drug-resistant Escherichia coli identifies natural products suppressing cephalosporin resistance. Nat. Commun. 2023, 14, 1976. [Google Scholar] [CrossRef]
- Gonzales, P.R.; Pesesky, M.W.; Bouley, R.; Ballard, A.; Biddy, B.A.; Suckow, M.A.; Wolter, W.R.; Schroeder, V.A.; Burnham, C.A.; Mobashery, S.; et al. Synergistic, collaterally sensitive beta-lactam combinations suppress resistance in MRSA. Nat. Chem. Biol. 2015, 11, 855–861. [Google Scholar] [CrossRef]
- Harrison, E.M.; Ba, X.; Coll, F.; Blane, B.; Restif, O.; Carvell, H.; Koser, C.U.; Jamrozy, D.; Reuter, S.; Lovering, A.; et al. Genomic identification of cryptic susceptibility to penicillins and beta-lactamase inhibitors in methicillin-resistant Staphylococcus aureus. Nat. Microbiol. 2019, 4, 1680–1691. [Google Scholar] [CrossRef]
- Trampari, E.; Holden, E.R.; Wickham, G.J.; Ravi, A.; Martins, L.d.O.; Savva, G.M.; Webber, M.A. Exposure of Salmonella biofilms to antibiotic concentrations rapidly selects resistance with collateral tradeoffs. npj Biofilms Microbiomes 2021, 7, 3. [Google Scholar] [CrossRef]
- Roemhild, R.; Andersson, D.I. Mechanisms and therapeutic potential of collateral sensitivity to antibiotics. PLoS Pathog. 2021, 17, e1009172. [Google Scholar] [CrossRef] [PubMed]
- Le, D.; Krasnopeeva, E.; Sinjab, F.; Pilizota, T.; Kim, M. Active efflux leads to heterogeneous dissipation of proton motive force by protonophores in bacteria. mBio 2021, 12, e0067621. [Google Scholar] [CrossRef] [PubMed]
- Radlinski, L.C.; Rowe, S.E.; Brzozowski, R.; Wilkinson, A.D.; Huang, R.; Eswara, P.; Conlon, B.P. Chemical induction of aminoglycoside uptake overcomes antibiotic tolerance and resistance in Staphylococcus aureus. Cell Chem. Biol. 2019, 26, 1355–1364.e1354. [Google Scholar] [CrossRef]
- Weston, N.; Sharma, P.; Ricci, V.; Piddock, L.J.V. Regulation of the AcrAB-TolC efflux pump in Enterobacteriaceae. Res. Microbiol. 2018, 169, 425–431. [Google Scholar] [CrossRef]
- Yamasaki, S.; Zwama, M.; Yoneda, T.; Hayashi-Nishino, M.; Nishino, K. Drug resistance and physiological roles of RND multidrug efflux pumps in Salmonella enterica, Escherichia coli and Pseudomonas aeruginosa. Microbiology 2023, 169, 001322. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Wang-Kan, X.; Neuberger, A.; van Veen, H.W.; Pos, K.M.; Piddock, L.J.V.; Luisi, B.F. Multidrug efflux pumps: Structure, function and regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. [Google Scholar] [CrossRef]
- Jang, S. AcrAB-TolC, a major efflux pump in Gram negative bacteria: Toward understanding its operation mechanism. BMB Rep. 2023, 56, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Webber, M.A.; Bailey, A.M.; Blair, J.M.A.; Morgan, E.; Stevens, M.P.; Hinton, J.C.D.; Ivens, A.; Wain, J.; Piddock, L.J.V. The global consequence of disruption of the AcrAB-TolC efflux pump in Salmonella enterica includes reduced expression of SPI-1 and other attributes required to infect the host. J. Bacteriol. 2009, 191, 4276–4285. [Google Scholar] [CrossRef]
- Klenotic, P.A.; Moseng, M.A.; Morgan, C.E.; Yu, E.W. Structural and functional diversity of resistance-nodulation-cell division transporters. Chem. Rev. 2021, 121, 5378–5416. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, V.K.; Pathania, R. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian J. Med. Res. 2019, 149, 129–145. [Google Scholar]
- Yu, E.W.; Aires, J.R.; McDermott, G.; Nikaido, H. A periplasmic drug-binding site of the AcrB multidrug efflux pump: A crystallographic and site-directed mutagenesis study. J. Bacteriol. 2005, 187, 6804–6815. [Google Scholar] [CrossRef] [PubMed]
- Vargiu, A.V.; Nikaido, H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc. Natl. Acad. Sci. USA 2012, 109, 20637–20642. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Plesiat, P.; Nikaido, H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef] [PubMed]
- Eaves, D.J.; Ricci, V.; Piddock, L.J.V. Expression of acrB, acrF, acrD, marA, and soxS in Salmonella enterica serovar Typhimurium: Role in multiple antibiotic resistance. Antimicrob. Agents Chemother. 2004, 48, 1145–1150. [Google Scholar] [CrossRef]
- Peng, B.; Su, Y.-b.; Li, H.; Han, Y.; Guo, C.; Tian, Y.-M.; Peng, X.-X. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 2015, 21, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Harikishore, A.; Mathiyazakan, V.; Pethe, K.; Grüber, G. Novel targets and inhibitors of the Mycobacterium tuberculosis cytochrome bd oxidase to foster anti-tuberculosis drug discovery. Expert Opin. Drug Discov. 2023, 18, 917–927. [Google Scholar] [CrossRef]
- Grossman, T.H. Tetracycline antibiotics and resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef]
- Zhou, G.; Wang, Q.; Wang, Y.; Wen, X.; Peng, H.; Peng, R.; Shi, Q.; Xie, X.; Li, L. Outer membrane porins contribute to antimicrobial resistance in Gram-negative bacteria. Microorganisms 2023, 11, 1690. [Google Scholar] [CrossRef]
- Uddin, M.J.; Jeon, G.; Ahn, J. Variability in the adaptive response of antibiotic-resistant Salmonella Typhimurium to environmental stresses. Microb. Drug Resist. 2019, 25, 182–192. [Google Scholar] [CrossRef]
- Bruni, G.N.; Kralj, J.M. Membrane voltage dysregulation driven by metabolic dysfunction underlies bactericidal activity of aminoglycosides. eLife 2020, 9, e58706. [Google Scholar] [CrossRef]
- Farha, M.A.; Verschoor, C.P.; Bowdish, D.; Brown, E.D. Collapsing the proton motive force to identify synergistic combinations against Staphylococcus aureus. Chem. Biol. 2013, 20, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-C.; Han, F.; Song, M.-R.; Chen, S.; Li, Q.; Zhang, Q.; Zhu, K.; Shen, J.-Z. Natural flavones from Morus alba against methicillin-resistant Staphylococcus aureus via targeting the proton motive force and membrane permeability. J. Agric. Food Chem. 2019, 67, 10222–10234. [Google Scholar] [CrossRef]
- Amaral, L.; Martins, A.; Spengler, G.; Molnar, J. Efflux pumps of Gram-negative bacteria: What they do, how they do it, with what and how to deal with them. Front. Pharmacol. 2014, 4, 168. [Google Scholar] [CrossRef] [PubMed]
- Mulkidjanian, A.Y. Proton in the well and through the desolvation barrier. Biochim. Byophys. Acta 2006, 1757, 415–427. [Google Scholar] [CrossRef]
- Black, P.A.; Warren, R.M.; Louw, G.E.; van Helden, P.D.; Victor, T.C.; Kana, B.D. Energy metabolism and drug efflux in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2014, 58, 2491–2503. [Google Scholar] [CrossRef] [PubMed]
- Capita, R.; Riesco-Pelaez, F.; Alonso-Hernando, A.; Alonso-Calleja, C. Exposure of Escherichia coli ATCC 12806 to sublethal concentrations of food-grade biocides influences its ability to form biofilm, resistance to antimicrobials, and ultrastructure. Appl. Environ. Microbiol. 2014, 80, 1268–1280. [Google Scholar] [CrossRef]
- CLSI. M07-A10. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. In Approved Method of Analysis of CLSI, 10th ed.; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2015. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Dawan, J.; Uddin, M.J.; Ahn, J. Development of de novo resistance in Salmonella Typhimurium treated with antibiotic combinations. FEMS Microbiol. Lett. 2019, 366, fnz127. [Google Scholar] [CrossRef]
- Uddin, M.J.; Ahn, J. Characterization of β-lactamase- and efflux pump-mediated multiple antibiotic resistance in Salmonella Typhimurium. Food Sci. Biotechnol. 2018, 27, 921–928. [Google Scholar] [CrossRef]
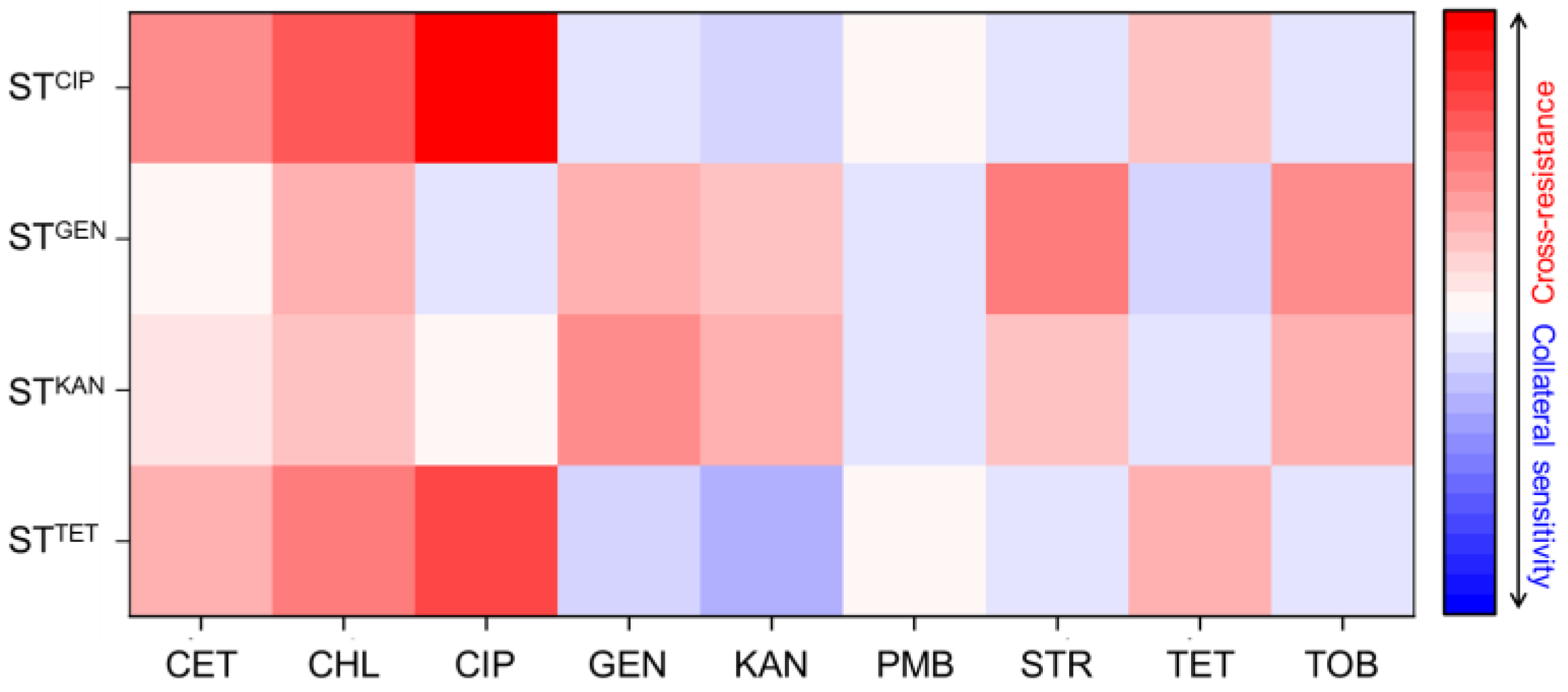
 ), gentamicin-resistant S. Typhimurium (STGEN,
), gentamicin-resistant S. Typhimurium (STGEN,  ), kanamycin-resistant S. Typhimurium (STKAN,
), kanamycin-resistant S. Typhimurium (STKAN,  ), and tetracycline-induced resistant S. Typhimurium (STTET,
), and tetracycline-induced resistant S. Typhimurium (STTET,  ).
).
 ), gentamicin-resistant S. Typhimurium (STGEN,
), gentamicin-resistant S. Typhimurium (STGEN,  ), kanamycin-resistant S. Typhimurium (STKAN,
), kanamycin-resistant S. Typhimurium (STKAN,  ), and tetracycline-induced resistant S. Typhimurium (STTET,
), and tetracycline-induced resistant S. Typhimurium (STTET,  ).
).
| Class | Antibiotic | Target Site | Antimicrobial Activity |
|---|---|---|---|
| Cephems | Cefotaxime | Cell wall | Bactericidal |
| Phenicols | Chloramphenicol | 50S ribosomal subunit | Bacteriostatic |
| Fluoroquinolones | Ciprofloxacin | DNA gyrase | Bactericidal |
| Aminoglycosides | Gentamycin | 30S ribosomal subunit | Bactericidal |
| Aminoglycosides | Kanamycin | 30S ribosomal subunit | Bactericidal |
| Aminoglycosides | Streptomycin | 30S ribosomal subunit | Bactericidal |
| Aminoglycosides | Tobramycin | 30S ribosomal subunit | Bactericidal |
| Glycopeptides | Polymyxin B | Cell membrane | Bactericidal |
| Tetracyclines | Tetracycline | 30S ribosomal subunit | Bacteriostatic |
| Antibiotic | STWT | STCIP | STGEN | STKAN | STTET |
|---|---|---|---|---|---|
| Cefotaxime | 0.0625 | 1 | 0.0625 | 0.125 | 0.5 |
| Chloramphenicol | 0.5 | 32 | 4 | 2 | 16 |
| Ciprofloxacin | 0.0156 | 16 | 0.0078 | 0.0156 | 2 |
| Gentamicin | 8 | 4 | 64 | 128 | 2 |
| Kanamycin | 32 | 8 | 128 | 256 | 4 |
| Polymyxin B | 4 | 4 | 2 | 2 | 4 |
| Streptomycin | 32 | 16 | 1024 | 128 | 16 |
| Tetracycline | 2 | 8 | 0.5 | 1 | 16 |
| Tobramycin | 16 | 8 | 256 | 128 | 8 |
| Strain | Antibiotic | Efflux Pump Inhibitor | ||
|---|---|---|---|---|
| No | CCCP | PAβN | ||
| STCIP | Ciprofloxacin | 16 | 16 | 2 |
| STGEN | Gentamicin | 64 | 64 | 64 |
| STKAN | Kanamycin | 256 | 256 | 256 |
| STTET | Tetracycline | 32 | 64 | 32 |
| Gene | Molecular Function | Primes Sequence | References |
|---|---|---|---|
| 16s rRNA | Reference gene | F: AGGCCTTCGGGTTGTAAAGT R: GTTAGCCGGTGCTTCTTCTG | [59] |
| acrA | Multidrug efflux pump | F: AAAACGGCAAAGCGAAGGT R: GTACCGGACTGCGGGAATT | [59] |
| acrB | Multidrug efflux pump | F: TGAAAAAAATGGAACCGTTCTTC R: CGAACGGCGTGGTGTCA | [59] |
| ompC | Outer membrane porins | F: TCGCAGCCTGCTGAACCAGAAC R: ACGGGTTGCGTTATAGGTCTGAG | [60] |
| ompF | Outer membrane porins | F: CGGAATTTATTGACGGCAGT R: GAGATAAAAAAACAGGACCG | [60] |
| ramA | Transcriptional activator | F: CCAGAAGGTGTATGATATTTGTCTCAAG R: GGTTGAACGTGCGGGTAAA | [60] |
| tolC | Multidrug efflux pump | F: GCCCGTGCGCAATATGAT R: CCGCGTTATCCAGGTTGTTG | [59] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasan, M.; Wang, J.; Ahn, J. Ciprofloxacin and Tetracycline Resistance Cause Collateral Sensitivity to Aminoglycosides in Salmonella Typhimurium. Antibiotics 2023, 12, 1335. https://doi.org/10.3390/antibiotics12081335
Hasan M, Wang J, Ahn J. Ciprofloxacin and Tetracycline Resistance Cause Collateral Sensitivity to Aminoglycosides in Salmonella Typhimurium. Antibiotics. 2023; 12(8):1335. https://doi.org/10.3390/antibiotics12081335
Chicago/Turabian StyleHasan, Mahadi, Jun Wang, and Juhee Ahn. 2023. "Ciprofloxacin and Tetracycline Resistance Cause Collateral Sensitivity to Aminoglycosides in Salmonella Typhimurium" Antibiotics 12, no. 8: 1335. https://doi.org/10.3390/antibiotics12081335
APA StyleHasan, M., Wang, J., & Ahn, J. (2023). Ciprofloxacin and Tetracycline Resistance Cause Collateral Sensitivity to Aminoglycosides in Salmonella Typhimurium. Antibiotics, 12(8), 1335. https://doi.org/10.3390/antibiotics12081335







