The Art of War with Pseudomonas aeruginosa: Targeting Mex Efflux Pumps Directly to Strategically Enhance Antipseudomonal Drug Efficacy
Abstract
1. Pseudomonas aeruginosa: The Battle-Worn Strategist and Its Intricately Orchestrated Defense Weaponry
2. RND Efflux Pumps in P. aeruginosa, the Typhons of Antibiotic Resistance
2.1. MexAB-OprM
2.2. MexXY
2.3. MexCD-OprJ
2.4. MexEF-OprN
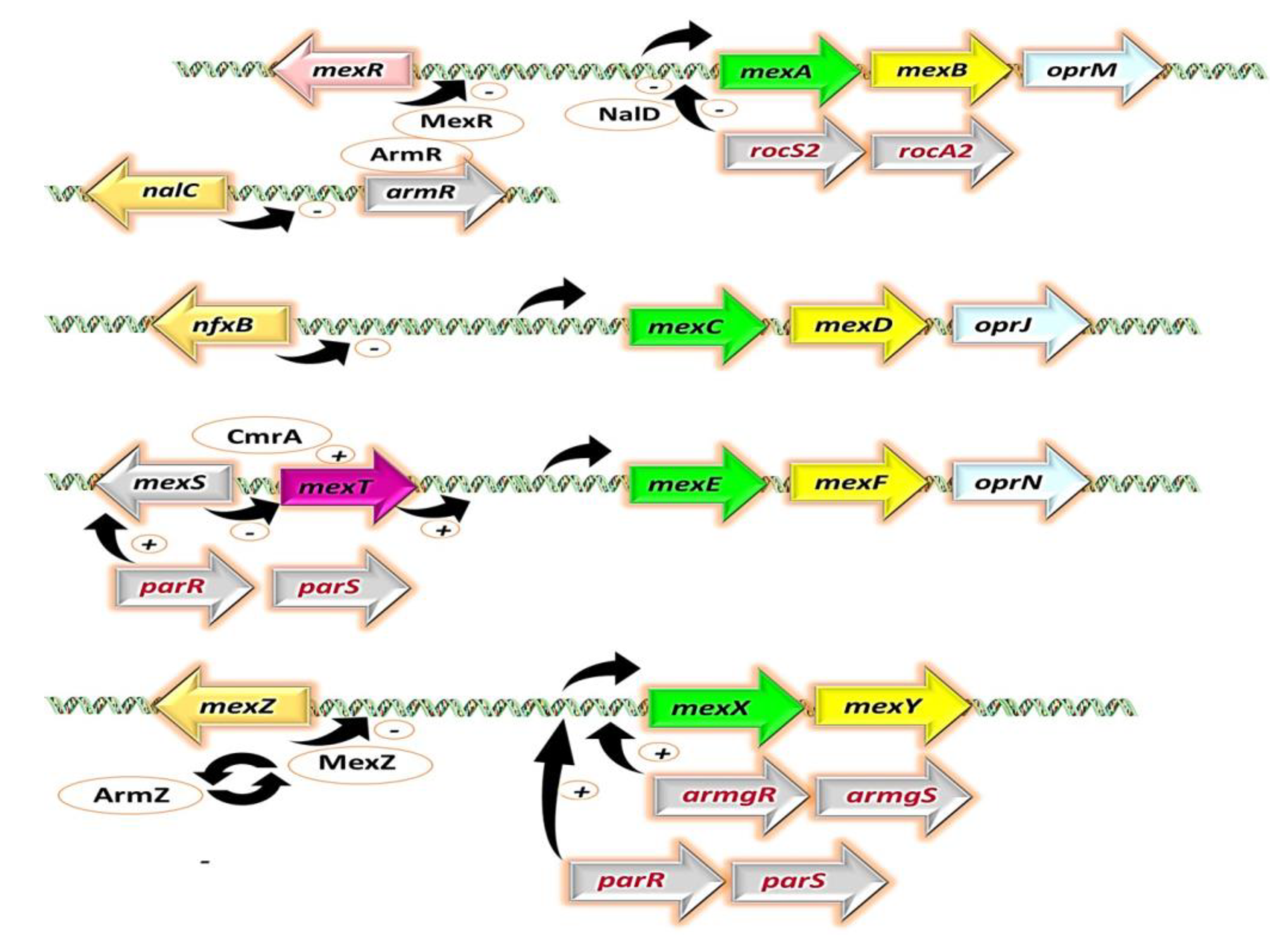
3. The Perilous Dance: Unveiling the Clinical Relevance of Mex Efflux Pumps in P. aeruginosa Isolates
4. Athena’s Wisdom: Non-Antibiotic Strategies against P. aeruginosa Efflux Pumps
4.1. Achilles’ Heel Exposed: EPIs Target P. aeruginosa MDR
4.1.1. Plant-Derived EPIs
- Alkaloids
- Conessine (Figure 6), a steroidal alkaloid derived from Holarrhena antidysenterica, has been studied for its effects on P. aeruginosa Mex pumps. Research conducted by Siriyong et al. in 2017 demonstrated that conessine can significantly reduce the MICs of various antipseudomonal antibiotics such as levofloxacin, cefotaxime, and tetracycline by at least 8-fold in a strain overexpressing the MexAB-OprM efflux pump. Interestingly, conessine does not appear to act as a membrane permeabilizer, as it does not cause the accumulation of 1-N-phenylnaphthylamine [251].
- Catharanthine (Figure 6), Dwivedi et al. (2018) evaluated catharanthine, a terpene indole alkaloid isolated from Catharanthus roseus, against MDR P. aeruginosa strains. They found that catharanthine exhibited no intrinsic antibacterial activity, but when combined with tetracycline and streptomycin, it significantly reduced their MICs, increasing P. aeruginosa’s susceptibility and reducing the emergence of tetracycline-resistant mutants. Furthermore, catharanthine showed efficacy comparable to EPIs such as PAβN and displayed activity against Mex pump-expressing P. aeruginosa strains [252].
- Berberine and palmatine (Figure 6), extracted from Berberis Vulgaris, exhibit EPI activity [253]. In a study by Aghayan et al. (2017) on P. aeruginosa isolates from burn infections, these compounds significantly reduced the MIC of antipseudomonal antibiotics, such as ciprofloxacin, by up to 8-fold in strains overexpressing MexAB-OprM [253]. Furthermore, Su et al. (2018) demonstrated the synergistic interaction of berberine with imipenem, effectively reversing imipenem resistance in P. aeruginosa by inhibiting the MexXY OprM efflux pump. These findings highlight the potential of berberine and palmatine as EPIs for combating MDR in P. aeruginosa [254].
- Theobromine (Figure 6), an alkaloid derived from Theobroma cacao, has been identified as an inhibitor of the P. aeruginosa MexAB-OprM efflux pump [239]. According to research by Piddock et al. (2010), it was suggested that compounds with small heterocyclic or nitrogen-containing structures have the potential to act as inhibitors of RND efflux pumps. In their study, a library of 26 compounds was evaluated, and among them, theobromine emerged as a highly potent plant-based EPI [255]. The compound demonstrated significant efficacy in reducing the MIC of ciprofloxacin in P. aeruginosa strains overexpressing the MexAB-OprM efflux pump. These findings support the notion that theobromine holds promise as an effective EPI in combating antibiotic resistance in P. aeruginosa [255].
- Phenolic compounds
- p-Coumaric acid (Figure 6) is a phenolic acid that can be extracted from a variety of edible plants such as Gnetum cleistostachyum [239,256]. In a recent study, Choudhury et al. (2016) investigated p-Coumaric acid and its derivative as a potential EPI of the MexAB-OprM in P. aeruginosa. These compounds showed promising results in preliminary screening, including significant docking scores and the ability to enhance the activity of ciprofloxacin in MexAB-OprM overexpressing strains of P. aeruginosa. The researchers suggest that these compounds could serve as lead molecules for the development of MexAB-OprM inhibitors, offering a potential solution to combat multidrug-resistant P. aeruginosa infections [256].
- Curcumin (Figure 6) is a polyphenol curcuminoid derived from the rhizomes of Curcuma longa. It has been investigated for its potential as an adjuvant to enhance the antimicrobial activities of commonly used antibiotics against MDR P. aeruginosa. In a study by Negi et al. (2019), curcumin (50 mg/L) was found to significantly decrease the MIC values of various antipseudomonal drugs, including meropenem, carbenicillin, ceftazidime, gentamicin, and ciprofloxacin, when used in combination with them against 170 clinical P. aeruginosa isolates. This synergistic effect suggests the promising role of curcumin in combating drug-resistant bacterial infections and its potential as an EPI [257].
- Resveratrol (Figure 6), a polyphenol stilbenoid found in various fruits and vegetables, including peanuts, blueberries, cranberries, and Japanese knotweed [258,259], has shown promising antimicrobial and antibiofilm properties when combined with colistin against colistin-resistant P. aeruginosa. Wang et al. (2023) demonstrated that resveratrol enhanced the activity of colistin against resistant P. aeruginosa in vitro and improved colistin efficacy in vivo [259]. However, further investigation is needed to evaluate the impact of resveratrol on Mex efflux pumps in P. aeruginosa.
4.1.2. EPIs of Synthetic Origin
- Peptidomimetic Compounds
- Replacement of aa1 and aa2: For the optimization of PAβN, it is crucial to consider the replacement of amino acids 1 and 2. These amino acids should possess both aromatic and basic properties, with the possibility of reversing their order [265,267]. Notably, substituting L-phenylalanine with L-homo-phenylalanine has been shown to significantly enhance EPI potency by two-fold. Furthermore, alternative basic amino side chains, such as ornithine or aminomethylproline, offer viable options for substitution, expanding the scope of EPI modifications [265,267] (Figure 7 and Figure 8).
- Modification of the cap moiety: The replacement of the naphthyl moiety with alternative fused rings, such as 5-aminoindan and 6-aminoquinoline, leads to a reduction in PAβN toxicity while enhancing the pharmacological properties of PAβN and its derivatives [265]. Notably, the incorporation of a 3-aminoquinoline moiety is crucial for mitigating cytotoxicity in mammalian cells during in vitro experiments [265] (Figure 7). These extensive SAR investigations have yielded a range of derivatives, including MC-02,595 and MC-04,124 [267,268,269], which exhibit potent EPI activities (Figure 8).
- 2.
- Arylpiperidines and Arylpiperazine Derivatives
- 3.
- Pyridopyrimidine and Pyranopyridine Derivatives
- 4.
- TXA Compounds
4.1.3. Microbial-Derived EPIs
- (i)
- Competitive inhibition: this involves EPIs that bind to the same site as the substrate and compete with it for access to the pump. This reduces the amount of substrate that can be transported out of the cell and increases its intracellular concentration [75,216]. An example of a competitive inhibitor is PAβN, which binds to the substrate-binding site of RND efflux pumps and inhibits their activity [123,221].
- (ii)
- Non-competitive substrate action: this mechanism involves EPIs that bind to a different site than the substrate on the efflux pump and prevent its transport by altering the conformation or function of the pump. These EPIs are also called substrate inhibitors because they act as substrates for the efflux pump but cannot be transported out of the cell [200,287]. An example of a non-competitive substrate inhibitor is D13-9001, which binds to a novel site on AcrB, a component of the RND efflux pump AcrAB-TolC, and inhibits its activity [221,272,273,274].
- (iii)
- Hindering functional movement: this strategy includes EPIs that bind to the efflux pump and interfere with its functional movement or rotation, which is essential for transporting substrates across the membrane [288,289]. These EPIs are also called allosteric inhibitors because they bind to a site other than the substrate-binding site and affect the activity of the efflux pump indirectly [290,291]. An example of an allosteric inhibitor is PAβN, which binds to MexB, a component of the RND efflux pump MexAB-OprM, and inhibits its activity by hindering its functional movement [221].
- (iv)
- Co-substrate action: this method incorporates EPIs that act as co-substrates for the efflux pump and require its energy-dependent transport to exert their inhibitory effect [292]. These EPIs are also called prodrugs, because they are converted into active inhibitors inside the bacterial cell after being transported by the efflux pump [293]. An example of a prodrug is MBX-3132, which is transported by RND efflux pumps and then oxidized into a reactive quinone-imine intermediate that covalently modifies and inhibits the pumps [294,295].
4.2. Prometheus’s Gift Unleashed: Antisense EPIs Tackle P. aeruginosa MDR
4.3. Perseus Unleashed: Phage-Based Therapeutics against P. aeruginosa MDR
5. MexB: The Zeus of Dominance—Tripartite Conquest in Bacterial Resistance
6. Unveiling the Mythological Enigma: MexB’s Multifaceted Binding Sites for Diverse Ligands
6.1. Multiple Binding Pockets and Ligand Interactions of MexB
6.2. Fluoroquinolone Antibiotic Binding Sites in MexB
6.3. EPI Binding Sites in MexB
7. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 2022, 7, 1–27. [Google Scholar]
- Labovská, S. Pseudomonas aeruginosa as a Cause of Nosocomial Infections. In Pseudomonas aeruginosa—Biofilm Formation, Infections and Treatments; IntechOpen: London, UK, 2021. [Google Scholar]
- Khan, H.A.; Ahmad, A.; Mehboob, R. Nosocomial infections and their control strategies. Asian Pac. J. Trop. Biomed. 2015, 5, 509–514. [Google Scholar] [CrossRef]
- Reynolds, D.; Kollef, M. The epidemiology and pathogenesis and treatment of Pseudomonas aeruginosa infections: An update. Drugs 2021, 81, 2117–2131. [Google Scholar] [CrossRef]
- Wood, S.J.; Kuzel, T.M.; Shafikhani, S.H. Pseudomonas aeruginosa: Infections, Animal Modeling, and Therapeutics. Cells 2023, 12, 199. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.J.; Goldufsky, J.W.; Seu, M.Y.; Dorafshar, A.H.; Shafikhani, S.H. Pseudomonas aeruginosa Cytotoxins: Mechanisms of Cytotoxicity and Impact on Inflammatory Responses. Cells 2023, 12, 195. [Google Scholar] [CrossRef]
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas aeruginosa: An audacious pathogen with an adaptable arsenal of virulence factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef]
- Rossi, E.; La Rosa, R.; Bartell, J.A.; Marvig, R.L.; Haagensen, J.A.; Sommer, L.M.; Molin, S.; Johansen, H.K. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat. Rev. Microbiol. 2021, 19, 331–342. [Google Scholar] [CrossRef]
- del Mar Cendra, M.; Torrents, E. Pseudomonas aeruginosa biofilms and their partners in crime. Biotechnol. Adv. 2021, 49, 107734. [Google Scholar] [CrossRef]
- Recio, R.; Mancheño, M.; Viedma, E.; Villa, J.; Orellana, M.Á.; Lora-Tamayo, J.; Chaves, F. Predictors of mortality in bloodstream infections caused by Pseudomonas aeruginosa and impact of antimicrobial resistance and bacterial virulence. Antimicrob. Agents Chemother. 2020, 64, e01759-19. [Google Scholar] [CrossRef]
- Sader, H.S.; Castanheira, M.; Duncan, L.R.; Flamm, R.K. Antimicrobial susceptibility of Enterobacteriaceae and Pseudomonas aeruginosa isolates from United States medical centers stratified by infection type: Results from the International Network for Optimal Resistance Monitoring (INFORM) surveillance program, 2015–2016. Diagn. Microbiol. Infect. Dis. 2018, 92, 69–74. [Google Scholar] [PubMed]
- Kunz Coyne, A.J.; El Ghali, A.; Holger, D.; Rebold, N.; Rybak, M.J. Therapeutic strategies for emerging multidrug-resistant Pseudomonas aeruginosa. Infect. Dis. Ther. 2022, 11, 661–682. [Google Scholar] [CrossRef]
- Idris, F.N.; Nadzir, M.M. Multi-drug resistant ESKAPE pathogens and the uses of plants as their antimicrobial agents. Arch. Microbiol. 2023, 205, 115. [Google Scholar] [CrossRef]
- Blomquist, K.C.; Nix, D.E. A critical evaluation of newer β-lactam antibiotics for treatment of Pseudomonas aeruginosa infections. Ann. Pharmacother. 2021, 55, 1010–1024. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.-J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Klockgether, J.; Cramer, N.; Wiehlmann, L.; Davenport, C.F.; Tümmler, B. Pseudomonas aeruginosa genomic structure and diversity. Front. Microbiol. 2011, 2, 150. [Google Scholar] [CrossRef]
- Stover, C.K.; Pham, X.Q.; Erwin, A.; Mizoguchi, S.; Warrener, P.; Hickey, M.; Brinkman, F.; Hufnagle, W.; Kowalik, D.; Lagrou, M. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef] [PubMed]
- Adejobi, A.; Ojo, O.; Alaka, O.; Odetoyin, B.; Onipede, A. Antibiotic resistance pattern of Pseudomonas spp. from patients in a tertiary hospital in South-West Nigeria. Germs 2021, 11, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Girlich, D.; Naas, T.; Nordmann, P. Biochemical characterization of the naturally occurring oxacillinase OXA-50 of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2004, 48, 2043–2048. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.-F.; Jayawardena, S.R.; Del Puerto, A.; Wiehlmann, L.; Laabs, U.; Tümmler, B.; Mathee, K. Characterization of poxB, a chromosomal-encoded Pseudomonas aeruginosa oxacillinase. Gene 2005, 358, 82–92. [Google Scholar] [CrossRef]
- Li, X.-Z.; Plésiat, P.; Nikaido, H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef] [PubMed]
- Breidenstein, E.B.; de la Fuente-Núñez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef]
- Fajardo, A.; Hernando-Amado, S.; Oliver, A.; Ball, G.; Filloux, A.; Martinez, J.L. Characterization of a novel Zn2+-dependent intrinsic imipenemase from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2014, 69, 2972–2978. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Ortega, C.; Wiegand, I.; Olivares, J.; Hancock, R.E.; Martínez, J.L. Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to β-lactam antibiotics. Antimicrob. Agents Chemother. 2010, 54, 4159–4167. [Google Scholar] [CrossRef]
- Doötsch, A.; Becker, T.; Pommerenke, C.; Magnowska, Z.; Jaänsch, L.; Haäussler, S. Genomewide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009, 53, 2522–2531. [Google Scholar] [CrossRef] [PubMed]
- Olivares, J.; Bernardini, A.; Garcia-Leon, G.; Corona, F.B.; Sanchez, M.; Martinez, J.L. The intrinsic resistome of bacterial pathogens. Front. Microbiol. 2013, 4, 103. [Google Scholar] [CrossRef]
- Juan, C.; Torrens, G.; González-Nicolau, M.; Oliver, A. Diversity and regulation of intrinsic β-lactamases from non-fermenting and other Gram-negative opportunistic pathogens. FEMS Microbiol. Rev. 2017, 41, 781–815. [Google Scholar] [CrossRef] [PubMed]
- Cabot, G.; Ocampo-Sosa, A.A.; Tubau, F.; Macia, M.D.; Rodríguez, C.; Moya, B.; Zamorano, L.; Suárez, C.; Peña, C.; Martínez-Martínez, L. Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa isolates from bloodstream infections: Prevalence and impact on resistance in a Spanish multicenter study. Antimicrob. Agents Chemother. 2011, 55, 1906–1911. [Google Scholar] [CrossRef]
- Moya, B.; Dötsch, A.; Juan, C.; Blázquez, J.; Zamorano, L.; Haussler, S.; Oliver, A. β-Lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog. 2009, 5, e1000353. [Google Scholar] [CrossRef]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef]
- Poole, K. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 2001, 3, 255–264. [Google Scholar]
- Riera, E.; Cabot, G.; Mulet, X.; García-Castillo, M.; del Campo, R.; Juan, C.; Cantón, R.; Oliver, A. Pseudomonas aeruginosa carbapenem resistance mechanisms in Spain: Impact on the activity of imipenem, meropenem and doripenem. J. Antimicrob. Chemother. 2011, 66, 2022–2027. [Google Scholar] [CrossRef] [PubMed]
- Moyá, B.; Beceiro, A.; Cabot, G.; Juan, C.; Zamorano, L.; Alberti, S.; Oliver, A. Pan-β-lactam resistance development in Pseudomonas aeruginosa clinical strains: Molecular mechanisms, penicillin-binding protein profiles, and binding affinities. Antimicrob. Agents Chemother. 2012, 56, 4771–4778. [Google Scholar] [CrossRef] [PubMed]
- Bruchmann, S.; Dötsch, A.; Nouri, B.; Chaberny, I.F.; Häussler, S. Quantitative contributions of target alteration and decreased drug accumulation to Pseudomonas aeruginosa fluoroquinolone resistance. Antimicrob. Agents Chemother. 2013, 57, 1361–1368. [Google Scholar] [CrossRef]
- Feng, Y.; Jonker, M.J.; Moustakas, I.; Brul, S.; Ter Kuile, B.H. Dynamics of mutations during development of resistance by Pseudomonas aeruginosa against five antibiotics. Antimicrob. Agents Chemother. 2016, 60, 4229–4236. [Google Scholar] [CrossRef]
- López-Causapé, C.; Cabot, G.; del Barrio-Tofiño, E.; Oliver, A. The versatile mutational resistome of Pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 685. [Google Scholar] [CrossRef]
- López-Causapé, C.; Sommer, L.M.; Cabot, G.; Rubio, R.; Ocampo-Sosa, A.A.; Johansen, H.K.; Figuerola, J.; Cantón, R.; Kidd, T.J.; Molin, S. Evolution of the Pseudomonas aeruginosa mutational resistome in an international cystic fibrosis clone. Sci. Rep. 2017, 7, 5555. [Google Scholar] [CrossRef]
- Greipel, L.; Fischer, S.; Klockgether, J.; Dorda, M.; Mielke, S.; Wiehlmann, L.; Cramer, N.; Tümmler, B. Molecular epidemiology of mutations in antimicrobial resistance loci of Pseudomonas aeruginosa isolates from airways of cystic fibrosis patients. Antimicrob. Agents Chemother. 2016, 60, 6726–6734. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Virulence Mech. Bact. Pathog. 2016, 22, 481–511. [Google Scholar] [CrossRef]
- Peterson, E.; Kaur, P. Antibiotic resistance mechanisms in bacteria: Relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef]
- Miller, S.I. Antibiotic resistance and regulation of the gram-negative bacterial outer membrane barrier by host innate immune molecules. MBio 2016, 7, e01541-16. [Google Scholar] [CrossRef] [PubMed]
- Mehla, J.; Malloci, G.; Mansbach, R.; López, C.A.; Tsivkovski, R.; Haynes, K.; Leus, I.V.; Grindstaff, S.B.; Cascella, R.H.; D’Cunha, N. Predictive rules of efflux inhibition and avoidance in Pseudomonas aeruginosa. MBio 2021, 12, e02785-20. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, G.; Leus, I.V.; Weeks, J.W.; Wolloscheck, D.; Rybenkov, V.V.; Zgurskaya, H.I. Synergy between active efflux and outer membrane diffusion defines rules of antibiotic permeation into Gram-negative bacteria. MBio 2017, 8, e01172-17. [Google Scholar] [CrossRef]
- Li, X.-Z.; Barré, N.; Poole, K. Influence of the MexA-MexB-OprM multidrug efflux system on expression of the MexC-MexD-OprJ and MexE-MexF-OprN multidrug efflux systems in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2000, 46, 885–893. [Google Scholar] [CrossRef]
- Westfall, D.A.; Krishnamoorthy, G.; Wolloscheck, D.; Sarkar, R.; Zgurskaya, H.I.; Rybenkov, V.V. Bifurcation kinetics of drug uptake by Gram-negative bacteria. PLoS ONE 2017, 12, e0184671. [Google Scholar] [CrossRef]
- Saha, P.; Sikdar, S.; Krishnamoorthy, G.; Zgurskaya, H.I.; Rybenkov, V.V. Drug permeation against efflux by two transporters. ACS Infect. Dis. 2020, 6, 747–758. [Google Scholar] [CrossRef]
- Nichols, W.W. Modeling the kinetics of the permeation of antibacterial agents into growing bacteria and its interplay with efflux. Antimicrob. Agents Chemother. 2017, 61, e02576-16. [Google Scholar] [CrossRef]
- Kojima, S.; Nikaido, H. Permeation rates of penicillins indicate that Escherichia coli porins function principally as nonspecific channels. Proc. Natl. Acad. Sci. 2013, 110, E2629–E2634. [Google Scholar] [CrossRef]
- Pool, K. Pseudomonas aeruginosa: Resistance to the max. Front. Microbiol 2011, 2, 65. [Google Scholar] [CrossRef]
- Dreier, J.; Ruggerone, P. Interaction of antibacterial compounds with RND efflux pumps in Pseudomonas aeruginosa. Front. Microbiol. 2015, 6, 660. [Google Scholar] [CrossRef]
- Amaral, L.; Martins, A.; Spengler, G.; Molnar, J. Efflux pumps of Gram-negative bacteria: What they do, how they do it, with what and how to deal with them. Front. Pharmacol. 2014, 4, 168. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Cook, D.N.; Hearst, J.E.; Nikaido, H. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 1994, 2, 489–493. [Google Scholar] [CrossRef]
- Sulavik, M.C.; Houseweart, C.; Cramer, C.; Jiwani, N.; Murgolo, N.; Greene, J.; DiDomenico, B.; Shaw, K.J.; Miller, G.H.; Hare, R. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 2001, 45, 1126–1136. [Google Scholar] [CrossRef]
- Elkins, C.A.; Nikaido, H. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominately by two large periplasmic loops. J. Bacteriol. 2002, 184, 6490–6498. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Yamaguchi, A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 2001, 183, 5803–5812. [Google Scholar] [CrossRef] [PubMed]
- Adamiak, J.W.; Jhawar, V.; Bonifay, V.; Chandler, C.E.; Leus, I.V.; Ernst, R.K.; Schweizer, H.P.; Zgurskaya, H.I. Loss of RND-type multidrug efflux pumps triggers iron starvation and lipid A modifications in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2021, 65, e00592-21. [Google Scholar] [CrossRef]
- Laudy, A.E. Non-antibiotics, efflux pumps and drug resistance of Gram-negative rods. Pol. J. Microbiol. 2018, 67, 129–135. [Google Scholar] [CrossRef]
- Lennen, R.M.; Politz, M.G.; Kruziki, M.A.; Pfleger, B.F. Identification of transport proteins involved in free fatty acid efflux in Escherichia coli. J. Bacteriol. 2013, 195, 135–144. [Google Scholar] [CrossRef]
- Li, X.-Z.; Nikaido, H.; Poole, K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1995, 39, 1948–1953. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Nakashima, R.; Sakurai, K. Structural basis of RND-type multidrug exporters. Front. Microbiol. 2015, 6, 327. [Google Scholar] [CrossRef]
- Alvarez-Ortega, C.; Olivares, J.; Martínez, J.L. RND multidrug efflux pumps: What are they good for? Front. Microbiol. 2013, 4, 7. [Google Scholar] [CrossRef]
- Fernando, D.M.; Kumar, A. Resistance-nodulation-division multidrug efflux pumps in gram-negative bacteria: Role in virulence. Antibiotics 2013, 2, 163–181. [Google Scholar] [CrossRef]
- Başkan, C.; Sırıken, B. Role of Efflux Pump in Biofilm Formation of Multidrug-Resistant Pseudomonas aeruginosa. Int. J. Mol. Sci. 2019, 23, 15779. [Google Scholar]
- Thanassi, D.G.; Cheng, L.W.; Nikaido, H. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 1997, 179, 2512–2518. [Google Scholar] [CrossRef] [PubMed]
- Teelucksingh, T.; Thompson, L.K.; Cox, G. The evolutionary conservation of Escherichia coli drug efflux pumps supports physiological functions. J. Bacteriol. 2020, 202, e00367-20. [Google Scholar] [CrossRef] [PubMed]
- Minagawa, S.; Inami, H.; Kato, T.; Sawada, S.; Yasuki, T.; Miyairi, S.; Horikawa, M.; Okuda, J.; Gotoh, N. RND type efflux pump system MexAB-OprM of Pseudomonas aeruginosa selects bacterial languages, 3-oxo-acyl-homoserine lactones, for cell-to-cell communication. BMC Microbiol. 2012, 12, 1–10. [Google Scholar] [CrossRef]
- Alcalde-Rico, M.; Olivares-Pacheco, J.; Halliday, N.; Cámara, M.; Martínez, J.L. The impaired quorum sensing response of Pseudomonas aeruginosa MexAB-OprM efflux pump overexpressing mutants is not due to non-physiological efflux of 3-oxo-C12-HSL. Environ. Microbiol. 2020, 22, 5167–5188. [Google Scholar] [CrossRef]
- Akinduti, P.A.; George, O.W.; Ohore, H.U.; Ariyo, O.E.; Popoola, S.T.; Adeleye, A.I.; Akinwande, K.S.; Popoola, J.O.; Rotimi, S.O.; Olufemi, F.O. Evaluation of Efflux-Mediated Resistance and Biofilm formation in Virulent Pseudomonas aeruginosa Associated with Healthcare Infections. Antibiotics 2023, 12, 626. [Google Scholar] [CrossRef]
- Nishino, K.; Latifi, T.; Groisman, E.A. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2006, 59, 126–141. [Google Scholar] [CrossRef]
- Moore-Machacek, A.s.; Gloe, A.; O’Leary, N.; Reen, F.J. Efflux, Signaling and Warfare in a Polymicrobial World. Antibiotics 2023, 12, 731. [Google Scholar] [CrossRef] [PubMed]
- Zgurskaya, H.I.; Adamiak, J.W.; Leus, I.V. Making sense of drug-efflux transporters in the physiological environment. Curr. Opin. Microbiol. 2022, 69, 102179. [Google Scholar] [CrossRef]
- Atassi, G.; Medernach, R.; Scheetz, M.; Nozick, S.; Rhodes, N.J.; Murphy-Belcaster, M.; Murphy, K.R.; Alisoltani, A.; Ozer, E.A.; Hauser, A.R. Genomics of aminoglycoside resistance in Pseudomonas aeruginosa bloodstream infections at a United States Academic Hospital. Microbiol. Spectr. 2023, 49, e05087-22. [Google Scholar] [CrossRef]
- Thy, M.; Timsit, J.-F.; de Montmollin, E. Aminoglycosides for the Treatment of Severe Infection Due to Resistant Gram-Negative Pathogens. Antibiotics 2023, 12, 860. [Google Scholar] [CrossRef]
- Nikaido, H.; Pagès, J.-M. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 2012, 36, 340–363. [Google Scholar] [CrossRef] [PubMed]
- Avrain, L.; Mertens, P.; Van Bambeke, F. RND efflux pumps in P. aeruginosa: An underestimated resistance mechanism. Antibiot. Susceptibility 2013, 26321, 26–28. [Google Scholar]
- Yamasaki, S.; Zwama, M.; Yoneda, T.; Hayashi-Nishino, M.; Nishino, K. Drug resistance and physiological roles of RND multidrug efflux pumps in Salmonella enterica, Escherichia coli and Pseudomonas aeruginosa. Microbiology 2023, 169, 001322. [Google Scholar] [CrossRef]
- Piddock, L.J. Multidrug-resistance efflux pumps? not just for resistance. Nat. Rev. Microbiol. 2006, 4, 629–636. [Google Scholar] [CrossRef]
- Sun, J.; Deng, Z.; Yan, A. Bacterial multidrug efflux pumps: Mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 2014, 453, 254–267. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, C.; Su, S.; Panmanee, W.; Lau, G.W.; Browne, T.; Cox, K.; Paul, A.T.; Ko, S.-H.B.; Mortensen, J.E.; Lam, J.S. A putative ABC transporter permease is necessary for resistance to acidified nitrite and EDTA in Pseudomonas aeruginosa under aerobic and anaerobic planktonic and biofilm conditions. Front. Microbiol. 2016, 7, 291. [Google Scholar] [CrossRef]
- Chen, L.; Duan, K. A PhoPQ-regulated ABC transporter system exports tetracycline in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 3016–3024. [Google Scholar] [CrossRef] [PubMed]
- Yero, D.; Díaz-Lobo, M.; Costenaro, L.; Conchillo-Solé, O.; Mayo, A.; Ferrer-Navarro, M.; Vilaseca, M.; Gibert, I.; Daura, X. The Pseudomonas aeruginosa substrate-binding protein Ttg2D functions as a general glycerophospholipid transporter across the periplasm. Commun. Biol. 2021, 4, 448. [Google Scholar] [CrossRef]
- Zhang, L.; Mah, T.-F. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J. Bacteriol. 2008, 190, 4447–4452. [Google Scholar] [CrossRef] [PubMed]
- Poudyal, B.; Sauer, K. The ABC of biofilm drug tolerance: The MerR-like regulator BrlR is an activator of ABC transport systems, with PA1874-77 contributing to the tolerance of Pseudomonas aeruginosa biofilms to tobramycin. Antimicrob. Agents Chemother. 2018, 62, e01981-17. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.W.; Zhang, L.; Mah, T.-F. PA3225 is a transcriptional repressor of antibiotic resistance mechanisms in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2017, 61, e02114-16. [Google Scholar] [CrossRef] [PubMed]
- Dulyayangkul, P.; Satapoomin, N.; Avison, M.B.; Charoenlap, N.; Vattanaviboon, P.; Mongkolsuk, S. Over-expression of hypochlorite inducible Major Facilitator Superfamily (MFS) pumps reduces antimicrobial drug susceptibility by increasing the production of MexXY Mediated by ArmZ in Pseudomonas aeruginosa. Front. Microbiol. 2021, 11, 592153. [Google Scholar] [CrossRef]
- Bock, L.J.; Ferguson, P.M.; Clarke, M.; Pumpitakkul, V.; Wand, M.E.; Fady, P.-E.; Allison, L.; Fleck, R.A.; Shepherd, M.J.; Mason, A.J. Pseudomonas aeruginosa adapts to octenidine via a combination of efflux and membrane remodelling. Commun. Biol. 2021, 4, 1058. [Google Scholar] [CrossRef]
- Bissonnette, L.; Champetier, S.; Buisson, J.; Roy, P. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: Similarity of the product to transmembrane transport proteins. J. Bacteriol. 1991, 173, 4493–4502. [Google Scholar] [CrossRef]
- Stokes, H.; Hall, R.M. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation. Plasmid 1991, 26, 10–19. [Google Scholar] [CrossRef]
- Li, X.-Z.; Poole, K.; Nikaido, H. Contributions of MexAB-OprM and an EmrE homolog to intrinsic resistance of Pseudomonas aeruginosa to aminoglycosides and dyes. Antimicrob. Agents Chemother. 2003, 47, 27–33. [Google Scholar] [CrossRef]
- Ninio, S.; Rotem, D.; Schuldiner, S. Functional analysis of novel multidrug transporters from human pathogens. J. Biol. Chem. 2001, 276, 48250–48256. [Google Scholar] [CrossRef]
- Duan, K.; Dammel, C.; Stein, J.; Rabin, H.; Surette, M.G. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 2003, 50, 1477–1491. [Google Scholar] [CrossRef] [PubMed]
- He, G.-X.; Kuroda, T.; Mima, T.; Morita, Y.; Mizushima, T.; Tsuchiya, T. An H+-coupled multidrug efflux pump, PmpM, a member of the MATE family of transporters, from Pseudomonas aeruginosa. J. Bacteriol. 2004, 186, 262–265. [Google Scholar] [CrossRef]
- Zhao, J.; Hellwig, N.; Djahanschiri, B.; Khera, R.; Morgner, N.; Ebersberger, I.; Wang, J.; Michel, H. Assembly and functional role of PACE transporter PA2880 from Pseudomonas aeruginosa. Microbiol. Spectr. 2022, 10, e01453-21. [Google Scholar] [CrossRef]
- Poole, K.; Krebes, K.; McNally, C.; Neshat, S. Multiple antibiotic resistance in Pseudomonas aeruginosa: Evidence for involvement of an efflux operon. J. Bacteriol. 1993, 175, 7363–7372. [Google Scholar] [CrossRef]
- Masuda, N.; Sakagawa, E.; Ohya, S.; Gotoh, N.; Tsujimoto, H.; Nishino, T. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2000, 44, 3322–3327. [Google Scholar] [CrossRef] [PubMed]
- Rahbar, M.; Hamidi-Farahani, R.; Asgari, A.; Esmailkhani, A.; Soleiman-Meigooni, S. Expression of RND efflux pumps mediated antibiotic resistance in Pseudomonas aeruginosa clinical strains. Microb. Pathog. 2021, 153, 104789. [Google Scholar]
- Pesingi, P.V.; Singh, B.R.; Pesingi, P.K.; Bhardwaj, M.; Singh, S.V.; Kumawat, M.; Sinha, D.K.; Gandham, R.K. MexAB-OprM efflux pump of Pseudomonas aeruginosa offers resistance to carvacrol: A herbal antimicrobial agent. Front. Microbiol. 2019, 10, 2664. [Google Scholar] [CrossRef]
- Poole, K.; Gotoh, N.; Tsujimoto, H.; Zhao, Q.; Wada, A.; Yamasaki, T.; Neshat, S.; Yamagishi, J.i.; Li, X.Z.; Nishino, T. Overexpression of the mexC–mexD–oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 1996, 21, 713–725. [Google Scholar] [CrossRef]
- Fraud, S.; Campigotto, A.J.; Chen, Z.; Poole, K. MexCD-OprJ multidrug efflux system of Pseudomonas aeruginosa: Involvement in chlorhexidine resistance and induction by membrane-damaging agents dependent upon the AlgU stress response sigma factor. Antimicrob. Agents Chemother. 2008, 52, 4478–4482. [Google Scholar] [CrossRef]
- Dong, N.; Zeng, Y.; Wang, Y.; Liu, C.; Lu, J.; Cai, C.; Liu, X.; Chen, Y.; Wu, Y.; Fang, Y. Distribution and spread of the mobilised RND efflux pump gene cluster tmexCD-toprJ in clinical Gram-negative bacteria: A molecular epidemiological study. Lancet Microbe 2022, 3, e846–e856. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Wan, M.; Wang, C.; Gao, X.; Yang, Q.; Partridge, S.R.; Wang, Y.; Zong, Z.; Doi, Y.; Shen, J. Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including tigecycline, in Klebsiella pneumoniae. MBio 2020, 11, e02930-19. [Google Scholar] [CrossRef] [PubMed]
- Köhler, T.; Michéa-Hamzehpour, M.; Henze, U.; Gotoh, N.; Kocjancic Curty, L.; Pechère, J.C. Characterization of MexE–MexF–OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 1997, 23, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Aendekerk, S.; Ghysels, B.; Cornelis, P.; Baysse, C. Characterization of a new efflux pump, MexGHI-OpmD, from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiology 2002, 148, 2371–2381. [Google Scholar] [CrossRef]
- Sakhtah, H.; Koyama, L.; Zhang, Y.; Morales, D.K.; Fields, B.L.; Price-Whelan, A.; Hogan, D.A.; Shepard, K.; Dietrich, L.E. The Pseudomonas aeruginosa efflux pump MexGHI-OpmD transports a natural phenazine that controls gene expression and biofilm development. Proc. Natl. Acad. Sci. USA 2016, 113, E3538–E3547. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, H.; Mima, T.; Morita, Y.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. Functional cloning and characterization of a multidrug efflux pump, mexHI-opmD, from a Pseudomonas aeruginosa mutant. Antimicrob. Agents Chemother. 2003, 47, 2990–2992. [Google Scholar] [CrossRef]
- Chuanchuen, R.; Narasaki, C.T.; Schweizer, H.P. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J. Bacteriol. 2002, 184, 5036–5044. [Google Scholar] [CrossRef]
- Mima, T.; Sekiya, H.; Mizushima, T.; Kuroda, T.; Tsuchiya, T. Gene cloning and properties of the RND-type multidrug efflux pumps MexPQ-OpmE and MexMN-OprM from Pseudomonas aeruginosa. Microbiol. Immunol. 2005, 49, 999–1002. [Google Scholar] [CrossRef]
- Ranjitkar, S.; Jones, A.K.; Mostafavi, M.; Zwirko, Z.; Iartchouk, O.; Barnes, S.W.; Walker, J.R.; Willis, T.W.; Lee, P.S.; Dean, C.R. Target (MexB)-and efflux-based mechanisms decreasing the effectiveness of the efflux pump inhibitor D13-9001 in Pseudomonas aeruginosa PAO1: Uncovering a new role for MexMN-OprM in efflux of β-lactams and a novel regulatory circuit (MmnRS) controlling MexMN expression. Antimicrob. Agents Chemother. 2019, 63, e01718-18. [Google Scholar]
- Li, Y.; Mima, T.; Komori, Y.; Morita, Y.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. A new member of the tripartite multidrug efflux pumps, MexVW–OprM, in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2003, 52, 572–575. [Google Scholar] [CrossRef]
- Mine, T.; Morita, Y.; Kataoka, A.; Mizushima, T.; Tsuchiya, T. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1999, 43, 415–417. [Google Scholar] [CrossRef]
- Singh, M.; Sykes, E.M.; Li, Y.; Kumar, A. MexXY RND pump of Pseudomonas aeruginosa PA7 effluxes bi-anionic β-lactams carbenicillin and sulbenicillin when it partners with the outer membrane factor OprA but not with OprM. Microbiology 2020, 166, 1095–1106. [Google Scholar] [CrossRef]
- Mima, T.; Kohira, N.; Li, Y.; Sekiya, H.; Ogawa, W.; Kuroda, T.; Tsuchiya, T. Gene cloning and characteristics of the RND-type multidrug efflux pump MuxABC-OpmB possessing two RND components in Pseudomonas aeruginosa. Microbiology 2009, 155, 3509–3517. [Google Scholar] [CrossRef] [PubMed]
- Mima, T.; Joshi, S.; Gomez-Escalada, M.; Schweizer, H.P. Identification and characterization of TriABC-OpmH, a triclosan efflux pump of Pseudomonas aeruginosa requiring two membrane fusion proteins. J. Bacteriol. 2007, 189, 7600–7609. [Google Scholar] [CrossRef] [PubMed]
- Winsor, G.L.; Griffiths, E.J.; Lo, R.; Dhillon, B.K.; Shay, J.A.; Brinkman, F.S. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016, 44, D646–D653. [Google Scholar] [CrossRef]
- Lorusso, A.B.; Carrara, J.A.; Barroso, C.D.N.; Tuon, F.F.; Faoro, H. Role of Efflux Pumps on Antimicrobial Resistance in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2022, 23, 15779. [Google Scholar] [CrossRef]
- McMurry, L.; Petrucci, R.E., Jr.; Levy, S.B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc. Natl. Acad. Sci. USA 1980, 77, 3974–3977. [Google Scholar] [CrossRef]
- Nikaido, H. Prevention of drug access to bacterial targets: Permeability barriers and active efflux. Science 1994, 264, 382–388. [Google Scholar] [CrossRef]
- Saier, M.H., Jr.; Paulsen, I.T.; Marek, K.s.; Pao, S.S.; Ronald, A.s.; Nikaido, H. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 1998, 12, 265–274. [Google Scholar]
- Stubenrauch, C.J.; Bamert, R.S.; Wang, J.; Lithgow, T. A noncanonical chaperone interacts with drug efflux pumps during their assembly into bacterial outer membranes. PLoS Biol. 2022, 20, e3001523. [Google Scholar] [CrossRef] [PubMed]
- Franklin, M.W.; Nepomnyachiy, S.; Feehan, R.; Ben-Tal, N.; Kolodny, R.; Slusky, J.S. Efflux pumps represent possible evolutionary convergence onto the β-barrel fold. Structure 2018, 26, 1266.e1262–1274.e1262. [Google Scholar] [CrossRef]
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial resistance in bacteria: Mechanisms, evolution, and persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef] [PubMed]
- AlMatar, M.; Albarri, O.; Makky, E.A.; Köksal, F. Efflux pump inhibitors: New updates. Pharmacol. Rep. 2021, 73, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L.J. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 2006, 19, 382–402. [Google Scholar] [CrossRef] [PubMed]
- Baugh, S.; Ekanayaka, A.S.; Piddock, L.J.; Webber, M.A. Loss of or inhibition of all multidrug resistance efflux pumps of Salmonella enterica serovar Typhimurium results in impaired ability to form a biofilm. J. Antimicrob. Chemother. 2012, 67, 2409–2417. [Google Scholar] [CrossRef]
- Miryala, S.K.; Anbarasu, A.; Ramaiah, S. Systems biology studies in Pseudomonas aeruginosa PA01 to understand their role in biofilm formation and multidrug efflux pumps. Microb. Pathog. 2019, 136, 103668. [Google Scholar] [CrossRef]
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020. [Google Scholar] [CrossRef]
- Pearson, J.P.; Van Delden, C.; Iglewski, B.H. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 1999, 181, 1203–1210. [Google Scholar] [CrossRef]
- Liang, Z.; Chen, Y.; Chen, Y.-F.; Cheng, Y.; Zhang, L. RND efflux pump and its interrelationship with quorum sensing system. Yi Chuan Hered. 2016, 38, 894–901. [Google Scholar]
- Poole, K. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 2007, 39, 162–176. [Google Scholar] [CrossRef]
- Sanz-García, F.; Hernando-Amado, S.; López-Causapé, C.; Oliver, A.; Martínez, J.L. Low Ciprofloxacin Concentrations Select Multidrug-Resistant Mutants Overproducing Efflux Pumps in Clinical Isolates of Pseudomonas aeruginosa. Microbiol. Spectr. 2022, 10, e00723-22. [Google Scholar] [CrossRef]
- Sanz-García, F.; Gil-Gil, T.; Laborda, P.; Ochoa-Sánchez, L.E.; Martínez, J.L.; Hernando-Amado, S. Coming from the wild: Multidrug resistant opportunistic pathogens presenting a primary, not human-linked, environmental habitat. Int. J. Mol. Sci. 2021, 22, 8080. [Google Scholar] [CrossRef]
- Nag, A.; Mehra, S. Involvement of the SCO3366 efflux pump from S. coelicolor in rifampicin resistance and its regulation by a TetR regulator. Appl. Microbiol. Biotechnol. 2022, 106, 2175–2190. [Google Scholar] [CrossRef]
- Yao, X.; Tao, F.; Zhang, K.; Tang, H.; Xu, P. Multiple roles for two efflux pumps in the polycyclic aromatic hydrocarbon-degrading Pseudomonas putida strain B6-2 (DSM 28064). Appl. Environ. Microbiol. 2017, 83, e01882-17. [Google Scholar] [CrossRef]
- Zwama, M.; Yamaguchi, A.; Nishino, K. Phylogenetic and functional characterisation of the Haemophilus influenzae multidrug efflux pump AcrB. Commun. Biol. 2019, 2, 340. [Google Scholar] [CrossRef] [PubMed]
- Compagne, N.; Vieira Da Cruz, A.; Müller, R.T.; Hartkoorn, R.C.; Flipo, M.; Pos, K.M. Update on the Discovery of Efflux Pump Inhibitors against Critical Priority Gram-Negative Bacteria. Antibiotics 2023, 12, 180. [Google Scholar] [CrossRef] [PubMed]
- Glavier, M.; Puvanendran, D.; Salvador, D.; Decossas, M.; Phan, G.; Garnier, C.; Frezza, E.; Cece, Q.; Schoehn, G.; Picard, M. Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex. Nat. Commun. 2020, 11, 4948. [Google Scholar] [CrossRef] [PubMed]
- Jeannot, K.; Elsen, S.; Koöhler, T.; Attree, I.; Van Delden, C.; Plésiat, P. Resistance and virulence of Pseudomonas aeruginosa clinical strains overproducing the MexCD-OprJ efflux pump. Antimicrob. Agents Chemother. 2008, 52, 2455–2462. [Google Scholar] [CrossRef]
- Llanes, C.; Köhler, T.; Patry, I.; Dehecq, B.; van Delden, C.; Plésiat, P. Role of the MexEF-OprN efflux system in low-level resistance of Pseudomonas aeruginosa to ciprofloxacin. Antimicrob. Agents Chemother. 2011, 55, 5676–5684. [Google Scholar] [CrossRef]
- Guénard, S.; Muller, C.; Monlezun, L.; Benas, P.; Broutin, I.; Jeannot, K.; Plésiat, P. Multiple mutations lead to MexXY-OprM-dependent aminoglycoside resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2014, 58, 221–228. [Google Scholar] [CrossRef]
- Venter, H.; Mowla, R.; Ohene-Agyei, T.; Ma, S. RND-type drug efflux pumps from Gram-negative bacteria: Molecular mechanism and inhibition. Front. Microbiol. 2015, 6, 377. [Google Scholar] [CrossRef]
- Jamshidi, S.; Sutton, J.M.; Rahman, K.M. Mapping the dynamic functions and structural features of acrb efflux pump transporter using accelerated molecular dynamics simulations. Sci. Rep. 2018, 8, 10470. [Google Scholar] [CrossRef]
- Wang, Z.; Fan, G.; Hryc, C.F.; Blaza, J.N.; Serysheva, I.I.; Schmid, M.F.; Chiu, W.; Luisi, B.F.; Du, D. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. Elife 2017, 6, e24905. [Google Scholar] [CrossRef]
- Nikaido, H.; Takatsuka, Y. Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2009, 1794, 769–781. [Google Scholar] [CrossRef]
- Murakami, S. Structures and transport mechanisms of RND efflux pumps. In Efflux-Mediated Antimicrobial Resistance in Bacteria: Mechanisms, Regulation and Clinical Implications; Springer Nature. Cham, Switzerland, 2016; pp. 3–28.
- McNeil, H.E.; Alav, I.; Torres, R.C.; Rossiter, A.E.; Laycock, E.; Legood, S.; Kaur, I.; Davies, M.; Wand, M.; Webber, M.A. Identification of binding residues between periplasmic adapter protein (PAP) and RND efflux pumps explains PAP-pump promiscuity and roles in antimicrobial resistance. PLoS Pathog. 2019, 15, e1008101. [Google Scholar] [CrossRef]
- Alenazy, R. Drug Efflux Pump Inhibitors: A Promising Approach to Counter Multidrug Resistance in Gram-Negative Pathogens by Targeting AcrB Protein from AcrAB-TolC Multidrug Efflux Pump from Escherichia coli. Biology 2022, 11, 1328. [Google Scholar] [CrossRef] [PubMed]
- Avakh, A.; Rezaei, K. Inhibition of the Mex Pumps of Pseudomonas aeruginosa with a Newly Characterized Member of Peptidomimetic Family. Br. Microbiol. Res. J. 2016, 16, 1–13. [Google Scholar] [CrossRef]
- López, C.A.; Travers, T.; Pos, K.M.; Zgurskaya, H.I.; Gnanakaran, S. Dynamics of intact MexAB-OprM efflux pump: Focusing on the MexA-OprM interface. Sci. Rep. 2017, 7, 16521. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Z.; Livermore, D.M.; Nikaido, H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: Resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob. Agents Chemother. 1994, 38, 1732–1741. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Z.; Ma, D.; Livermore, D.M.; Nikaido, H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: Active efflux as a contributing factor to beta-lactam resistance. Antimicrob. Agents Chemother. 1994, 38, 1742–1752. [Google Scholar] [CrossRef] [PubMed]
- Tsutsumi, K.; Yonehara, R.; Ishizaka-Ikeda, E.; Miyazaki, N.; Maeda, S.; Iwasaki, K.; Nakagawa, A.; Yamashita, E. Structures of the wild-type MexAB–OprM tripartite pump reveal its complex formation and drug efflux mechanism. Nat. Commun. 2019, 10, 1520. [Google Scholar] [CrossRef]
- Poole, K.; Tetro, K.; Zhao, Q.; Neshat, S.; Heinrichs, D.E.; Bianco, N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: MexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 1996, 40, 2021–2028. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Srikumar, R.; Poole, K. MexAB-OprM hyperexpression in NalC-type multidrug-resistant Pseudomonas aeruginosa: Identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol. Microbiol. 2004, 53, 1423–1436. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Cao, L.; Gould, V.C.; Avison, M.B.; Poole, K. nalD encodes a second repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 8649–8654. [Google Scholar] [CrossRef] [PubMed]
- Tafti, F.A.; Eslami, G.; Zandi, H.; Barzegar, K. Mutations in nalc gene of Mex AB-OprM efflux pump in carbapenem resistant Pseudomonas aeruginosa isolated from burn wounds in Yazd, Iran. Iran. J. Microbiol. 2020, 12, 32. [Google Scholar] [PubMed]
- Ziha-Zarifi, I.; Llanes, C.; Koöhler, T.; Pechere, J.-C.; Plesiat, P. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 1999, 43, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Horna, G.; López, M.; Guerra, H.; Saénz, Y.; Ruiz, J. Interplay between MexAB-OprM and MexEF-OprN in clinical isolates of Pseudomonas aeruginosa. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, D.; Ghosh, A.; Dhar Chanda, D.; Das Talukdar, A.; Dutta Choudhury, M.; Paul, D.; Maurya, A.P.; Chakravorty, A.; Bhattacharjee, A. Premature termination of MexR leads to overexpression of MexAB-OprM efflux pump in Pseudomonas aeruginosa in a tertiary referral hospital in India. PLoS ONE 2016, 11, e0149156. [Google Scholar]
- Boutoille, D.; Corvec, S.; Caroff, N.; Giraudeau, C.; Espaze, E.; Caillon, J.; Plésiat, P.; Reynaud, A. Detection of an IS21 insertion sequence in the mexR gene of Pseudomonas aeruginosa increasing β-lactam resistance. FEMS Microbiol. Lett. 2004, 230, 143–146. [Google Scholar] [CrossRef]
- Ma, Z.; Xu, C.; Zhang, X.; Wang, D.; Pan, X.; Liu, H.; Zhu, G.; Bai, F.; Cheng, Z.; Wu, W. A MexR mutation which confers aztreonam resistance to Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 659808. [Google Scholar] [CrossRef]
- Aguilar-Rodea, P.; Zúñiga, G.; Cerritos, R.; Rodríguez-Espino, B.A.; Gomez-Ramirez, U.; Nolasco-Romero, C.G.; López-Marceliano, B.; Rodea, G.E.; Mendoza-Elizalde, S.; Reyes-López, A. Nucleotide substitutions in the mexR, nalC and nalD regulator genes of the MexAB-OprM efflux pump are maintained in Pseudomonas aeruginosa g enetic lineages. PLoS ONE 2022, 17, e0266742. [Google Scholar] [CrossRef]
- Suresh, M.; Nithya, N.; Jayasree, P.; Vimal, K.; Manish Kumar, P. Mutational analyses of regulatory genes, mexR, nalC, nalD and mexZ of mexAB-oprM and mexXY operons, in efflux pump hyperexpressing multidrug-resistant clinical isolates of Pseudomonas aeruginosa. World J. Microbiol. Biotechnol. 2018, 34, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Beig, M.; Taheri, M.; Arabestani, M.R. Expression of MexAB-OprM efflux pump and OprD porin in carbapenemase producing Pseudomonas aeruginosa clinical isolates. Gene Rep. 2020, 20, 100744. [Google Scholar] [CrossRef]
- Grosjean, M.; Tazrout, S.; Bour, M.; Triponey, P.; Muller, C.; Jeannot, K.; Plésiat, P. Reassessment of the cooperativity between efflux system MexAB-OprM and cephalosporinase AmpC in the resistance of Pseudomonas aeruginosa to β-lactams. J. Antimicrob. Chemother. 2021, 76, 536–539. [Google Scholar] [CrossRef]
- Roy, P.H.; Tetu, S.G.; Larouche, A.; Elbourne, L.; Tremblay, S.; Ren, Q.; Dodson, R.; Harkins, D.; Shay, R.; Watkins, K. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS ONE 2010, 5, e8842. [Google Scholar] [CrossRef]
- Morita, Y.; Tomida, J.; Kawamura, Y. Primary mechanisms mediating aminoglycoside resistance in the multidrug-resistant Pseudomonas aeruginosa clinical isolate PA7. Microbiology 2012, 158, 1071–1083. [Google Scholar] [CrossRef]
- Housseini B Issa, K.; Phan, G.; Broutin, I. Functional mechanism of the efflux pumps transcription regulators from Pseudomonas aeruginosa based on 3D structures. Front. Mol. Biosci. 2018, 5, 57. [Google Scholar] [CrossRef] [PubMed]
- Muller, C.; Plésiat, P.; Jeannot, K. A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and β-lactams in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2011, 55, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Hay, T.; Fraud, S.; Lau, C.H.-F.; Gilmour, C.; Poole, K. Antibiotic inducibility of the mexXY multidrug efflux operon of Pseudomonas aeruginosa: Involvement of the MexZ anti-repressor ArmZ. PLoS ONE 2013, 8, e56858. [Google Scholar] [CrossRef]
- Kawalek, A.; Modrzejewska, M.; Zieniuk, B.; Bartosik, A.A.; Jagura-Burdzy, G. Interaction of ArmZ with the DNA-binding domain of MexZ induces expression of mexXY multidrug efflux pump genes and antimicrobial resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019, 63, e01199-19. [Google Scholar] [CrossRef]
- Seupt, A.; Schniederjans, M.; Tomasch, J.; Häussler, S. Expression of the MexXY aminoglycoside efflux pump and presence of an aminoglycoside-modifying enzyme in clinical Pseudomonas aeruginosa isolates are highly correlated. Antimicrob. Agents Chemother. 2020, 65, e01166-20. [Google Scholar] [CrossRef]
- Singh, M.; Yau, Y.C.; Wang, S.; Waters, V.; Kumar, A. MexXY efflux pump overexpression and aminoglycoside resistance in cystic fibrosis isolates of Pseudomonas aeruginosa from chronic infections. Can. J. Microbiol. 2017, 63, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Westbrock-Wadman, S.; Sherman, D.R.; Hickey, M.J.; Coulter, S.N.; Zhu, Y.Q.; Warrener, P.; Nguyen, L.Y.; Shawar, R.M.; Folger, K.R.; Stover, C.K. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 1999, 43, 2975–2983. [Google Scholar] [CrossRef]
- Vettoretti, L.; Plésiat, P.; Muller, C.; El Garch, F.; Phan, G.; Attrée, I.; Ducruix, A.; Llanes, C. Efflux unbalance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 2009, 53, 1987–1997. [Google Scholar] [CrossRef]
- Shigemura, K.; Osawa, K.; Kato, A.; Tokimatsu, I.; Arakawa, S.; Shirakawa, T.; Fujisawa, M. Association of overexpression of efflux pump genes with antibiotic resistance in Pseudomonas aeruginosa strains clinically isolated from urinary tract infection patients. J. Antibiot. 2015, 68, 568–572. [Google Scholar] [CrossRef] [PubMed]
- Gomis-Font, M.A.; Pitart, C.; del Barrio-Tofiño, E.; Zboromyrska, Y.; Cortes-Lara, S.; Mulet, X.; Marco, F.; Vila, J.; López-Causapé, C.; Oliver, A. Emergence of Resistance to Novel Cephalosporin–β-lactamase Inhibitor Combinations through the Modification of the Pseudomonas aeruginosa MexCD-OprJ Efflux Pump. Antimicrob. Agents Chemother. 2021, 65, e00089-21. [Google Scholar] [CrossRef]
- Morita, Y.; Komori, Y.; Mima, T.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. Construction of a series of mutants lacking all of the four major mex operons for multidrug efflux pumps or possessing each one of the operons from Pseudomonas aeruginosa PAO1: MexCD-OprJ is an inducible pump. FEMS Microbiol. Lett. 2001, 202, 139–143. [Google Scholar] [CrossRef]
- Srikumar, R.; Kon, T.; Gotoh, N.; Poole, K. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexC-MexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob. Agents Chemother. 1998, 42, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Masuda, N.; Gotoh, N.; Ohya, S.; Nishino, T. Quantitative correlation between susceptibility and OprJ production in NfxB mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1996, 40, 909–913. [Google Scholar] [CrossRef]
- Mao, W.; Warren, M.S.; Black, D.S.; Satou, T.; Murata, T.; Nishino, T.; Gotoh, N.; Lomovskaya, O. On the mechanism of substrate specificity by resistance nodulation division (RND)-type multidrug resistance pumps: The large periplasmic loops of MexD from Pseudomonas aeruginosa are involved in substrate recognition. Mol. Microbiol. 2002, 46, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Cazares, A.; Moore, M.P.; Hall, J.P.; Wright, L.L.; Grimes, M.; Emond-Rhéault, J.-G.; Pongchaikul, P.; Santanirand, P.; Levesque, R.C.; Fothergill, J.L. A megaplasmid family driving dissemination of multidrug resistance in Pseudomonas. Nat. Commun. 2020, 11, 1370. [Google Scholar] [CrossRef]
- Fetar, H.; Gilmour, C.; Klinoski, R.; Daigle, D.M.; Dean, C.R.; Poole, K. mexEF-oprN multidrug efflux operon of Pseudomonas aeruginosa: Regulation by the MexT activator in response to nitrosative stress and chloramphenicol. Antimicrob. Agents Chemother. 2011, 55, 508–514. [Google Scholar] [CrossRef]
- Koöhler, T.; Epp, S.F.; Curty, L.K.; Pechère, J.-C. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 1999, 181, 6300–6305. [Google Scholar] [CrossRef]
- Sobel, M.L.; Neshat, S.; Poole, K. Mutations in PA2491 (mexS) promote MexT-dependent mexEF-oprN expression and multidrug resistance in a clinical strain of Pseudomonas aeruginosa. J. Bacteriol. 2005, 187, 1246–1253. [Google Scholar] [CrossRef]
- Maseda, H.; Saito, K.; Nakajima, A.; Nakae, T. Variation of the mexT gene, a regulator of the MexEF-oprN efflux pump expression in wild-type strains of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2000, 192, 107–112. [Google Scholar] [CrossRef]
- Morita, Y.; Tomida, J.; Kawamura, Y. Efflux-mediated fluoroquinolone resistance in the multidrug-resistant Pseudomonas aeruginosa clinical isolate PA7: Identification of a novel MexS variant involved in upregulation of the mexEF-oprN multidrug efflux operon. Front. Microbiol. 2015, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Richardot, C.; Juarez, P.; Jeannot, K.; Patry, I.; Plésiat, P.; Llanes, C. Amino acid substitutions account for most MexS alterations in clinical nfxC mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 2302–2310. [Google Scholar] [CrossRef] [PubMed]
- Ochs, M.M.; McCusker, M.P.; Bains, M.; Hancock, R.E. Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob. Agents Chemother. 1999, 43, 1085–1090. [Google Scholar] [CrossRef] [PubMed]
- Mlynarcik, P.; Kolar, M. Molecular mechanisms of polymyxin resistance and detection of mcr genes. Biomed. Pap. Med. Fac. Palacky Univ. Olomouc 2019, 163, 28–38. [Google Scholar] [CrossRef]
- Hocquet, D.; Berthelot, P.; Roussel-Delvallez, M.; Favre, R.; Jeannot, K.; Bajolet, O.; Marty, N.; Grattard, F.; Mariani-Kurkdjian, P.; Bingen, E. Pseudomonas aeruginosa may accumulate drug resistance mechanisms without losing its ability to cause bloodstream infections. Antimicrob. Agents Chemother. 2007, 51, 3531–3536. [Google Scholar] [CrossRef]
- Solé, M.; Fabrega, A.; Cobos-Trigueros, N.; Zamorano, L.; Ferrer-Navarro, M.; Ballesté-Delpierre, C.; Reustle, A.; Castro, P.; Nicolás, J.M.; Oliver, A. In vivo evolution of resistance of Pseudomonas aeruginosa strains isolated from patients admitted to an intensive care unit: Mechanisms of resistance and antimicrobial exposure. J. Antimicrob. Chemother. 2015, 70, 3004–3013. [Google Scholar] [CrossRef]
- Goli, H.R.; Nahaei, M.R.; Rezaee, M.A.; Hasani, A.; Kafil, H.S.; Aghazadeh, M.; Sheikhalizadeh, V. Contribution of mexAB-oprM and mexXY (-oprA) efflux operons in antibiotic resistance of clinical Pseudomonas aeruginosa isolates in Tabriz, Iran. Infect. Genet. Evol. 2016, 45, 75–82. [Google Scholar] [CrossRef]
- Shuaib, S.L.; Gul, A.; Ahmed, J.; Rehman, N.; Ali, L.; Mumtaz, S.; Muhammad, A. Frequency of mexa gene in Pseudomonas aeruginosa isolated from clinical samples of a Tertiary Care Hospital in Pakistan. Prof. Med. J. 2020, 27, 2389–2393. [Google Scholar] [CrossRef]
- Talebi-Taher, M.; Gholami, A.; Rasouli-Kouhi, S.; Adabi, M. Role of efflux pump inhibitor in decreasing antibiotic cross-resistance of Pseudomonas aeruginosa in a burn hospital in Iran. J. Infect. Dev. Ctries. 2016, 10, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Xavier, D.E.; Picão, R.C.; Girardello, R.; Fehlberg, L.C.; Gales, A.C. Efflux pumps expression and its association with porin down-regulation and β-lactamase production among Pseudomonas aeruginosa causing bloodstream infections in Brazil. BMC Microbiol. 2010, 10, 217. [Google Scholar] [CrossRef]
- Ozer, B.; Duran, N.; Onlen, Y.; Savas, L. Efflux pump genes and antimicrobial resistance of Pseudomonas aeruginosa strains isolated from lower respiratory tract infections acquired in an intensive care unit. J. Antibiot. 2012, 65, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Terzi, H.A.; Kulah, C.; Ciftci, I.H. The effects of active efflux pumps on antibiotic resistance in Pseudomonas aeruginosa. World J. Microbiol. Biotechnol. 2014, 30, 2681–2687. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.-X.; Yi, X.-X.; Cho, A.; O’Gara, F.; Wang, Y.-P. CpxR activates MexAB-OprM efflux pump expression and enhances antibiotic resistance in both laboratory and clinical nalB-type isolates of Pseudomonas aeruginosa. PLoS Pathog. 2016, 12, e1005932. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, V.K.; Pathania, R. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian J. Med. Res. 2019, 149, 129. [Google Scholar]
- Gil-Gil, T.; Laborda, P.; Ochoa-Sánchez, L.E.; Martínez, J.L.; Hernando-Amado, S. Efflux in Gram-negative bacteria: What are the latest opportunities for drug discovery? Expert Opin. Drug Discov. 2023, 5, 671–686. [Google Scholar] [CrossRef]
- Gaurav, A.; Bakht, P.; Saini, M.; Pandey, S.; Pathania, R. Role of bacterial efflux pumps in antibiotic resistance, virulence, and strategies to discover novel efflux pump inhibitors. Microbiology 2023, 169, 001333. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, S.; Klinger-Strobel, M.; Bohnert, J.A.; Wendler, S.; Rödel, J.; Pletz, M.W.; Löffler, B.; Tuchscherr, L. Clinically approved drugs inhibit the Staphylococcus aureus multidrug NorA efflux pump and reduce biofilm formation. Front. Microbiol. 2019, 10, 2762. [Google Scholar] [CrossRef] [PubMed]
- Reza, A.; Sutton, J.M.; Rahman, K.M. Effectiveness of efflux pump inhibitors as biofilm disruptors and resistance breakers in gram-negative (ESKAPEE) bacteria. Antibiotics 2019, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wen, Y.M. The role of bacterial biofilm in persistent infections and control strategies. Int. J. Oral Sci. 2011, 3, 66–73. [Google Scholar] [CrossRef]
- Zhao, A.; Sun, J.; Liu, Y. Understanding bacterial biofilms: From definition to treatment strategies. Front. Cell Infect. Microbiol. 2023, 13, 1137947. [Google Scholar] [CrossRef]
- Sionov, R.V.; Steinberg, D. Targeting the Holy Triangle of Quorum Sensing, Biofilm Formation, and Antibiotic Resistance in Pathogenic Bacteria. Microorganisms 2022, 10, 1239. [Google Scholar] [CrossRef]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef]
- Subhadra, B.; Kim, D.H.; Woo, K.; Surendran, S.; Choi, C.H. Control of Biofilm Formation in Healthcare: Recent Advances Exploiting Quorum-Sensing Interference Strategies and Multidrug Efflux Pump Inhibitors. Materials 2018, 11, 1676. [Google Scholar] [CrossRef]
- Wang, W.Q.; Feng, X.C.; Shi, H.T.; Wang, Y.M.; Jiang, C.Y.; Xiao, Z.J.; Xu, Y.J.; Zhang, X.; Yuan, Y.; Ren, N.Q. Biofilm inhibition based on controlling the transmembrane transport and extracellular accumulation of quorum sensing signals. Env. Res 2023, 221, 115218. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Wang, G.; Xie, Y.; Tang, W.; Liu, B.; Zhang, J. Efflux pump inhibitor combined with ofloxacin decreases MRSA biofilm formation by regulating the gene expression of NorA and quorum sensing. RSC Adv. 2023, 13, 2707–2717. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, Y.; Huang, Y.; Jin, Q.; Ji, J. Inhibiting Quorum Sensing by Active Targeted pH-Sensitive Nanoparticles for Enhanced Antibiotic Therapy of Biofilm-Associated Bacterial Infections. ACS Nano 2023, 11, 10019–10032. [Google Scholar] [CrossRef] [PubMed]
- Bernal, P.; Molina-Santiago, C.; Daddaoua, A.; Llamas, M.A. Antibiotic adjuvants: Identification and clinical use. Microb. Biotechnol. 2013, 6, 445. [Google Scholar] [CrossRef] [PubMed]
- Bolla, J.-M.; Alibert-Franco, S.; Handzlik, J.; Chevalier, J.; Mahamoud, A.; Boyer, G.; Kieć-Kononowicz, K.; Pagès, J.-M. Strategies for bypassing the membrane barrier in multidrug resistant Gram-negative bacteria. FEBS Lett. 2011, 585, 1682–1690. [Google Scholar] [CrossRef]
- Lomovskaya, O.; Warren, M.S.; Lee, A.; Galazzo, J.; Fronko, R.; Lee, M.; Blais, J.; Cho, D.; Chamberland, S.; Renau, T. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrob. Agents Chemother. 2001, 45, 105–116. [Google Scholar] [CrossRef]
- Pagès, J.-M.; Amaral, L. Mechanisms of drug efflux and strategies to combat them: Challenging the efflux pump of Gram-negative bacteria. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2009, 1794, 826–833. [Google Scholar] [CrossRef]
- Seukep, A.J.; Fokoua-Maxime, C.D.; Mbuntcha, H.G.; Chen, G.; Assob, J.C.N.; Tenniswood, M.; Sarker, S.D.; Kuete, V.; Ming-Quan, G. Bacterial drug efflux pump inhibitors from plants. In Antimicrobial Resistance: Underlying Mechanisms and Therapeutic Approaches; Springer: Berlin/Heidelberg, Germany, 2022; pp. 487–532. [Google Scholar]
- Lomovskaya, O.; Bostian, K.A. Practical applications and feasibility of efflux pump inhibitors in the clinic—A vision for applied use. Biochem. Pharmacol. 2006, 71, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Lomovskaya, O.; Zgurskaya, H.I.; Totrov, M.; Watkins, W.J. Waltzing transporters and ‘the dance macabre’ between humans and bacteria. Nat. Rev. Drug Discov. 2007, 6, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Poole, K.; Heinrichs, D.E.; Neshat, S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: Regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol. Microbiol. 1993, 10, 529–544. [Google Scholar] [CrossRef]
- Opperman, T.J.; Nguyen, S.T. Recent advances toward a molecular mechanism of efflux pump inhibition. Front. Microbiol. 2015, 6, 421. [Google Scholar] [CrossRef]
- Schweizer, H.P. Understanding efflux in Gram-negative bacteria: Opportunities for drug discovery. Expert Opin. Drug Discov. 2012, 7, 633–642. [Google Scholar] [CrossRef]
- Ramaswamy, V.K.; Cacciotto, P.; Malloci, G.; Ruggerone, P.; Vargiu, A.V. Multidrug efflux pumps and their inhibitors characterized by computational modeling. Efflux-Mediat. Antimicrob. Resist. Bact. Mech. Regul. Clin. Implic. 2016, 11, 797–831. [Google Scholar]
- K Bhardwaj, A.; Mohanty, P. Bacterial efflux pumps involved in multidrug resistance and their inhibitors: Rejuvinating the antimicrobial chemotherapy. Recent Pat. Anti-Infect. Drug Discov. 2012, 7, 73–89. [Google Scholar] [CrossRef]
- Cheesman, M.J.; Ilanko, A.; Blonk, B.; Cock, I.E. Developing new antimicrobial therapies: Are synergistic combinations of plant extracts/compounds with conventional antibiotics the solution? Pharmacogn. Rev. 2017, 11, 57. [Google Scholar]
- Stermitz, F.R.; Lorenz, P.; Tawara, J.N.; Zenewicz, L.A.; Lewis, K. Synergy in a medicinal plant: Antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc. Natl. Acad. Sci. USA 2000, 97, 1433–1437. [Google Scholar] [CrossRef]
- Stavri, M.; Piddock, L.J.; Gibbons, S. Bacterial efflux pump inhibitors from natural sources. J. Antimicrob. Chemother. 2007, 59, 1247–1260. [Google Scholar] [CrossRef]
- Gibbons, S.; Oluwatuyi, M.; Kaatz, G.W. A novel inhibitor of multidrug efflux pumps in Staphylococcus aureus. J. Antimicrob. Chemother. 2003, 51, 13–17. [Google Scholar] [CrossRef]
- Pfeifer, H.; Greenblatt, D.; Koch-Wester, J. Clinical toxicity of reserpine in hospitalized patients: A report from the Boston Collaborative Drug Surveillance Program. Am. J. Med. Sci. 1976, 271, 269–276. [Google Scholar] [CrossRef]
- Kumar, A.; Khan, I.A.; Koul, S.; Koul, J.L.; Taneja, S.C.; Ali, I.; Ali, F.; Sharma, S.; Mirza, Z.M.; Kumar, M. Novel structural analogues of piperine as inhibitors of the NorA efflux pump of Staphylococcus aureus. J. Antimicrob. Chemother. 2008, 61, 1270–1276. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kumar, M.; Sharma, S.; Nargotra, A.; Koul, S.; Khan, I.A. Piperine as an inhibitor of Rv1258c, a putative multidrug efflux pump of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2010, 65, 1694–1701. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.C.; Ip, M.; Lau, C.B.; Lui, S.; Jolivalt, C.; Ganem-Elbaz, C.; Litaudon, M.; Reiner, N.E.; Gong, H.; See, R.H. Synergistic effects of baicalein with ciprofloxacin against NorA over-expressed methicillin-resistant Staphylococcus aureus (MRSA) and inhibition of MRSA pyruvate kinase. J. Ethnopharmacol. 2011, 137, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Shiota, S.; Kuroda, T.; Hatano, T.; Yoshida, T.; Mizushima, T.; Tsuchiya, T. Remarkable synergies between baicalein and tetracycline, and baicalein and β-lactams against methicillin-resistant Staphylococcus aureus. Microbiol. Immunol. 2005, 49, 391–396. [Google Scholar] [CrossRef]
- Morel, C.; Stermitz, F.R.; Tegos, G.; Lewis, K. Isoflavones as potentiators of antibacterial activity. J. Agric. Food Chem. 2003, 51, 5677–5679. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, S.; Moser, E.; Kaatz, G.W. Catechin gallates inhibit multidrug resistance (MDR) in Staphylococcus aureus. Planta Med. 2004, 70, 1240–1242. [Google Scholar] [CrossRef]
- Sudano Roccaro, A.; Blanco, A.R.; Giuliano, F.; Rusciano, D.; Enea, V. Epigallocatechin-gallate enhances the activity of tetracycline in staphylococci by inhibiting its efflux from bacterial cells. Antimicrob. Agents Chemother. 2004, 48, 1968–1973. [Google Scholar] [CrossRef] [PubMed]
- Oluwatuyi, M.; Kaatz, G.W.; Gibbons, S. Antibacterial and resistance modifying activity of Rosmarinus officinalis. Phytochemistry 2004, 65, 3249–3254. [Google Scholar] [CrossRef] [PubMed]
- Lorenzi, V.; Muselli, A.; Bernardini, A.F.; Berti, L.; Pagès, J.-M.; Amaral, L.; Bolla, J.-M. Geraniol restores antibiotic activities against multidrug-resistant isolates from gram-negative species. Antimicrob. Agents Chemother. 2009, 53, 2209–2211. [Google Scholar] [CrossRef]
- Seukep, A.J.; Kuete, V.; Nahar, L.; Sarker, S.D.; Guo, M. Plant-derived secondary metabolites as the main source of efflux pump inhibitors and methods for identification. J. Pharm. Anal. 2020, 10, 277–290. [Google Scholar] [CrossRef]
- Rindi, L. Efflux Pump Inhibitors Against Nontuberculous Mycobacteria. Int. J. Mol. Sci. 2020, 21, 4191. [Google Scholar] [CrossRef]
- Lamut, A.; Peterlin Mašič, L.; Kikelj, D.; Tomašič, T. Efflux pump inhibitors of clinically relevant multidrug resistant bacteria. Med. Res. Rev. 2019, 39, 2460–2504. [Google Scholar] [CrossRef]
- Nayim, P.; Mbaveng, A.T.; Wamba, B.E.N.; Fankam, A.G.; Dzotam, J.K.; Kuete, V. Antibacterial and Antibiotic-Potentiating Activities of Thirteen Cameroonian Edible Plants against Gram-Negative Resistant Phenotypes. Sci. World J. 2018, 2018, 4020294. [Google Scholar] [CrossRef]
- Biswas, S.S.; Browne, R.B.; Borah, V.V.; Roy, J.D. In Silico Approach for Phytocompound-Based Drug Designing to Fight Efflux Pump-Mediated Multidrug-Resistant Mycobacterium tuberculosis. Appl. Biochem. Biotechnol. 2021, 193, 1757–1779. [Google Scholar] [CrossRef]
- Tegos, G.P.; Haynes, M.; Strouse, J.J.; Khan, M.M.; Bologa, C.G.; Oprea, T.I.; Sklar, L.A. Microbial efflux pump inhibition: Tactics and strategies. Curr. Pharm. Des. 2011, 17, 1291–1302. [Google Scholar] [CrossRef]
- Prasch, S.; Bucar, F. Plant derived inhibitors of bacterial efflux pumps: An update. Phytochem. Rev. 2015, 14, 961–974. [Google Scholar] [CrossRef]
- Tegos, G.; Stermitz, F.R.; Lomovskaya, O.; Lewis, K. Multidrug pump inhibitors uncover remarkable activity of plant antimicrobials. Antimicrob. Agents Chemother. 2002, 46, 3133–3141. [Google Scholar] [CrossRef]
- González-Lamothe, R.; Mitchell, G.; Gattuso, M.; Diarra, M.S.; Malouin, F.; Bouarab, K. Plant antimicrobial agents and their effects on plant and human pathogens. Int. J. Mol. Sci. 2009, 10, 3400–3419. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, Y.; Nishimura, T.; Harada, M.; Kashiwagi, R.; Yamamoto, M.; Noutoshi, Y.; Toyoda, K.; Taguchi, F.; Takemoto, D.; Matsui, H. Role of Two Sets of RND-Type Multidrug Efflux Pump Transporter Genes, mexAB-oprM and mexEF-oprN, in Virulence of Pseudomonas syringae pv. tabaci 6605. Plant Pathol. J. 2020, 36, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Simões, M.; Bennett, R.N.; Rosa, E.A. Understanding antimicrobial activities of phytochemicals against multidrug resistant bacteria and biofilms. Nat. Prod. Rep. 2009, 26, 746–757. [Google Scholar] [CrossRef]
- Rana, T.; Singh, S.; Kaur, N.; Pathania, K.; Farooq, U. A review on efflux pump inhibitors of medically important bacteria from plant sources. Int. J. Pharm. Sci. Rev. Res. 2014, 26, 101–111. [Google Scholar]
- Siriyong, T.; Srimanote, P.; Chusri, S.; Yingyongnarongkul, B.-e.; Suaisom, C.; Tipmanee, V.; Voravuthikunchai, S.P. Conessine as a novel inhibitor of multidrug efflux pump systems in Pseudomonas aeruginosa. BMC Complement. Altern. Med. 2017, 17, 1–7. [Google Scholar] [CrossRef]
- Dwivedi, G.R.; Tyagi, R.; Sanchita, T.S.; Pati, S.; Srivastava, S.K.; Darokar, M.P.; Sharma, A. Antibiotics potentiating potential of catharanthine against superbug Pseudomonas aeruginosa. J. Biomol. Struct. Dyn. 2018, 36, 4270–4284. [Google Scholar] [CrossRef] [PubMed]
- Aghayan, S.S.; Mogadam, H.K.; Fazli, M.; Darban-Sarokhalil, D.; Khoramrooz, S.S.; Jabalameli, F.; Yaslianifard, S.; Mirzaii, M. The effects of berberine and palmatine on efflux pumps inhibition with different gene patterns in Pseudomonas aeruginosa isolated from burn infections. Avicenna J. Med. Biotechnol. 2017, 9, 2. [Google Scholar]
- Su, F.; Wang, J. Berberine inhibits the MexXY-OprM efflux pump to reverse imipenem resistance in a clinical carbapenem-resistant Pseudomonas aeruginosa isolate in a planktonic state. Exp. Ther. Med. 2018, 15, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L.J.; Garvey, M.I.; Rahman, M.M.; Gibbons, S. Natural and synthetic compounds such as trimethoprim behave as inhibitors of efflux in Gram-negative bacteria. J. Antimicrob. Chemother. 2010, 65, 1215–1223. [Google Scholar] [CrossRef]
- Choudhury, D.; Das Talukdar, A.; Chetia, P.; Bhattacharjee, A.; Dutta Choudhury, M. Screening of natural products and derivatives for the identification of RND efflux pump inhibitors. Comb. Chem. High Throughput Screen. 2016, 19, 705–713. [Google Scholar] [CrossRef]
- Negi, N.; Prakash, P.; Gupta, M.L.; Mohapatra, T.M. Possible role of curcumin as an efflux pump inhibitor in multi drug resistant clinical isolates of Pseudomonas aeruginosa. J. Clin. Diagn. Res. JCDR 2014, 8, DC04. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Jang, J.-H.; Li, M.-H.; Surh, Y.-J. Resveratrol upregulates heme oxygenase-1 expression via activation of NF-E2-related factor 2 in PC12 cells. Biochem. Biophys. Res. Commun. 2005, 331, 993–1000. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Y.; Lin, Y.; Cao, J.; Xu, C.; Chen, L.; Wang, Y.; Sun, Y.; Zheng, X.; Liu, Y. Resveratrol Increases Sensitivity of Clinical Colistin-Resistant Pseudomonas aeruginosa to Colistin In Vitro and In Vivo. Microbiol. Spectr. 2023, 11, e01992-22. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Koehn, F.E.; Carter, G.T. The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 2005, 4, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.B.; Tiwari, V.K. Natural products: An evolving role in future drug discovery. Eur. J. Med. Chem. 2011, 46, 4769–4807. [Google Scholar] [CrossRef]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef]
- Vidaillac, C. Pharmacomodulations en Série Pyrrolo [1, 2-a] Quinoxaline: Application à la mise au Point D′inhibiteurs de Pompes D’efflux. Ph.D. Thesis, Université de Bordeaux, Bordeaux, France, 2007. [Google Scholar]
- Renau, T.E.; Léger, R.; Flamme, E.M.; Sangalang, J.; She, M.W.; Yen, R.; Gannon, C.L.; Griffith, D.; Chamberland, S.; Lomovskaya, O. Inhibitors of efflux pumps in pseudomonas a eruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J. Med. Chem. 1999, 42, 4928–4931. [Google Scholar] [CrossRef]
- Renau, T.E.; Léger, R.; Flamme, E.M.; She, M.W.; Gannon, C.L.; Mathias, K.M.; Lomovskaya, O.; Chamberland, S.; Lee, V.J.; Ohta, T. Addressing the stability of C-capped dipeptide efflux pump inhibitors that potentiate the activity of levofloxacin in Pseudomonas aeruginosa. Bioorg. Med. Chem. Lett. 2001, 11, 663–667. [Google Scholar] [CrossRef]
- Renau, T.E.; Léger, R.; Filonova, L.; Flamme, E.M.; Wang, M.; Yen, R.; Madsen, D.; Griffith, D.; Chamberland, S.; Dudley, M.N. Conformationally-restricted analogues of efflux pump inhibitors that potentiate the activity of levofloxacin in Pseudomonas aeruginosa. Bioorg. Med. Chem. Lett. 2003, 13, 2755–2758. [Google Scholar] [CrossRef] [PubMed]
- Renau, T.E.; Léger, R.; Yen, R.; She, M.W.; Flamme, E.M.; Sangalang, J.; Gannon, C.L.; Chamberland, S.; Lomovskaya, O.; Lee, V.J. Peptidomimetics of efflux pump inhibitors potentiate the activity of levofloxacin in Pseudomonas aeruginosa. Bioorg. Med. Chem. Lett. 2002, 12, 763–766. [Google Scholar] [CrossRef] [PubMed]
- Watkins, W.J.; Landaverry, Y.; Léger, R.; Litman, R.; Renau, T.E.; Williams, N.; Yen, R.; Zhang, J.Z.; Chamberland, S.; Madsen, D. The relationship between physicochemical properties, in vitro activity and pharmacokinetic profiles of analogues of diamine-containing efflux pump inhibitors. Bioorg. Med. Chem. Lett. 2003, 13, 4241–4244. [Google Scholar] [CrossRef] [PubMed]
- Bohnert, J.r.A.; Kern, W.V. Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob. Agents Chemother. 2005, 49, 849–852. [Google Scholar] [CrossRef]
- Ferrer-Espada, R.; Shahrour, H.; Pitts, B.; Stewart, P.S.; Sánchez-Gómez, S.; Martínez-de-Tejada, G. A permeability-increasing drug synergizes with bacterial efflux pump inhibitors and restores susceptibility to antibiotics in multi-drug resistant Pseudomonas aeruginosa strains. Sci. Rep. 2019, 9, 3452. [Google Scholar] [CrossRef]
- Y Mahmood, H.; Jamshidi, S.; Mark Sutton, J.; M Rahman, K. Current advances in developing inhibitors of bacterial multidrug efflux pumps. Curr. Med. Chem. 2016, 23, 1062–1081. [Google Scholar] [CrossRef]
- Moir, D.T.; Opperman, T.J.; Aron, Z.D.; Bowlin, T.L. Adjunctive therapy for multidrug-resistant bacterial infections: Type III secretion system and efflux inhibitors. Drug Discov. Today 2021, 26, 2173–2181. [Google Scholar] [CrossRef]
- Yoshida, K.-i.; Nakayama, K.; Ohtsuka, M.; Kuru, N.; Yokomizo, Y.; Sakamoto, A.; Takemura, M.; Hoshino, K.; Kanda, H.; Nitanai, H. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 7: Highly soluble and in vivo active quaternary ammonium analogue D13-9001, a potential preclinical candidate. Bioorg. Med. Chem. 2007, 15, 7087–7097. [Google Scholar] [CrossRef] [PubMed]
- Opperman, T.J.; Kwasny, S.M.; Kim, H.-S.; Nguyen, S.T.; Houseweart, C.; D’Souza, S.; Walker, G.C.; Peet, N.P.; Nikaido, H.; Bowlin, T.L. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob. Agents Chemother. 2014, 58, 722–733. [Google Scholar] [CrossRef]
- Vargiu, A.V.; Ruggerone, P.; Opperman, T.J.; Nguyen, S.T.; Nikaido, H. Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob. Agents Chemother. 2014, 58, 6224–6234. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.T.; Kwasny, S.M.; Ding, X.; Cardinale, S.C.; McCarthy, C.T.; Kim, H.-S.; Nikaido, H.; Peet, N.P.; Williams, J.D.; Bowlin, T.L. Structure–activity relationships of a novel pyranopyridine series of Gram-negative bacterial efflux pump inhibitors. Bioorg. Med. Chem. 2015, 23, 2024–2034. [Google Scholar] [CrossRef]
- Vaara, M.; Vaara, T. Sensitization of Gram-negative bacteria to antibiotics and complement by a nontoxic oligopeptide. Nature 1983, 303, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Aron, Z.; Opperman, T.J. Optimization of a novel series of pyranopyridine RND efflux pump inhibitors. Curr. Opin. Microbiol. 2016, 33, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Rosado-Lugo, J.D.; Datta, P.; Sun, Y.; Cao, Y.; Banerjee, A.; Yuan, Y.; Parhi, A.K. Evaluation of a Conformationally Constrained Indole Carboxamide as a Potential Efflux Pump Inhibitor in Pseudomonas aeruginosa. Antibiotics 2022, 11, 716. [Google Scholar] [CrossRef]
- Yuan, Y.; Rosado-Lugo, J.D.; Zhang, Y.; Datta, P.; Sun, Y.; Cao, Y.; Banerjee, A.; Parhi, A.K. Evaluation of heterocyclic carboxamides as potential efflux pump inhibitors in Pseudomonas aeruginosa. Antibiotics 2022, 11, 30. [Google Scholar] [CrossRef]
- Cohen, S.P.; McMurry, L.; Hooper, D.; Wolfson, J.; Levy, S. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: Decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob. Agents Chemother. 1989, 33, 1318–1325. [Google Scholar] [CrossRef]
- Ma, D.; Cook, D.; Alberti, M.; Pon, N.; Nikaido, H.; Hearst, J. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 1993, 175, 6299–6313. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.D.; Galazzo, J.L.; Staley, A.L.; Lee, J.C.; Warren, M.S.; Fuernkranz, H.; Chamberland, S.; Lomovskaya, O.; Miller, G.H. Microbial fermentation-derived inhibitors of efflux-pump-mediated drug resistance. IL Farm. 2001, 56, 81–85. [Google Scholar] [CrossRef]
- Wang, C.H.; Hsieh, Y.H.; Powers, Z.M.; Kao, C.Y. Defeating Antibiotic-Resistant Bacteria: Exploring Alternative Therapies for a Post-Antibiotic Era. Int. J. Mol. Sci. 2020, 21, 1061. [Google Scholar] [CrossRef]
- Tambat, R.; Mahey, N.; Chandal, N.; Verma, D.K.; Jangra, M.; Thakur, K.G.; Nandanwar, H. A Microbe-Derived Efflux Pump Inhibitor of the Resistance-Nodulation-Cell Division Protein Restores Antibiotic Susceptibility in Escherichia coli and Pseudomonas aeruginosa. ACS Infect. Dis. 2022, 8, 255–270. [Google Scholar] [CrossRef]
- Molnár, J.; Gyémánt, N.; Tanaka, M.; Hohmann, J.; Bergmann-Leitner, E.; Molnár, P.; Deli, J.; Didiziapetris, R.; Ferreira, M.J. Inhibition of multidrug resistance of cancer cells by natural diterpenes, triterpenes and carotenoids. Curr. Pharm. Des. 2006, 12, 287–311. [Google Scholar] [CrossRef] [PubMed]
- Ruggerone, P.; Murakami, S.; M Pos, K.; Vargiu, A.V. RND efflux pumps: Structural information translated into function and inhibition mechanisms. Curr. Top. Med. Chem. 2013, 13, 3079–3100. [Google Scholar] [CrossRef] [PubMed]
- Thakur, V.; Uniyal, A.; Tiwari, V. A comprehensive review on pharmacology of efflux pumps and their inhibitors in antibiotic resistance. Eur. J. Pharmacol. 2021, 903, 174151. [Google Scholar] [CrossRef]
- Plé, C.; Tam, H.-K.; Vieira Da Cruz, A.; Compagne, N.; Jiménez-Castellanos, J.-C.; Müller, R.T.; Pradel, E.; Foong, W.E.; Malloci, G.; Ballée, A. Pyridylpiperazine-based allosteric inhibitors of RND-type multidrug efflux pumps. Nat. Commun. 2022, 13, 115. [Google Scholar] [CrossRef]
- Kabra, R.; Chauhan, N.; Kumar, A.; Ingale, P.; Singh, S. Efflux pumps and antimicrobial resistance: Paradoxical components in systems genomics. Prog. Biophys. Mol. Biol. 2019, 141, 15–24. [Google Scholar] [CrossRef]
- Zárate, S.G.; Morales, P.; Świderek, K.; Bolanos-Garcia, V.M.; Bastida, A. A molecular modeling approach to identify novel inhibitors of the major facilitator superfamily of efflux pump transporters. Antibiotics 2019, 8, 25. [Google Scholar] [CrossRef]
- Nishino, K.; Yamasaki, S.; Nakashima, R.; Zwama, M.; Hayashi-Nishino, M. Function and inhibitory mechanisms of multidrug efflux pumps. Front. Microbiol. 2021, 12, 737288. [Google Scholar] [CrossRef]
- Si, Z.; Pethe, K.; Chan-Park, M.B. Chemical basis of combination therapy to combat antibiotic resistance. JACS Au 2023, 3, 276–292. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Lupoli, T.J. Counteracting antibiotic resistance enzymes and efflux pumps. Curr. Opin. Microbiol. 2023, 75, 102334. [Google Scholar] [CrossRef]
- Wang, H.; Meng, J.; Jia, M.; Ma, X.; He, G.; Yu, J.; Wang, R.; Bai, H.; Hou, Z.; Luo, X. oprM as a new target for reversion of multidrug resistance in Pseudomonas aeruginosa by antisense phosphorothioate oligodeoxynucleotides. FEMS Immunol. Med. Microbiol. 2010, 60, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Muzzi, A.; Masignani, V.; Rappuoli, R. The pan-genome: Towards a knowledge-based discovery of novel targets for vaccines and antibacterials. Drug Discov. Today 2007, 12, 429–439. [Google Scholar] [CrossRef]
- Geller, B.L. Antibacterial antisense. Curr. Opin. Mol. Ther. 2005, 7, 109–113. [Google Scholar]
- Danis-Wlodarczyk, K.; Vandenheuvel, D.; Jang, H.B.; Briers, Y.; Olszak, T.; Arabski, M.; Wasik, S.; Drabik, M.; Higgins, G.; Tyrrell, J. A proposed integrated approach for the preclinical evaluation of phage therapy in Pseudomonas infections. Sci. Rep. 2016, 6, 1–13. [Google Scholar]
- Chegini, Z.; Khoshbayan, A.; Taati Moghadam, M.; Farahani, I.; Jazireian, P.; Shariati, A. Bacteriophage therapy against Pseudomonas aeruginosa biofilms: A review. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 1–17. [Google Scholar] [CrossRef]
- Kakasis, A.; Panitsa, G. Bacteriophage therapy as an alternative treatment for human infections. A comprehensive review. Int. J. Antimicrob. Agents 2019, 53, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Forti, F.; Roach, D.R.; Cafora, M.; Pasini, M.E.; Horner, D.S.; Fiscarelli, E.V.; Rossitto, M.; Cariani, L.; Briani, F.; Debarbieux, L. Design of a broad-range bacteriophage cocktail that reduces Pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrob. Agents Chemother. 2018, 62, e02573-17. [Google Scholar] [CrossRef]
- Kellenberger, G.; Kellenberger, E. Electron microscopical studies of phage multiplication: III. Observation of single cell bursts. Virology 1957, 3, 275–285. [Google Scholar] [CrossRef]
- Oechslin, F.; Piccardi, P.; Mancini, S.; Gabard, J.; Moreillon, P.; Entenza, J.M.; Resch, G.; Que, Y.-A. Synergistic interaction between phage therapy and antibiotics clears Pseudomonas aeruginosa infection in endocarditis and reduces virulence. J. Infect. Dis. 2017, 215, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Turner, P.E.; Kim, S.; Mojibian, H.R.; Elefteriades, J.A.; Narayan, D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 2018, 60–66. [Google Scholar] [CrossRef]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef]
- Sennhauser, G.; Bukowska, M.A.; Briand, C.; Grütter, M.G. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J. Mol. Biol. 2009, 389, 134–145. [Google Scholar] [CrossRef]
- Guan, L.; Nakae, T. Identification of essential charged residues in transmembrane segments of the multidrug transporter MexB of Pseudomonas aeruginosa. J. Bacteriol. 2001, 183, 1734–1739. [Google Scholar] [CrossRef]
- Yu, E.W.; McDermott, G.; Zgurskaya, H.I.; Nikaido, H.; Koshland, D.E., Jr. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 2003, 300, 976–980. [Google Scholar] [CrossRef]
- Kobylka, J.; Kuth, M.S.; Müller, R.T.; Geertsma, E.R.; Pos, K.M. AcrB: A mean, keen, drug efflux machine. Ann. N. Y. Acad. Sci. 2020, 1459, 38–68. [Google Scholar] [CrossRef] [PubMed]
- Tikhonova, E.B.; Wang, Q.; Zgurskaya, H.I. Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J. Bacteriol. 2002, 184, 6499–6507. [Google Scholar] [CrossRef]
- Krishnamoorthy, G.; Tikhonova, E.B.; Zgurskaya, H.I. Fitting periplasmic membrane fusion proteins to inner membrane transporters: Mutations that enable Escherichia coli AcrA to function with Pseudomonas aeruginosa MexB. J. Bacteriol. 2008, 190, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Aires, J.R.; Kohler, T.; Nikaido, H.; Plésiat, P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 1999, 43, 2624–2628. [Google Scholar] [CrossRef]
- Paulsen, I.T.; Nguyen, L.; Sliwinski, M.K.; Rabus, R.; Saier, M.H., Jr. Microbial genome analyses: Comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 2000, 301, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Gervasoni, S.; Malloci, G.; Bosin, A.; Vargiu, A.V.; Zgurskaya, H.I.; Ruggerone, P. Recognition of quinolone antibiotics by the multidrug efflux transporter MexB of Pseudomonas aeruginosa. Phys. Chem. Chem. Phys. 2022, 24, 16566–16575. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, R.; Sakurai, K.; Yamasaki, S.; Nishino, K.; Yamaguchi, A. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 2011, 480, 565–569. [Google Scholar] [CrossRef]
- Alav, I.; Kobylka, J.; Kuth, M.S.; Pos, K.M.; Picard, M.; Blair, J.M.; Bavro, V.N. Structure, assembly, and function of tripartite efflux and type 1 secretion systems in gram-negative bacteria. Chem. Rev. 2021, 121, 5479–5596. [Google Scholar] [CrossRef]
- Nakashima, R.; Sakurai, K.; Yamasaki, S.; Hayashi, K.; Nagata, C.; Hoshino, K.; Onodera, Y.; Nishino, K.; Yamaguchi, A. Structural basis for the inhibition of bacterial multidrug exporters. Nature 2013, 500, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Müller, R.T.; Travers, T.; Cha, H.-j.; Phillips, J.L.; Gnanakaran, S.; Pos, K.M. Switch loop flexibility affects substrate transport of the AcrB efflux pump. J. Mol. Biol. 2017, 429, 3863–3874. [Google Scholar] [CrossRef] [PubMed]
- Sjuts, H.; Vargiu, A.V.; Kwasny, S.M.; Nguyen, S.T.; Kim, H.-S.; Ding, X.; Ornik, A.R.; Ruggerone, P.; Bowlin, T.L.; Nikaido, H. Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc. Natl. Acad. Sci. USA 2016, 113, 3509–3514. [Google Scholar] [CrossRef] [PubMed]
- Oswald, C.; Tam, H.-K.; Pos, K.M. Transport of lipophilic carboxylates is mediated by transmembrane helix 2 in multidrug transporter AcrB. Nat. Commun. 2016, 7, 1–10. [Google Scholar] [CrossRef]
- Sakurai, K.; Yamasaki, S.; Nakao, K.; Nishino, K.; Yamaguchi, A.; Nakashima, R. Crystal structures of multidrug efflux pump MexB bound with high-molecular-mass compounds. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Tang, X.; Wang, Z.; Lei, T.; Zhou, W.; Chang, S.; Li, D. Importance of protein flexibility on molecular recognition: Modeling binding mechanisms of aminopyrazine inhibitors to Nek2. Phys. Chem. Chem. Phys. 2018, 20, 5591–5605. [Google Scholar] [CrossRef]
- Ezelarab, H.A.; Abbas, S.H.; Hassan, H.A.; Abuo-Rahma, G.E.D.A. Recent updates of fluoroquinolones as antibacterial agents. Arch. Pharm. 2018, 351, 1800141. [Google Scholar] [CrossRef]
- Ornik-Cha, A.; Wilhelm, J.; Kobylka, J.; Sjuts, H.; Vargiu, A.V.; Malloci, G.; Reitz, J.; Seybert, A.; Frangakis, A.S.; Pos, K.M. Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms. Nat. Commun. 2021, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Dey, D.; Kavanaugh, L.G.; Conn, G.L. Antibiotic substrate selectivity of Pseudomonas aeruginosa MexY and MexB efflux systems is determined by a Goldilocks affinity. Antimicrob. Agents Chemother. 2020, 64, e00496-20. [Google Scholar] [CrossRef]
- Kumar Roy, R.; Patra, N. Configuration Flipping in Distal Pocket of Multidrug Transporter MexB Impacts the Efflux Inhibitory Mechanism. ChemPhysChem 2020, 21, 2516–2524. [Google Scholar] [CrossRef]
- Zgurskaya, H.I.; Malloci, G.; Chandar, B.; Vargiu, A.V.; Ruggerone, P. Bacterial efflux transporters’ polyspecificity—A gift and a curse? Curr. Opin. Microbiol. 2021, 61, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, V.K.; Vargiu, A.V.; Malloci, G.; Dreier, J.; Ruggerone, P. Molecular determinants of the promiscuity of MexB and MexY multidrug transporters of Pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 1144. [Google Scholar] [CrossRef]
- Catte, A.; K Ramaswamy, V.; Vargiu, A.V.; Malloci, G.; Bosin, A.; Ruggerone, P. Common recognition topology of mex transporters of Pseudomonas aeruginosa revealed by molecular modelling. Front. Pharmacol. 2022, 13, 4732. [Google Scholar] [CrossRef] [PubMed]
- Tamura, N.; Murakami, S.; Oyama, Y.; Ishiguro, M.; Yamaguchi, A. Direct interaction of multidrug efflux transporter AcrB and outer membrane channel TolC detected via site-directed disulfide cross-linking. Biochemistry 2005, 44, 11115–11121. [Google Scholar] [CrossRef]
- Murakami, S.; Nakashima, R.; Yamashita, E.; Matsumoto, T.; Yamaguchi, A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 2006, 443, 173–179. [Google Scholar] [CrossRef]
- Pos, K.M. Drug transport mechanism of the AcrB efflux pump. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2009, 1794, 782–793. [Google Scholar] [CrossRef]
- Seeger, M.A.; Schiefner, A.; Eicher, T.; Verrey, F.; Diederichs, K.; Pos, K.M. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 2006, 313, 1295–1298. [Google Scholar] [CrossRef]
- Seeger, M.A.; Von Ballmoos, C.; Eicher, T.; Brandstätter, L.; Verrey, F.; Diederichs, K.; Pos, K.M. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat. Struct. Mol. Biol. 2008, 15, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Nakashima, R.; Yamashita, E.; Yamaguchi, A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 2002, 419, 587–593. [Google Scholar] [CrossRef]
- Sennhauser, G.; Amstutz, P.; Briand, C.; Storchenegger, O.; Grütter, M.G. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007, 5, e7. [Google Scholar] [CrossRef] [PubMed]
- Aron, Z.; Opperman, T.J. The hydrophobic trap—The Achilles heel of RND efflux pumps. Res. Microbiol. 2018, 169, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Atzori, A.; Malloci, G.; Cardamone, F.; Bosin, A.; Vargiu, A.V.; Ruggerone, P. Molecular interactions of carbapenem antibiotics with the multidrug efflux transporter AcrB of Escherichia coli. Int. J. Mol. Sci. 2020, 21, 860. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Halim, H.; Al Dajani, A.a.; Abdelhalim, A.; Abdelmalek, S. The search of potential inhibitors of the AcrAB–TolC system of multidrug-resistant Escherichia coli: An in silico approach. Appl. Microbiol. Biotechnol. 2019, 103, 6309–6318. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.; Weng, J.; Wang, W. Insights into the inhibitory mechanism of D13-9001 to the multidrug transporter AcrB through molecular dynamics simulations. J. Phys. Chem. B 2016, 120, 2145–2154. [Google Scholar] [CrossRef] [PubMed]
- Salmaso, V.; Moro, S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: An overview. Front. Pharmacol. 2018, 9, 923. [Google Scholar] [CrossRef]
- Blanco, P.; Sanz-García, F.; Hernando-Amado, S.; Martínez, J.L.; Alcalde-Rico, M. The development of efflux pump inhibitors to treat Gram-negative infections. Expert Opin. Drug Discov. 2018, 13, 919–931. [Google Scholar] [CrossRef]
- Samreen, Q.F.A.; Ahmad, I. In silico screening and in vitro validation of phytocompounds as multidrug efflux pump inhibitor against E. coli. J. Biomol. Struct. Dyn. 2022, 41, 2189–2201. [Google Scholar]
- Di, L.; Kerns, E.H. Drug-Like Properties: Concepts, Structure Design and Methods from ADME to Toxicity Optimization; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Kerns, E.H.; Di, L.; Kerns, E.H. Drug-Like Properties: Concepts, Structure Design and Methods; Academic Press: New York, NY, USA, 2008; Volume 10. [Google Scholar]
- Yu, E.W.; Aires, J.R.; McDermott, G.; Nikaido, H. A periplasmic drug-binding site of the AcrB multidrug efflux pump: A crystallographic and site-directed mutagenesis study. J. Bacteriol. 2005, 187, 6804–6815. [Google Scholar] [CrossRef]
- Su, C.-C.; Li, M.; Gu, R.; Takatsuka, Y.; McDermott, G.; Nikaido, H.; Yu, E.W. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J. Bacteriol. 2006, 188, 7290–7296. [Google Scholar] [CrossRef]
- Vergalli, J.; Bodrenko, I.V.; Masi, M.; Moynié, L.; Acosta-Gutierrez, S.; Naismith, J.H.; Davin-Regli, A.; Ceccarelli, M.; Van den Berg, B.; Winterhalter, M. Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat. Rev. Microbiol. 2020, 18, 164–176. [Google Scholar] [CrossRef]
- Masi, M.; Winterhalter, M.; Pagès, J.-M. Outer membrane porins. Bact. Cell Walls Membr. 2019, 14, 1159912. [Google Scholar]
- Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef]
- Fairman, J.W.; Noinaj, N.; Buchanan, S.K. The structural biology of β-barrel membrane proteins: A summary of recent reports. Curr. Opin. Struct. Biol. 2011, 21, 523–531. [Google Scholar] [CrossRef]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2009, 1794, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Zgurskaya, H.I.; Nikaido, H. Multidrug resistance mechanisms: Drug efflux across two membranes. Mol. Microbiol. 2000, 37, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Chugh, A.; Eudes, F.; Shim, Y.S. Cell-penetrating peptides: Nanocarrier for macromolecule delivery in living cells. IUBMB Life 2010, 62, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 2000, 44, 235–249. [Google Scholar] [CrossRef]
- Yıldırım, M.A.; Goh, K.-I.; Cusick, M.E.; Barabási, A.-L.; Vidal, M. Drug—Target network. Nat. Biotechnol. 2007, 25, 1119–1126. [Google Scholar] [CrossRef]
- Föger, F. Strategies to overcome efflux pumps. In Oral Delivery of Macromolecular Drugs: Barriers, Strategies and Future Trends; Springer Science + Business Media, LLC: New York, NY, USA, 2009; pp. 123–136. [Google Scholar]
- Abdel-Karim, S.A.-A.M.; El-Ganiny, A.M.A.; El-Sayed, M.A.; Abbas, H.A.A. Promising FDA-approved drugs with efflux pump inhibitory activities against clinical isolates of Staphylococcus aureus. PLoS ONE 2022, 17, e0272417. [Google Scholar] [CrossRef]
- Xiang, J.; Zhao, R.; Wang, B.; Sun, X.; Guo, X.; Tan, S.; Liu, W. Advanced nano-carriers for anti-tumor drug loading. Front. Oncol. 2021, 11, 758143. [Google Scholar] [CrossRef] [PubMed]
- Chandrakala, V.; Aruna, V.; Angajala, G. Review on metal nanoparticles as nanocarriers: Current challenges and perspectives in drug delivery systems. Emergent Mater. 2022, 5, 1593–1615. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Tiwari, M.; Tiwari, V. Strategies to combat bacterial antimicrobial resistance: A focus on mechanism of the efflux pumps inhibitors. SN Compr. Clin. Med. 2021, 3, 510–527. [Google Scholar] [CrossRef]
- Papkou, A.; Hedge, J.; Kapel, N.; Young, B.; MacLean, R.C. Efflux pump activity potentiates the evolution of antibiotic resistance across S. aureus isolates. Nat. Commun. 2020, 11, 3970. [Google Scholar] [CrossRef] [PubMed]
- Shriram, V.; Khare, T.; Bhagwat, R.; Shukla, R.; Kumar, V. Inhibiting bacterial drug efflux pumps via phyto-therapeutics to combat threatening antimicrobial resistance. Front. Microbiol. 2018, 9, 2990. [Google Scholar] [CrossRef] [PubMed]
- Vila, J.; Martí, S.; Sanchez-Céspedes, J. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2007, 59, 1210–1215. [Google Scholar] [CrossRef]
- Nicolas-Chanoine, M.-H.; Mayer, N.; Guyot, K.; Dumont, E.; Pagès, J.-M. Interplay between membrane permeability and enzymatic barrier leads to antibiotic-dependent resistance in Klebsiella pneumoniae. Front. Microbiol. 2018, 9, 1422. [Google Scholar] [CrossRef]
- Ruscito, A.; DeRosa, M.C. Small-molecule binding aptamers: Selection strategies, characterization, and applications. Front. Chem. 2016, 4, 14. [Google Scholar] [CrossRef]
- Pfeiffer, F.; Mayer, G. Selection and biosensor application of aptamers for small molecules. Front. Chem. 2016, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Walmagh, M.; Van Puyenbroeck, V.; Cornelissen, A.; Cenens, W.; Aertsen, A.; Oliveira, H.; Azeredo, J.; Verween, G.; Pirnay, J.-P. Engineered endolysin-based “Artilysins” to combat multidrug-resistant gram-negative pathogens. MBio 2014, 5, 10–128. [Google Scholar] [CrossRef]
- Cramariuc, O.; Rog, T.; Javanainen, M.; Monticelli, L.; Polishchuk, A.V.; Vattulainen, I. Mechanism for translocation of fluoroquinolones across lipid membranes. Biochim. Biophys. Acta (BBA) Biomembr. 2012, 1818, 2563–2571. [Google Scholar] [CrossRef]
- Cooper, C.J.; Krishnamoorthy, G.; Wolloscheck, D.; Walker, J.K.; Rybenkov, V.V.; Parks, J.M.; Zgurskaya, H.I. Molecular properties that define the activities of antibiotics in Escherichia coli and Pseudomonas aeruginosa. ACS Infect. Dis. 2018, 4, 1223–1234. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, B.V.; Babu, T.M.C.; Reddy, N.V.; Rajendra, W. Homology modeling, molecular dynamics, and virtual screening of NorA efflux pump inhibitors of Staphylococcus aureus. Drug Des. Dev. Ther. 2016, 3237–3252. [Google Scholar] [CrossRef]
- Fiorentino, F.; Rotili, D.; Mai, A. Native mass spectrometry-directed drug discovery: Recent advances in investigating protein function and modulation. Drug Discov. Today 2023, 103548. [Google Scholar] [CrossRef]
- Oluwole, A.; Shutin, D.; Bolla, J.R. Mass spectrometry of intact membrane proteins: Shifting towards a more native-like context. Essays Biochem. 2023, 67, 201–213. [Google Scholar]
- Olsen, J.G.; Teilum, K.; Kragelund, B.B. Behaviour of intrinsically disordered proteins in protein–protein complexes with an emphasis on fuzziness. Cell. Mol. Life Sci. 2017, 74, 3175–3183. [Google Scholar] [CrossRef]
- Orts, J.; Riek, R. Protein—Ligand structure determination with the NMR molecular replacement tool, N MR2. J. Biomol. NMR 2020, 74, 633–642. [Google Scholar] [CrossRef]
- Li, Q.; Kang, C. A practical perspective on the roles of solution NMR spectroscopy in drug discovery. Molecules 2020, 25, 2974. [Google Scholar] [CrossRef]
- Ganesan, A.; Arimondo, P.B.; Rots, M.G.; Jeronimo, C.; Berdasco, M. The timeline of epigenetic drug discovery: From reality to dreams. Clin. Epigenet. 2019, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
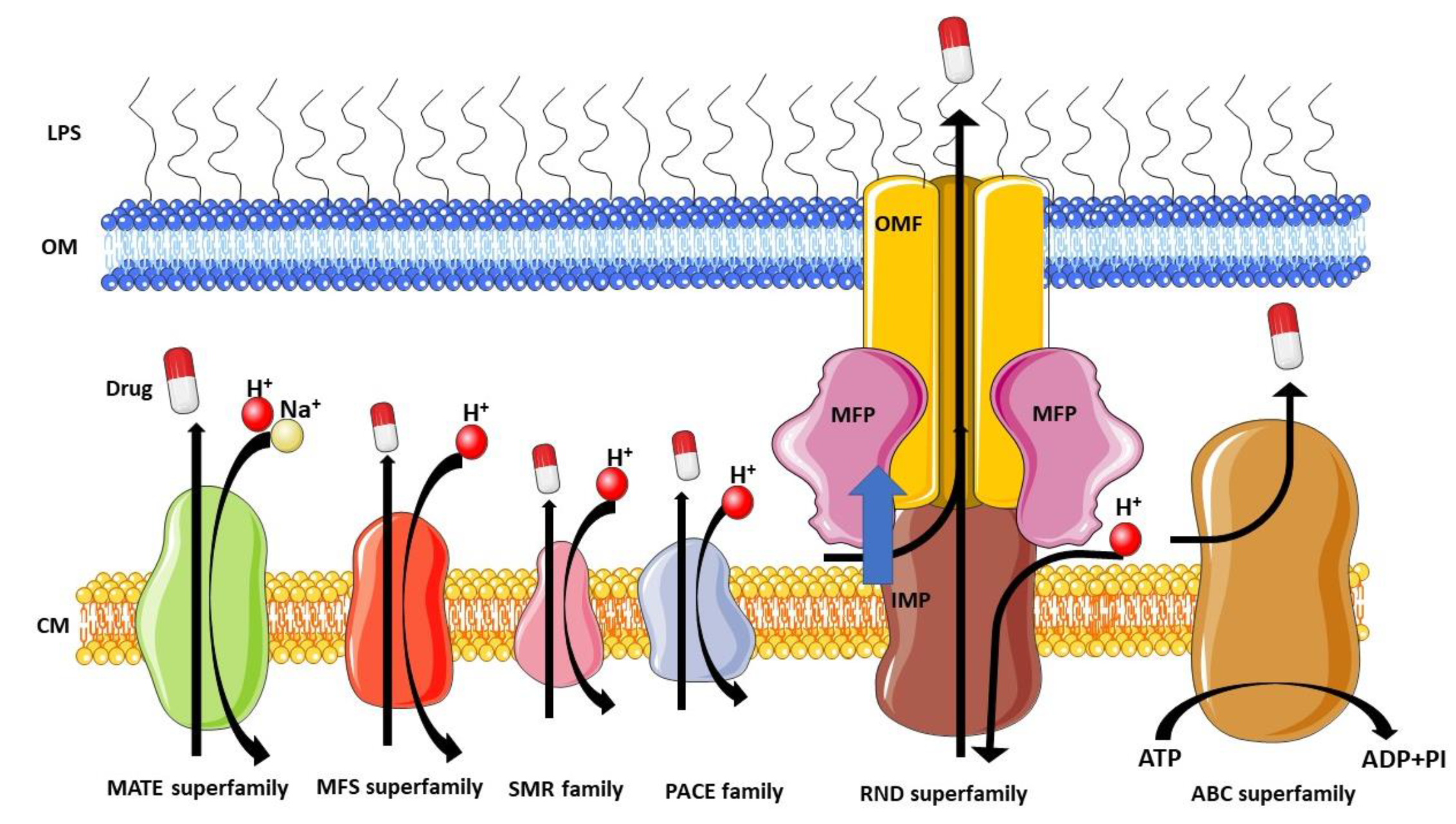
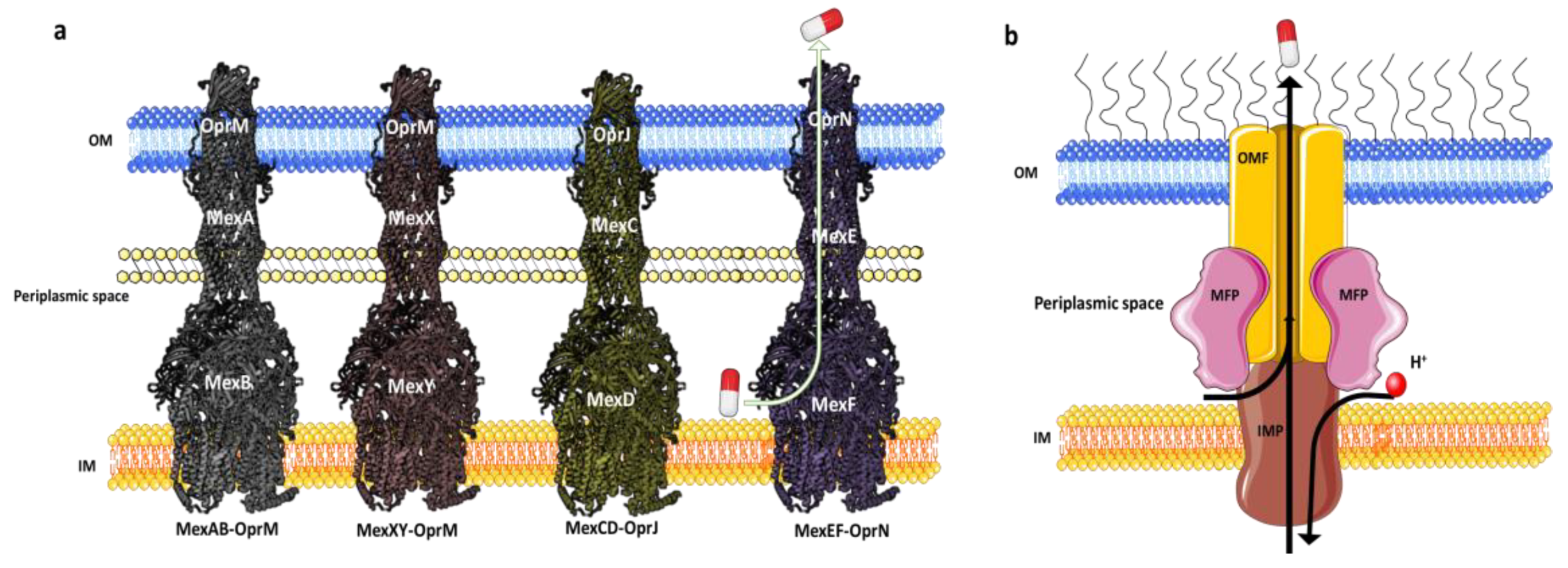





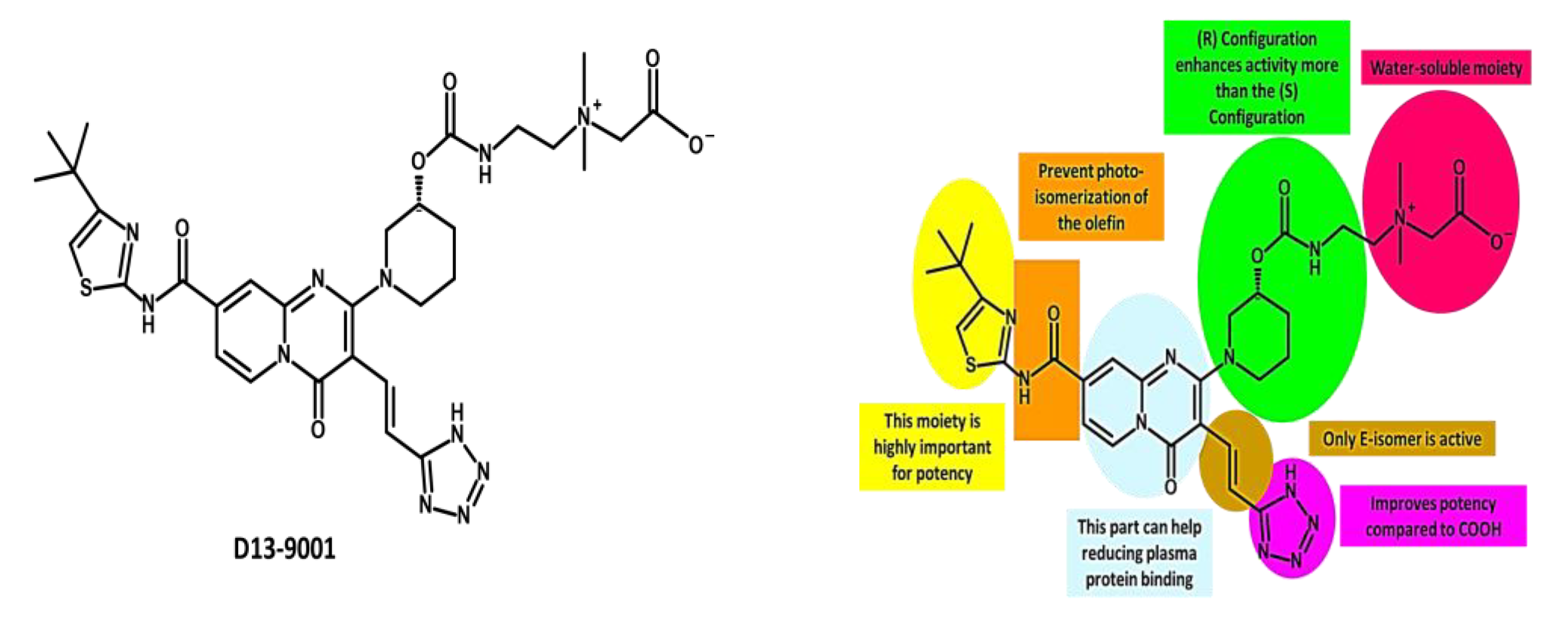
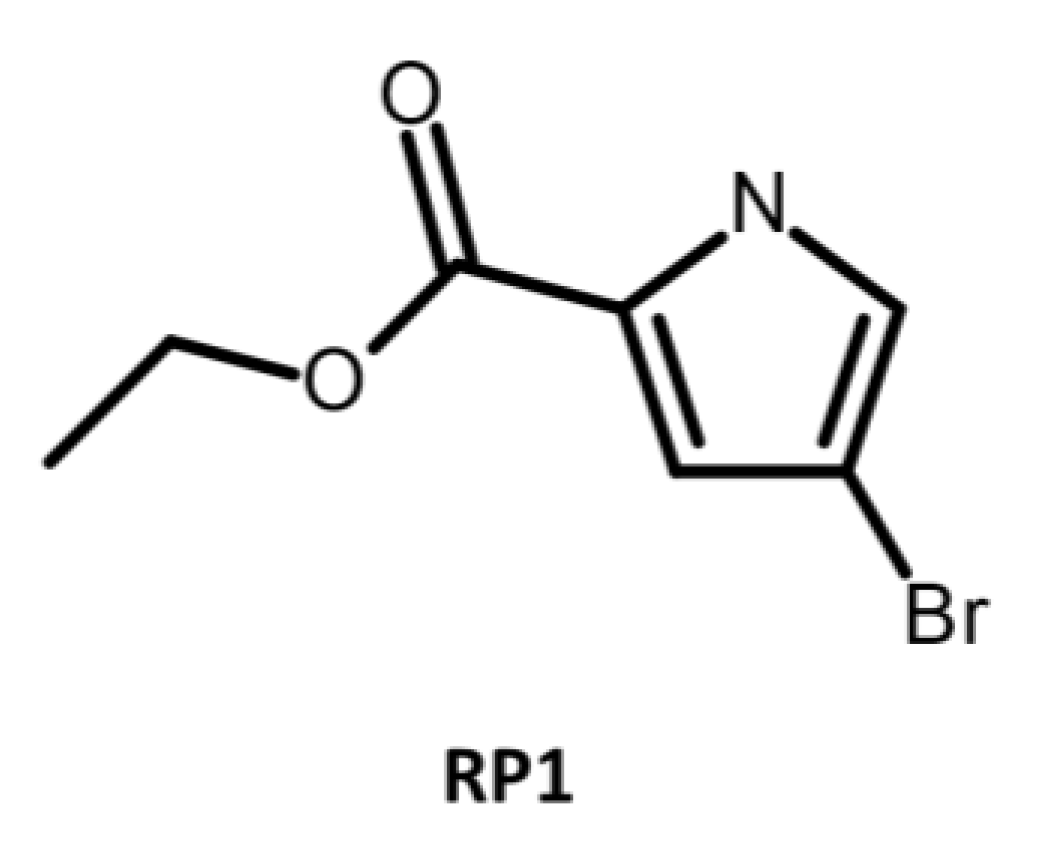

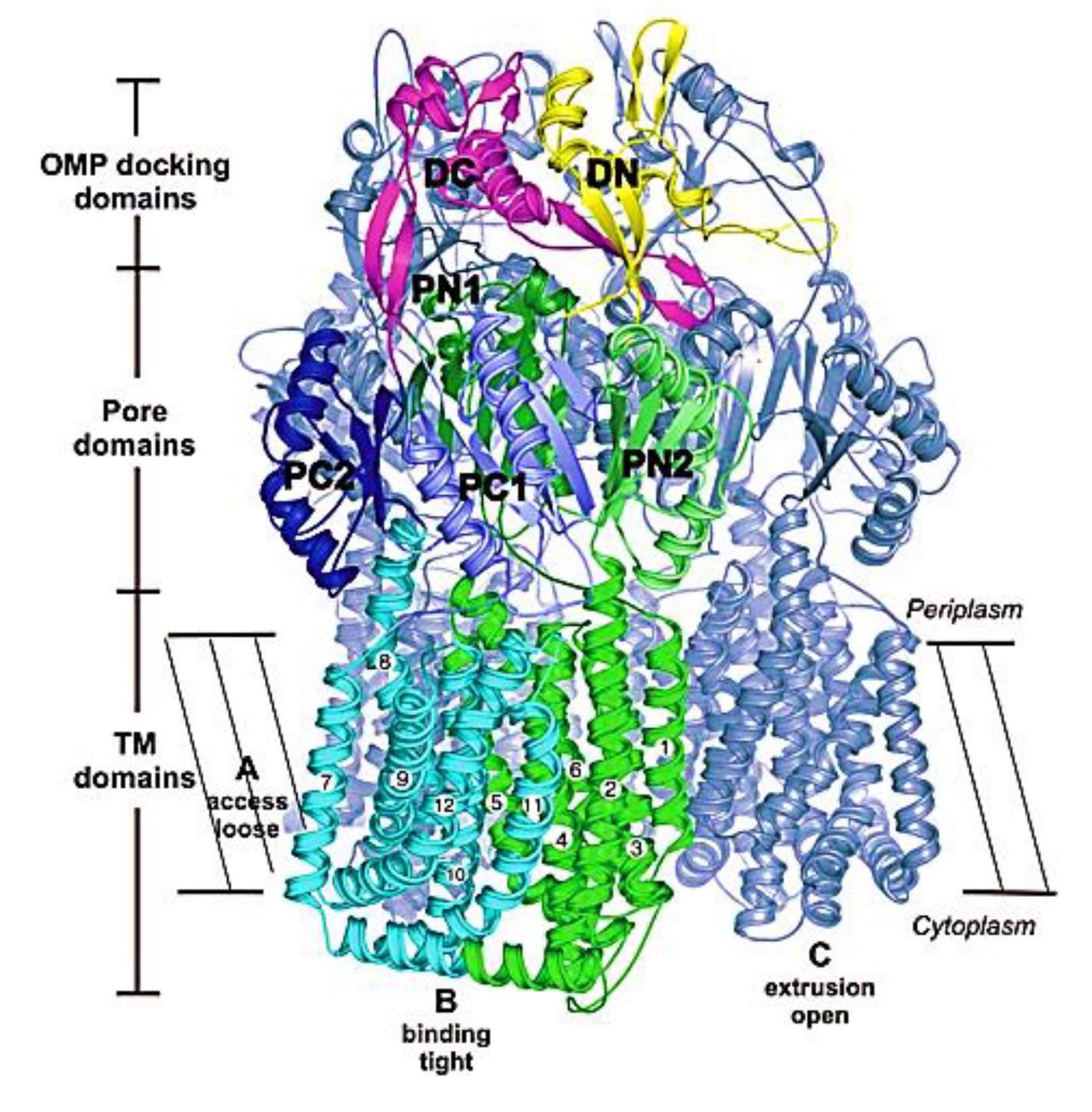
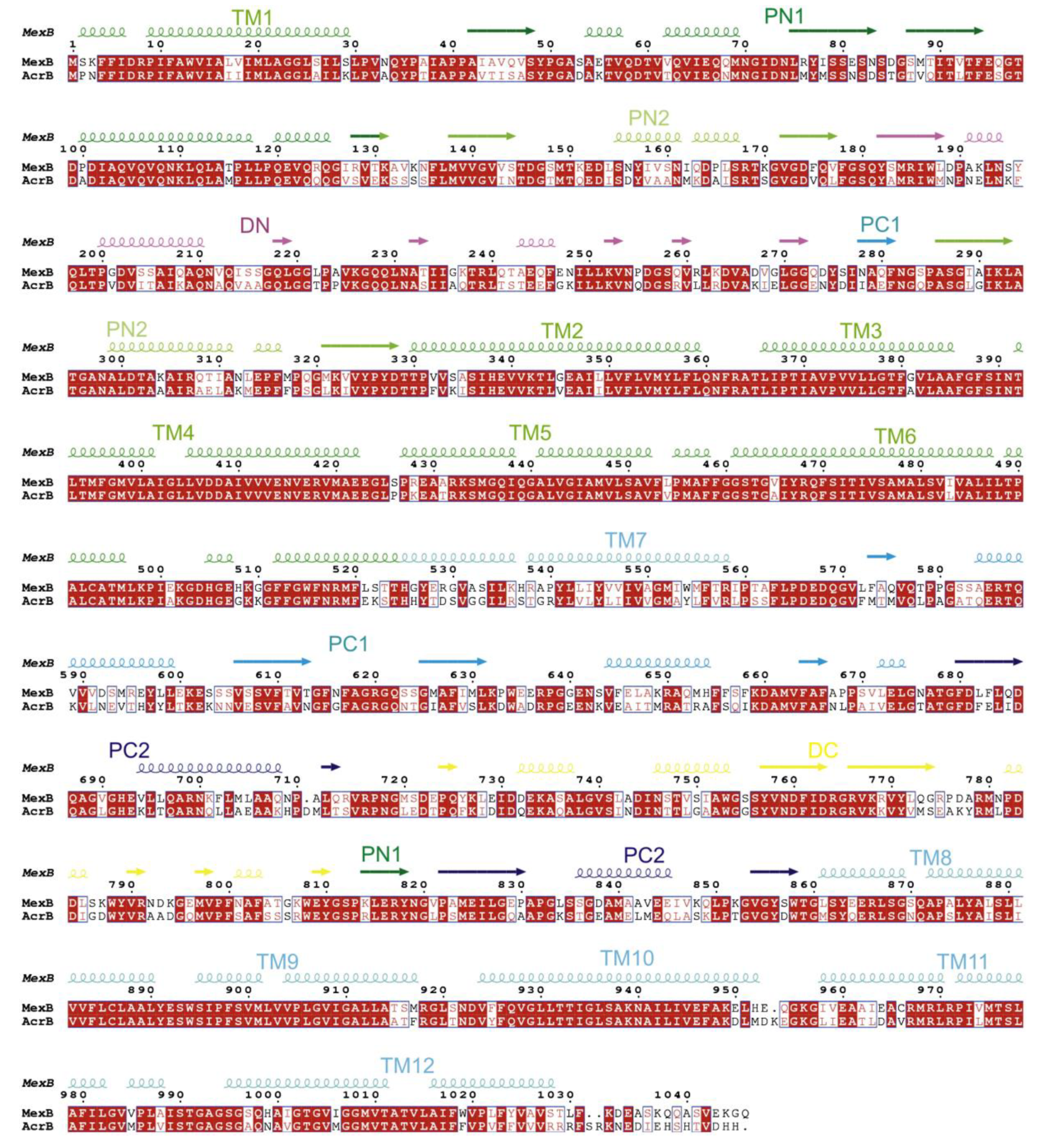
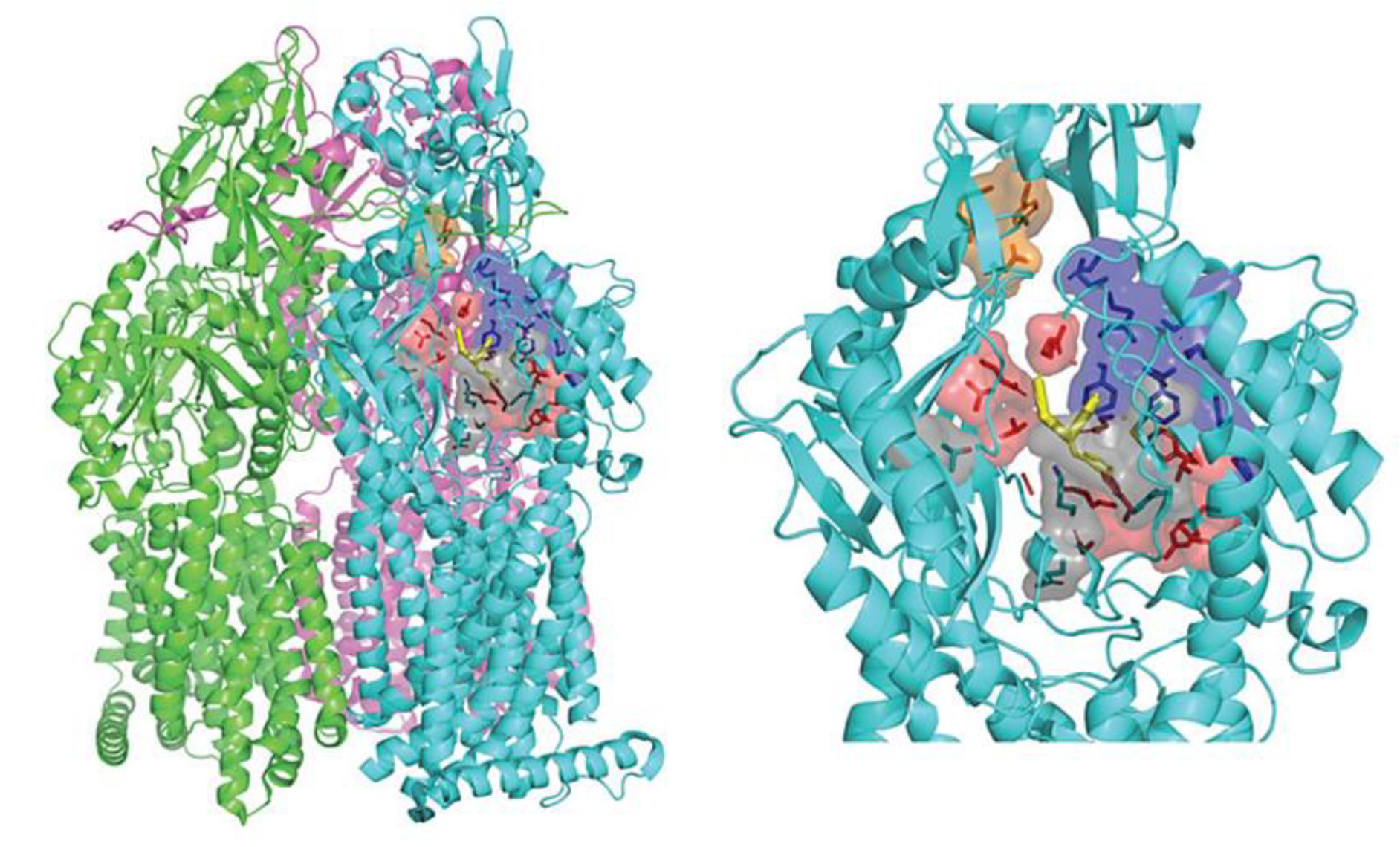
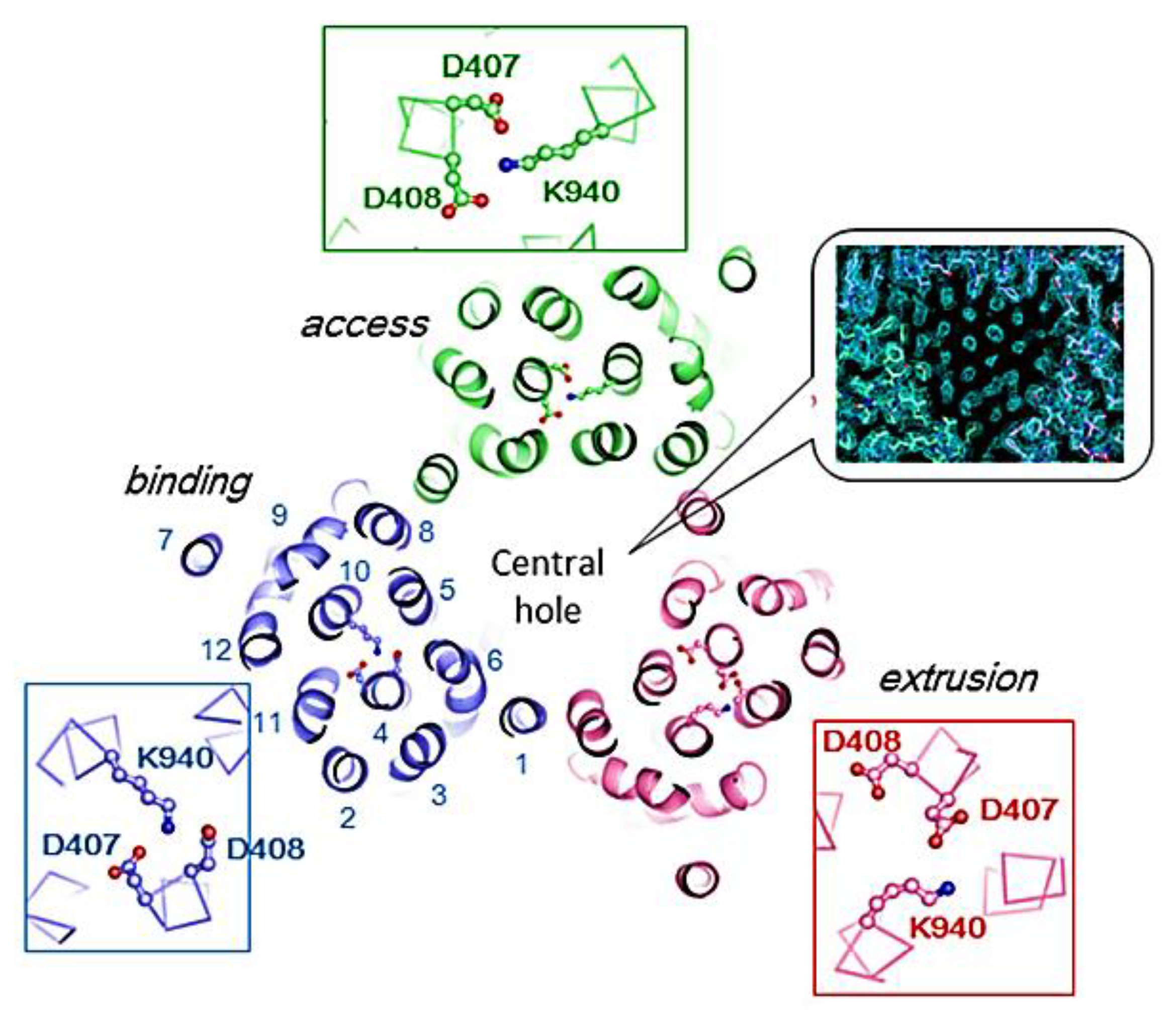
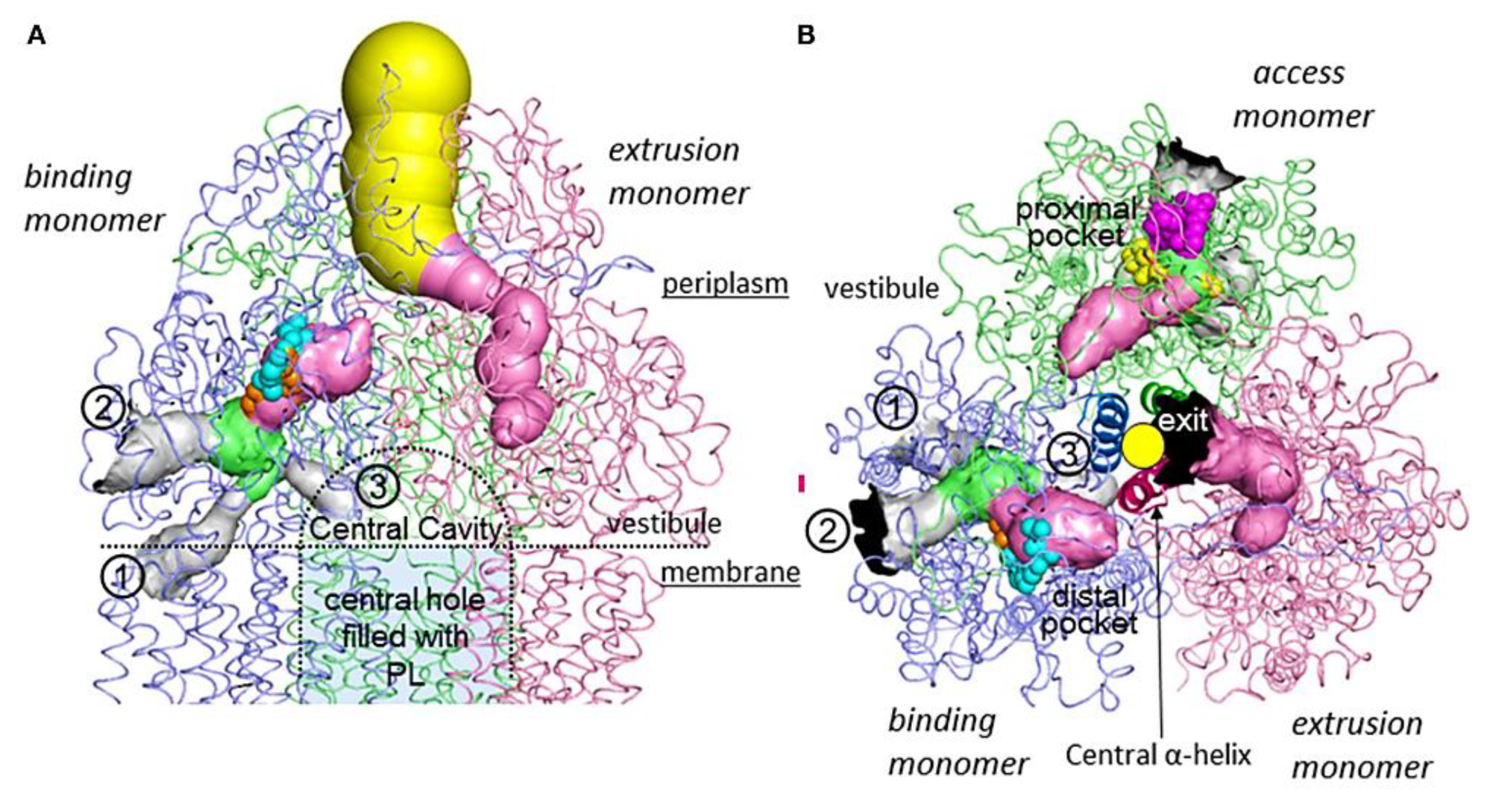
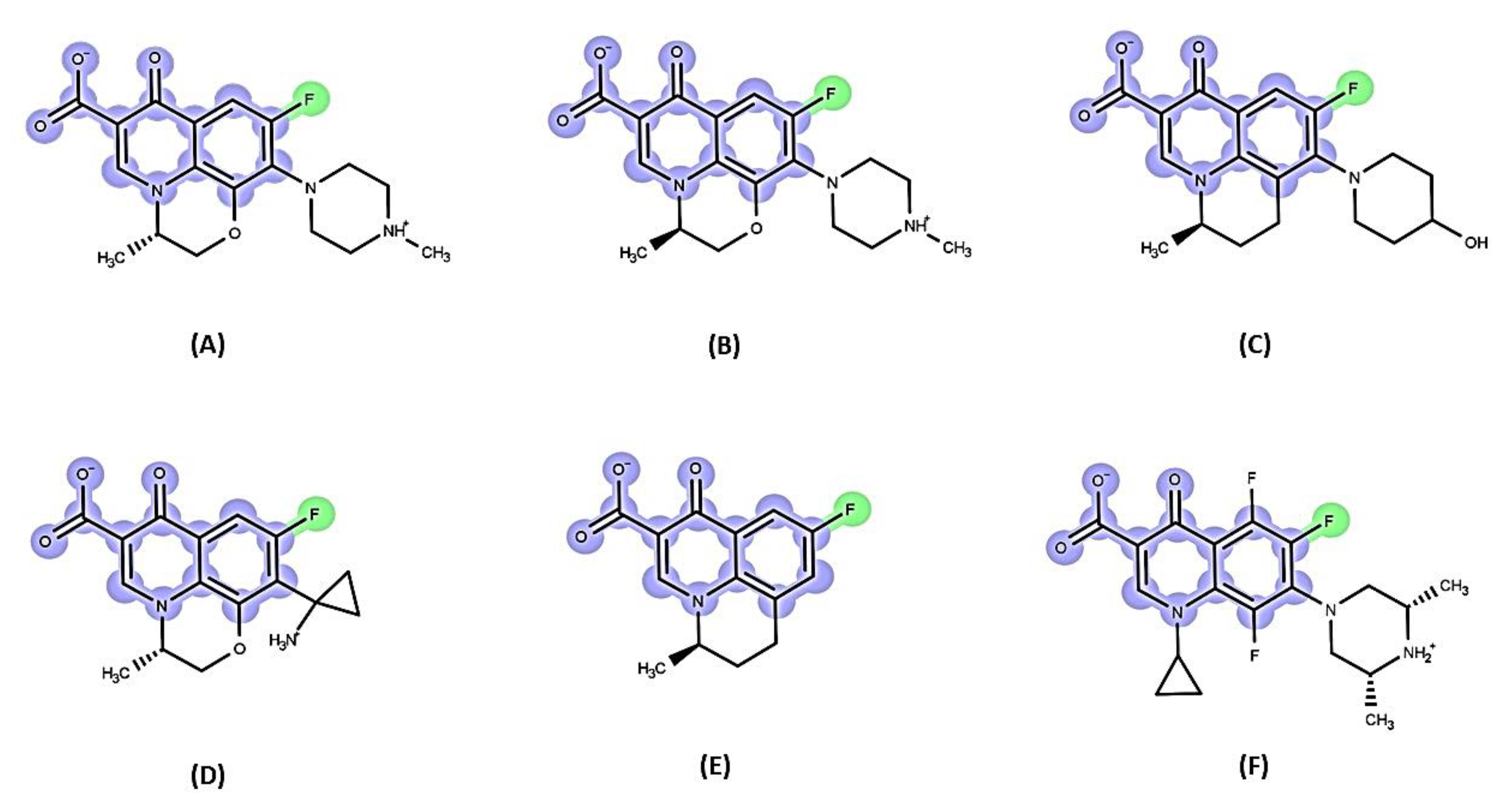
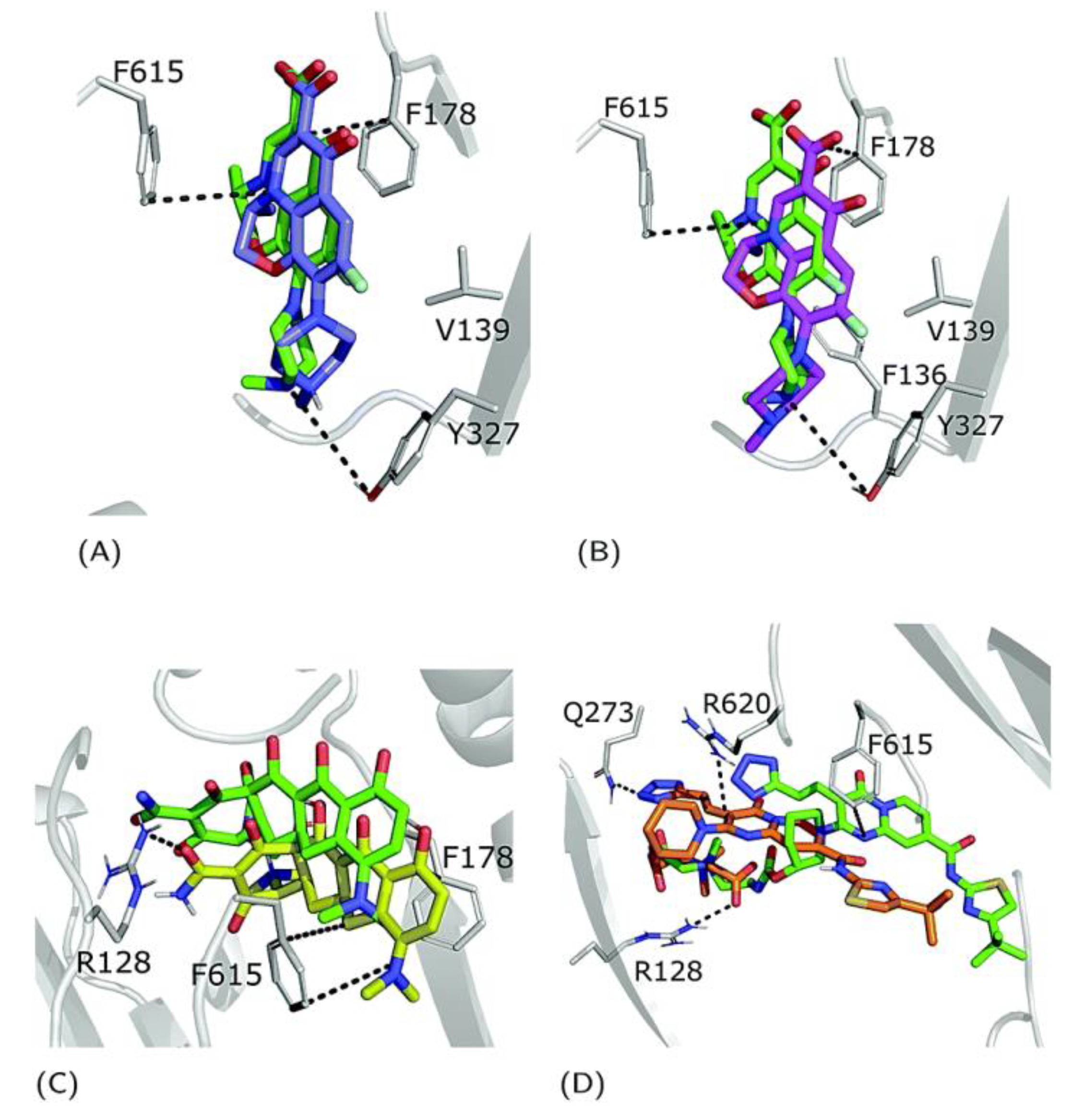

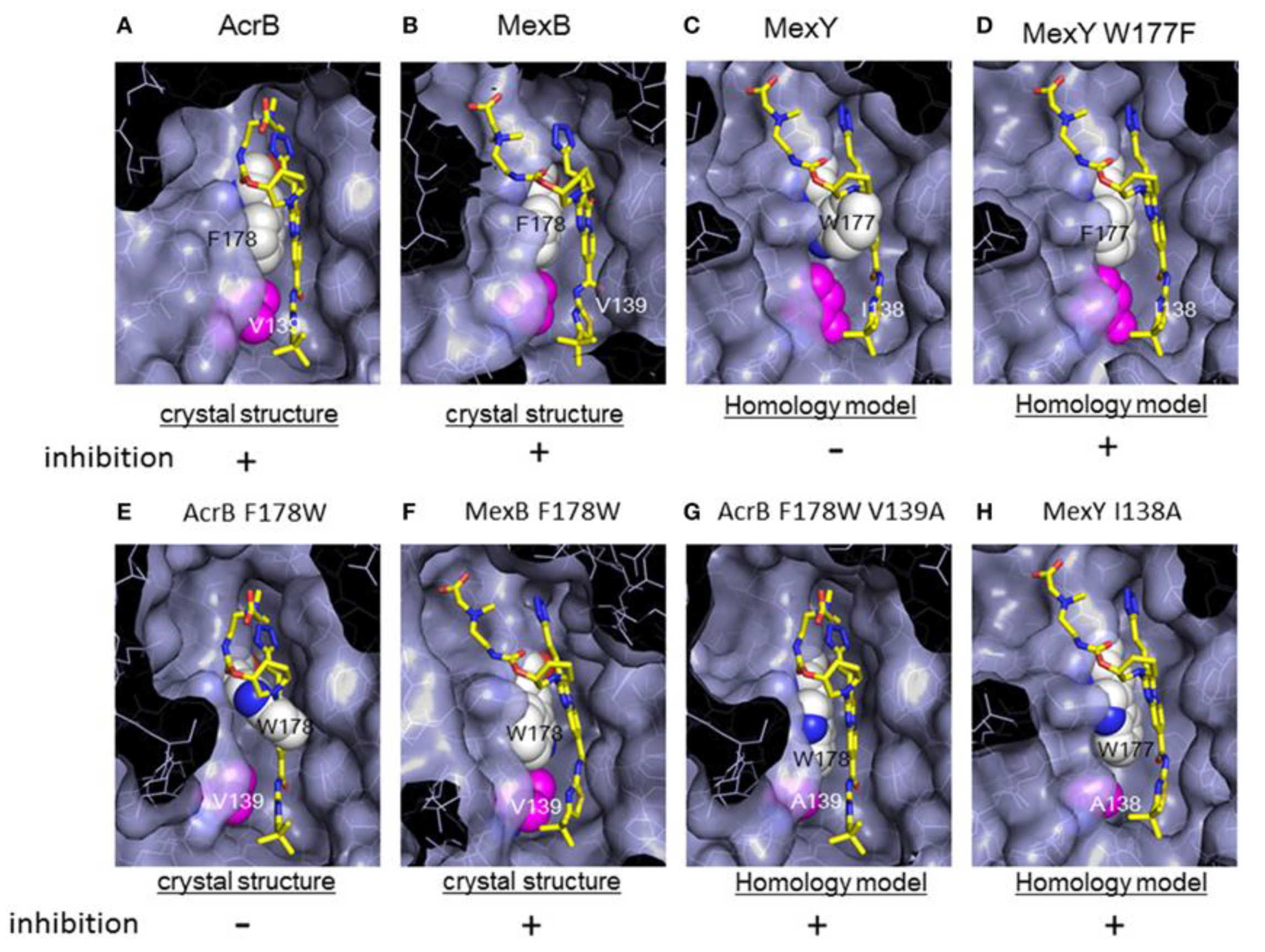
| Family 1 | Efflux Pump | Gene Ids 2 | Substrates 3 | References |
|---|---|---|---|---|
| ABC | Ttg2 (Mla) | PA4456-PA4455-PA4454-PA4453-PA4452 | CHL, CIP, COL, DMF, DOX, LVX, MIN, OFX, TET, TGC, TOB, TMP | [80,81,82] |
| PA1874-77 | PA1874-PA1875-PA1876-PA1877 | CIP, GEN, NOR, TOB | [83,84] | |
| PA3228 | PA3228 | CAR, LVX, NOR | [85] | |
| MFS | Mfs1 | PA1262 | PQT | [86] |
| Mfs2/SmvA | PA1282 | OCT, PQT | [86,87] | |
| CmlA1 | GNT62_RS22140 | CHL | [88,89] | |
| SMR | PASmr/EmrEPae | PA4990 | ACR, EtBr, GEN, KAN, NEO | [90,91] |
| SugE subfamily SMR | PA1882 | Further research is needed | [92] | |
| MATE | PmpM | PA1361 | ACR, BZK, CIP, EtBr, NOR, OFX, TPPCL | [93] |
| PACE | PA2880 | PA2880 | CHX | [94] |
| RND | MexAB-OprM | PA0425-PA0426-PA0427 | AMI, AMX, ATM, CAR, CR, MA, FEP, CFP, CFSL, CTX, FOX, CZOP, CPO, CES, CAZ, CZX, CRO, CXM, CHL, CTET, CIN, CIP, CLX, DP, DOR, ENX, ERY, FMOX, GEN, IPM, LVX, CLM, MEM, MOX, NAF, NAL, NOR, NOV, OFX, OMC, OTC, PG, PPA, PIP, PTZ, PMA, SPX, SPI, STN, SUL, TZB, TET, TIC, TOS | [95,96,97,98] |
| MexCD-OprJ | PA4599-PA4598-PA4597 | AMX, MA, FEP, CFP, CFSL, CTX, FOX, CZOP, CPO, CES, CZX, CRO, CXM, CHL, CHX, CTET, CIN, CIP, CLX, DOR, ENX, ERY, FMOX, LVX, CLM, MEM, NAF, NAL, NOR, NOV, OFX, OMC, OTC, PG, PPA, PIP, PMA, SPX, SPI, TET, TOS | [96,97,99,100] | |
| MexCD-TOprJ | LSG45_RS29735-LSG45_RS29740-LSG45_RS29745 | FEP, CEQ, CAZ, CTET, CIP, DOX, ERV, FLO, GEN, MIN, NAL, OTC, STR, TET, TGC | [101,102] | |
| MexEF-OprN | PA2493-PA2494-PA2495 | CHL, QN, TET, TMP | [103] | |
| MexGHI-OpmD | PA4205-PA4206-PA4207-PA4208 | 5-Me-PCA, ACR, EtBr, NOR, R6G, TET, V | [104,105,106] | |
| MexJK-OprM | PA3677-PA3676-PA0427 | ERY, TET, TCS | [107] | |
| MexMN-OprM | PA1435-PA1436-PA0427 | BAL30072, ATM, BIPM, CAR, CMN, CAZ, CFT, CHL, MEM, MET, MOX, NOV, PIP, SUL, TMC, TP, TIC | [108,109] | |
| MexPQ-OpmE | PA3523-PA3522-PA3521 | Hoechst 33342, CHL, CIP, ERY, KIT, NOR, RKM, TET, TPPCL | [108] | |
| MexVW-OprM | PA4374-PA4375-PA0427 | ACR, CPO, CHL, ERY, EtBr, NOR, OFX, TET | [110] | |
| MexXY-OprM (-OprA) | PA2019-PA2018-PA0427 (-PSPA7_3271) | ACR, AMI, AMX, MA, FEP, CFP, CFSL, CTX, FOX, CZOP, CPO, CAZ, CZX, CRO, CXM, CHL, CTET, CIN, CIP, CLX, DOR, ENX, ERY, EtBr, FMOX, GEN, IPM, KAN, LVX, CLM, MEM, NAF, NAL, NEO, NOR, OFX, OMC, OTC, PG, PPA, PIP, PTZ, PMA, SPX, SPI, STR, TZB, TET, TIC, TOB, TOS, CAR (only with OprA), SUL (only with OprA) | [96,97,111,112] | |
| MuxABC-OpmB | PA2528-PA2527-PA2526-PA2525 | ATM, ERY, KIT, NOV, RKM, TET | [113] | |
| TriABC-OpmH | PA0156-PA0157-PA0158-PA4974 | TCS | [114] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avakh, A.; Grant, G.D.; Cheesman, M.J.; Kalkundri, T.; Hall, S. The Art of War with Pseudomonas aeruginosa: Targeting Mex Efflux Pumps Directly to Strategically Enhance Antipseudomonal Drug Efficacy. Antibiotics 2023, 12, 1304. https://doi.org/10.3390/antibiotics12081304
Avakh A, Grant GD, Cheesman MJ, Kalkundri T, Hall S. The Art of War with Pseudomonas aeruginosa: Targeting Mex Efflux Pumps Directly to Strategically Enhance Antipseudomonal Drug Efficacy. Antibiotics. 2023; 12(8):1304. https://doi.org/10.3390/antibiotics12081304
Chicago/Turabian StyleAvakh, Asiyeh, Gary D. Grant, Matthew J. Cheesman, Tejaswini Kalkundri, and Susan Hall. 2023. "The Art of War with Pseudomonas aeruginosa: Targeting Mex Efflux Pumps Directly to Strategically Enhance Antipseudomonal Drug Efficacy" Antibiotics 12, no. 8: 1304. https://doi.org/10.3390/antibiotics12081304
APA StyleAvakh, A., Grant, G. D., Cheesman, M. J., Kalkundri, T., & Hall, S. (2023). The Art of War with Pseudomonas aeruginosa: Targeting Mex Efflux Pumps Directly to Strategically Enhance Antipseudomonal Drug Efficacy. Antibiotics, 12(8), 1304. https://doi.org/10.3390/antibiotics12081304








