Antimicrobial Efficacy of 7-Hydroxyflavone Derived from Amycolatopsis sp. HSN-02 and Its Biocontrol Potential on Cercospora Leaf Spot Disease in Tomato Plants
Abstract
:1. Introduction
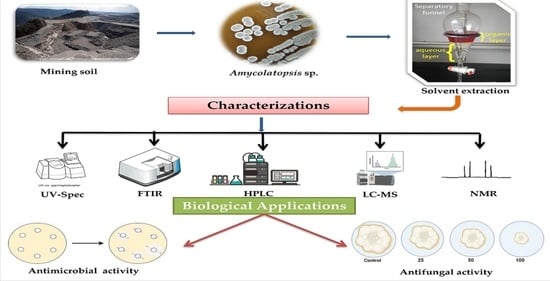
2. Results and Discussion
2.1. Isolation and Screening
2.2. Phylogenetic Analysis of Amycolatopsis sp. HSN-02
2.3. Characterizations of Amycolatopsis sp. HSN-02
2.4. Extraction of Secondary Metabolites
2.5. Purification of Ethyl Acetate Extract
2.6. Identification and Structure Elucidation
2.7. Antimicrobial Activity and Minimum Inhibitory Concentration (MIC) Assay of 7-Hydroxyflavone
2.8. Antifungal Assay
2.9. Suppression of Cercospora Leaf Spot in Tomato Plants by Purified Compound
2.10. Toxicity against Eukaryotic Cells
3. Materials and Methods
3.1. Pathogens Used in the Study
3.2. Isolation and Screening
3.3. Taxonomic Characterization
3.4. Morphological, Physiological, and Biochemical Characterizations
3.5. Fermentation and Extraction
3.6. Purification and Characterization
3.7. Identification of the Compound
3.8. Antimicrobial Activity and MIC Assay
3.9. Antifungal Assay
3.10. Effect of Purified Compound on Cercospora Leaf Spot in Tomato Plants
3.11. Toxicity against Eukaryotic Cells
3.12. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Demain, A.L.; Sanchez, S. Microbial drug discovery: 80 years of progress. J. Antibiot. 2009, 62, 5–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berdy, J. Bioactive microbial metabolites: A personal view. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, K.S. New Aspects of natural products in drug discovery. Trends. Microbiol. 2007, 15, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.R.; Mincer, T.J.; Williams, P.G.; Fenical, W. Marine actinomycete diversity and natural product discovery. Antonie Leeuwenhoek 2005, 87, 43–48. [Google Scholar] [CrossRef]
- Bull, A.T.; Stach, J.E.M. Marine Actinobacteria: New opportunities for natural product search and discovery. Trends. Microbiol. 2007, 15, 491–499. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clement, C.; Ouhdouch, Y.; Van Wezel, G.P. Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; Dangi, P.; Choudhary, M. Actinomycetes: Source, identification, and their applications. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 801–832. [Google Scholar]
- Genilloud, O. Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 2017, 34, 1203–1232. [Google Scholar] [CrossRef]
- Chen, S.; Wu, Q.; Shen, Q.; Wang, H. Progress in understanding the genetic information and biosynthetic pathways behind Amycolatopsis antibiotics, with implications for the continued discovery of novel drugs. Chem.Bio.Chem 2016, 17, 119–128. [Google Scholar] [CrossRef]
- Tang, B.; Xie, F.; Zhao, W.; Wang, J.; Dai, S.; Zheng, H.; Ding, X.; Cen, X.; Liu, H.; Yu, Y.; et al. A Systematic study of the whole genome sequence of Amycolatopsis methanolica strain 239 T provides an insight into its physiological and taxonomic properties which correlate with its position in the genus. Synth. Syst. Biotechnol. 2016, 1, 169–186. [Google Scholar] [CrossRef] [Green Version]
- Lechevalier, H.A.; Lechevalier, M.P.; Gerber, N.N. Chemical composition as a criterion in the classification of actinomycetes. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherlands, 1971; Volume 14, pp. 47–72. ISBN 978-0-12-002614-2. [Google Scholar]
- Takahashi, Y. Family Pseudonocardiaceae. In Identification Manual of Actinomycetes; The Society for Actinomycetes Japan, Ed.; The Business Centre for Academic Societies: Tokyo, Japan, 2001. [Google Scholar]
- Kriaa, M.; Hammami, I.; Sahnoun, M.; Azebou, M.C.; Triki, M.A.; Kammoun, R. Biocontrol of tomato plant diseases caused by Fusarium solani using a new isolated Aspergillus tubingensis CTM 507 glucose oxidase. Comptes. Rendus. Biol. 2015, 338, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.P.; Srinivasulu, A.; Babu, K.R. Symptomology of major fungal diseases on tomato and its management. J. Pharm. Phytochem. 2018, 7, 1817–1821. [Google Scholar]
- Bonaterra, A.; Badosa, E.; Daranas, N.; Francés, J.; Roselló, G.; Montesinos, E. Bacteria as biological control agents of plant diseases. Microorganisms 2022, 10, 1759. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, S.; Srinivas, V.; Naresh, N.; Alekhya, G.; Sharma, R. Exploiting plant growth-promoting Amycolatopsis sp. for bio-control of charcoal rot of sorghum (Sorghum bicolor L.) caused by Macrophomina phaseolina (Tassi) Goid. Arch. Phytopathol. Plant. Prot. 2019, 52, 543–559. [Google Scholar] [CrossRef]
- Alipour Kafi, S.; Karimi, E.; Akhlaghi Motlagh, M.; Amini, Z.; Mohammadi, A.; Sadeghi, A. Isolation and identification of Amycolatopsis sp. strain 1119 with potential to improve cucumber fruit yield and induce plant defense responses in commercial greenhouse. Plant Soil 2021, 468, 125–145. [Google Scholar] [CrossRef]
- Basavarajappa, D.S.; Kumar, R.S.; Nayaka, S. Formulation-based antagonistic endophyte Amycolatopsis sp. SND-1 triggers defense response in Vigna radiata (L.) R. Wilczek. (Mung Bean) against Cercospora leaf spot disease. Arch. Microbiol. 2023, 205, 77. [Google Scholar] [CrossRef]
- Ebada, S.S.; Eze, P.; Okoye, F.B.C.; Esimone, C.O.; Proksch, P. The Fungal endophyte Nigrospora oryzae produces quercetin monoglycosides previously known only from plants. ChemistrySelect 2016, 1, 2767–2771. [Google Scholar] [CrossRef]
- Kang, H.R.; Lee, D.; Benndorf, R.; Jung, W.H.; Beemelmanns, C.; Kang, K.S.; Kim, K.H. Termisoflavones A–C, Isoflavonoid glycosides from termite-associated Streptomyces sp. RB1. J. Nat. Prod. 2016, 79, 3072–3078. [Google Scholar] [CrossRef]
- Kostrzewa-Susłow, E.; Dmochowska-Gładysz, J.; Białońska, A.; Ciunik, Z. Microbial transformations of flavanone by Aspergillus niger and Penicillium chermesinum cultures. J. Mol. Catal. B Enzym. 2008, 52–53, 34–39. [Google Scholar] [CrossRef]
- Wu, C.; Zhu, H.; Van Wezel, G.P.; Choi, Y.H. Metabolomics-guided analysis of isocoumarin production by Streptomyces species MBT76 and biotransformation of flavonoids and phenylpropanoids. Metabolomics 2016, 12, 90. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.-J.; Zhang, S.-Y.; Ye, Y.-H.; Yu, Z.; Qi, H.; Zhang, H.; Xue, Z.-L.; Wang, J.-D.; Wu, M. Three new isoflavonoid glycosides from the mangrove-derived actinomycete Micromonospora aurantiaca 110B. Mar. Drugs 2019, 17, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parthiban, A.; Sachithanandam, V.; Lalitha, P.; Elumalai, D.; Asha, R.N.; Jeyakumar, T.C.; Muthukumaran, J.; Jain, M.; Jayabal, K.; Mageswaran, T.; et al. Isolation and biological evaluation 7-hydroxy flavone from Avicennia officinalis L: Insights from extensive in vitro, DFT, molecular docking and molecular dynamics simulation studies. J. Biomol. Struct. Dyn. 2023, 41, 2848–2860. [Google Scholar] [CrossRef]
- EL-Hefny, M.; Abd El-Kareem, M.S.M.; Salem, M.Z.M. GC-MS and HPLC analyses of phytochemical compounds from Withania somnifera L. leaves extract. Alex. J. Agric. Sci. 2022, 67, 10–17. [Google Scholar] [CrossRef]
- Ait Assou, S.; Anissi, J.; Sendide, K.; El Hassouni, M. Diversity and antimicrobial activities of actinobacteria isolated from mining soils in Midelt Region, Morocco. Sci. World J. 2023, 2023, 6106673. [Google Scholar] [CrossRef] [PubMed]
- Sripairoj, P.; Suwanborirux, K.; Tanasupawat, S. Characterization and antimicrobial activity of Amycolatopsis strains isolated from Thai soils. J. Appl. Polym. Sci. 2013, 3, 11–16. [Google Scholar]
- Zucchi, T.D.; Tan, G.Y.A.; Goodfellow, M. Amycolatopsis thermophila sp. nov. and Amycolatopsis viridis sp. nov., thermophilic actinomycetes isolated from arid soil. Int. J. Syst. Evol. Microbiol. 2012, 62, 168–172. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.D. Amycolatopsis jejuensis sp. nov. and Amycolatopsis halotolerans sp. nov., Novel actinomycetes isolated from a natural cave. Int. J. Syst. Evol. Microbiol. 2006, 56, 549–553. [Google Scholar] [CrossRef] [Green Version]
- Bian, J.; Li, Y.; Wang, J.; Song, F.-H.; Liu, M.; Dai, H.-Q.; Ren, B.; Gao, H.; Hu, X.; Liu, Z.-H.; et al. Amycolatopsis marina sp. nov., an actinomycete isolated from an ocean sediment. Int. J. Syst. Evol. Microbiol. 2009, 59, 477–481. [Google Scholar] [CrossRef] [Green Version]
- Goodfellow, M.; Kim, S.B.; Minnikin, D.E.; Whitehead, D.; Zhou, Z.H.; Mattinson-Rose, A.D. Amycolatopsis sacchari sp. nov., a moderately thermophilic actinomycete isolated from vegetable matter. Int. J. Syst. Evol. Microbiol. 2001, 51, 187–193. [Google Scholar] [CrossRef] [Green Version]
- Duangmal, K.; Mingma, R.; Pathom-aree, W.; Thamchaipenet, A.; Inahashi, Y.; Matsumoto, A.; Takahashi, Y. Amycolatopsis samaneae sp. nov., isolated from roots of Samanea saman (Jacq.) Merr. Int. J. Syst. Evol. Microbiol. 2011, 61, 951–955. [Google Scholar] [CrossRef] [Green Version]
- Labeda, D.P.; Donahue, J.M.; Williams, N.M.; Sells, S.F.; Henton, M.M. Amycolatopsis kentuckyensis sp. nov., Amycolatopsis lexingtonensis sp. nov. and Amycolatopsis pretoriensis sp. nov., isolated from equine placentas. Int. J. Syst. Evol. Microbiol. 2003, 53, 1601–1605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, M.J.; Cookson, W.O.C.M.; Moffatt, M.F. Sequencing the human microbiome in health and disease. Hum. Mol. Genet. 2013, 22, R88–R94. [Google Scholar] [CrossRef] [PubMed]
- Okoro, C.K.; Brown, R.; Jones, A.L.; Andrews, B.A.; Asenjo, J.A.; Goodfellow, M.; Bull, A.T. Diversity of Culturable Actinomycetes in Hyper-Arid Soils of the Atacama Desert, Chile. Antonie Leeuwenhoek 2009, 95, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Thakur, D.; Bora, T.C.; Bordoloi, G.N.; Mazumdar, S. Influence of nutrition and culturing conditions for optimum growth and antimicrobial metabolite production by Streptomyces sp. 201. J. Mycol. Médicale 2009, 19, 161–167. [Google Scholar] [CrossRef]
- Subathra Devi, C.; Saini, A.; Rastogi, S.; Jemimah Naine, S.; Mohanasrinivasan, V. Strain improvement and optimization studies for enhanced production of erythromycin in bagasse based medium using Saccharopolyspora erythraea MTCC 1103. 3 Biotech 2015, 5, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Pal, A.; Chattopadhyay, A.; Paul, A.K. Diversity and antimicrobial spectrum of endophytic bacteria isolated from Paederia foetida L. Int. J. Curr. Pharm. Res. 2012, 4, 123–127. [Google Scholar]
- Gousterova, A.; Paskaleva, D.; Vasileva-Tonkova, E. Characterization of culturable thermophilic actinobacteria from Livingston Island, Antarctica. Int. Res. J. Biol. Sci. 2014, 3, 30–36. [Google Scholar]
- Davison, J. Genetic Exchange between Bacteria in the Environment. Plasmid 1999, 42, 73–91. [Google Scholar] [CrossRef]
- Ser, H.-L.; Palanisamy, U.D.; Yin, W.-F.; Chan, K.-G.; Goh, B.-H.; Lee, L.-H. Streptomyces malaysiense sp. nov.: A novel malaysian mangrove soil actinobacterium with antioxidative activity and cytotoxic potential against human cancer cell lines. Sci. Rep. 2016, 6, 24247. [Google Scholar] [CrossRef] [Green Version]
- Desai, P.P.; Prabhurajeshwar, C.; Kelmani Chandrakant, R. Molecular Genotyping and antimicrobial activities of secondary metabolites from Streptomyces sp: Taxonomy, Extraction and Purification. J. Biol. Act. Prod. Nat. 2016, 6, 282–298. [Google Scholar] [CrossRef]
- Wang, X.; Huang, L.; Kang, Z.; Buchenauer, H.; Gao, X. Optimization of the fermentation process of actinomycete strain Hhs.015 T. J. Biomed. Biotechnol. 2010, 2010, 141876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.-H.; Fang, X.-L.; Li, Y.-P.; Zhang, X. Effects of constant and shifting dissolved oxygen concentration on the growth and antibiotic activity of Xenorhabdus nematophila. Bioresour. Technol. 2010, 101, 7529–7536. [Google Scholar] [CrossRef] [PubMed]
- Kemp, W. Infrared spectroscopy. In Organic Spectroscopy; Macmillan Press Ltd.: London, UK, 1991; pp. 19–56. [Google Scholar]
- Zheng, K.-X.; Jiang, Y.; Jiang, J.-X.; Huang, R.; He, J.; Wu, S.-H. A New phthalazinone derivative and a new isoflavonoid glycoside from lichen-associated Amycolatopsis sp. Fitoterapia 2019, 135, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, S.S.; Moni, S.S.; Sultan, M.H.; Ali Bakkari, M.; Madkhali, O.A.; Alshahrani, S.; Makeen, H.A.; Joseph Menachery, S.; Ur Rehman, Z.; Shamsher Alam, M.; et al. Potential bioactive secondary metabolites of Actinomycetes sp. isolated from rocky soils of the heritage village rijal, Alma, Saudi Arabia. Arab. J. Chem. 2022, 15, 103793. [Google Scholar] [CrossRef]
- Mathur, N.; Paliwal, A.; Kumar, M.; Sharma, P.; Bhatnagar, P. Antimicrobial activity of actinomycetes strain isolated from soils of unusual ecological niches. Int. J. Chem. Sci. 2012, 10, 2237–2247. [Google Scholar]
- Kostrzewa-Susłow, E.; Janeczko, T. Microbial transformations of 7-Hydroxyflavanone. Sci. World J. 2012, 2012, 254929. [Google Scholar] [CrossRef] [Green Version]
- Ding, W.-J.; Zhang, S.-Q.; Wang, J.-H.; Lin, Y.-X.; Liang, Q.-X.; Zhao, W.-J.; Li, C.-Y. A New Di-O-Prenylated flavone from an actinomycete Streptomyces sp. MA-12. J. Asian Nat. Prod. Res. 2013, 15, 209–214. [Google Scholar] [CrossRef]
- Alam, M.; Jha, D.K. Optimization of culture conditions for antimetabolite production by a rare tea garden actinobacterial isolate, Amycolatopsis sp. ST-28. Afr. J. Clin. Exp. Microbiol. 2019, 20, 209. [Google Scholar] [CrossRef]
- Nadkarni, S.R.; Patel, M.V.; Chatterjee, S.; Vijayakumar, E.K.S.; Desikan, K.R.; Blumbach, J.; Ganguli, B.N. Balhimycin, a new glycopeptide antibiotic produced by Amycolatopsis sp. Y-86,21022. Taxonomy, production, isolation and biological activity. J. Antibiot. 1994, 47, 334–341. [Google Scholar] [CrossRef] [Green Version]
- Martini, H.; Weidenborner, M.; Adams, S.; Kunz, B. Increased antifungal activity of 3- and 7-hydroxyflavone against Cladosporium herbarum and Penicillium glabrum through ester formation. Mycol. Res. 1997, 101, 920–922. [Google Scholar] [CrossRef]
- Campoccia, D.; Ravaioli, S.; Santi, S.; Mariani, V.; Santarcangelo, C.; De Filippis, A.; Montanaro, L.; Arciola, C.R.; Daglia, M. Exploring the anticancer effects of standardized extracts of poplar-type Propolis: In vitro cytotoxicity toward cancer and normal cell lines. Biomed Pharmacother. 2021, 141, 111895. [Google Scholar] [CrossRef] [PubMed]
- Muthuraj, R.; Meghashyama, P.B.; Dhanyakumar, S.B.; Halaswamy, H.M.; Shashiraj, K.N.; Bidhayak, C.; Pallavi, S.S.; Sreenivasa, N. Isolation, identification and characterization of antimicrobial activity exhibiting actinomycete Streptomyces paradoxus strain KUASN-7 from soil. Int. J. Curr. Microbiol. Appl. Sci. 2021, 10, 164–176. [Google Scholar] [CrossRef]
- Sreenivasa, N.; Halaswamy, H.; Bidhayak, C.; Pallavi, S.S.; Dhanyakumara, S.B.; Shashiraj, K.N.; Muthuraj, R.; Meghashyama, P.B.; Dattatraya, A.; Meenakshi, S.M. Efficacy of antibiotic sensitivity and antimicrobial activity of Streptomyces cinereoruber RSA-14 isolated from rhizosphere soil of Alternanthera sessilis (L.) R. Br. ex DC. J. Appl. Biol. Biotech. 2020, 8, 1–6. [Google Scholar] [CrossRef]
- Chakraborty, B.; Kumar, R.S.; Almansour, A.I.; Perumal, K.; Nayaka, S.; Brindhadevi, K. Streptomyces filamentosus strain KS17 isolated from microbiologically unexplored marine ecosystems exhibited a broad spectrum of antimicrobial activity against human pathogens. Process Biochem. 2022, 117, 42–52. [Google Scholar] [CrossRef]
- Saadoun, I.; Muhana, A. Optimal production conditions, extraction, partial purification and characterization of inhibitory compound(s) produced by Streptomyces Ds-104 isolate against multi-drug resistant Candida albicans. Curr. Trends Biotechnol. Pharm. 2008, 2, 402–432. [Google Scholar]
- El-Bilawy, E.H.; Al-Mansori, A.-N.A.; Alotibi, F.O.; Al-Askar, A.A.; Arishi, A.A.; Teiba, I.I.; Sabry, A.E.-N.; Elsharkawy, M.M.; Heflish, A.A.; Behiry, S.I.; et al. Antiviral and antifungal of Ulva fasciata extract: HPLC analysis of polyphenolic compounds. Sustainability 2022, 14, 12799. [Google Scholar] [CrossRef]
- Abdullah, Z.K.; Kihara, J.; Gondo, Y.; Ganphung, R.; Yokoyama, Y.; Ueno, M. Suppressive effect of secondary metabolites from Streptomyces plumbeusi isolate F31D against Fusarium oxysporum f. sp. lycopersici, the causal agent of fusarium wilt of tomato. J. Gen. Plant Pathol. 2021, 87, 335–343. [Google Scholar] [CrossRef]
- Sreenivasa, N.; Meghashyama, B.P.; Pallavi, S.S.; Bidhayak, C.; Dattatraya, A.; Muthuraj, R.; Shashiraj, K.N.; Halaswamy, H.; Dhanyakumara, D.; Vaishnavi, M.D. Biogenic synthesis of silver nanoparticles using Paenibacillus sp. in-vitro and their antibacterial, anticancer activity assessment against human colon tumor cell line. J. Environ. Biol. 2021, 42, 118–127. [Google Scholar] [CrossRef]















| Actinomycetes Isolates | B. cereus (MTCC 11778) | S. aureus (MTCC 6908) | S. flexneri (MTCC 1457) | C. glabrata (MTCC 3019) |
|---|---|---|---|---|
| HM-01 | +++ | +++ | + | + |
| HM-02 | − | ++ | − | ++ |
| HM-03 | − | + | +++ | − |
| HM-04 | − | +++ | ++ | − |
| HM-05 | ++ | − | − | + |
| HM-06 | − | − | + | ++ |
| HM-07 | ++ | +++ | − | ++ |
| HSN-01 | +++ | ++ | ++ | +++ |
| HSN-02 | ++ | +++ | +++ | ++ |
| HSN-03 | − | ++ | − | − |
| HSN-04 | ++ | − | + | − |
| HSN-05 | +++ | + | + | ++ |
| HSN-06 | + | ++ | +++ | − |
| HS-01 | +++ | − | + | − |
| HS-02 | − | + | − | ++ |
| HS-03 | ++ | − | + | − |
| HS-04 | + | +++ | ++ | +++ |
| HS-05 | +++ | − | − | + |
| HS-06 | − | − | +++ | − |
| HS-08 | + | − | +++ | − |
| HS-09 | ++ | + | ++ | + |
| Morphological Characterizations | ||||
|---|---|---|---|---|
| Colony characters | Aerial mycelia | Substrate mycelia | ||
| Color | Whitish to grey | Pale yellow | ||
| Gram staining | Gram-positive | |||
| Margin | Filiform | |||
| Elevation | Flat | |||
| Texture | Powdery and dry | |||
| Pigmentation | Yellow colored pigment | |||
| Physiological Characterizations | ||||
| Growth at different temperature | Growth in different pH | |||
| 20 °C | − | pH 5.0 | − | |
| 25 °C | w | pH 6.0 | − | |
| 30 °C | +++ | pH 7.0 | +++ | |
| 35 °C | ++ | pH 8.0 | w | |
| 40 °C | w | pH 9.0 | − | |
| 45 °C | w | pH 10.0 | − | |
| Tests | Results | Tests | Results |
|---|---|---|---|
| BETA-XYLOSIDASE | + | D-MANNITOL | − |
| L-Lysine-ARYLAMIDASE | + | D-MANNOSE | + |
| L-Aspartate ARYLAMIDASE | + | D-MELEZITOSE | − |
| Leucine ARYLAMIDASE | + | N-ACETYL-D-GLUCOSAMINE | − |
| Phenylalanine ARYLAMIDASE | + | PALATINOSE | − |
| L-Proline ARYLAMIDASE | − | L-RHAMNOSE | − |
| BETA-GALACTOSIDASE | − | BETA-GLUCOSIDASE | + |
| L-Pyrrolidonyl-ARYLAMIDASE | + | BETA-MANNOSIDASE | − |
| ALPHA-GALACTOSIDASE | − | PHOSPHORYL CHOLINE | − |
| Alanine ARYLAMIDASE | + | PYRUVATE | − |
| Tyrosine ARYLAMIDASE | + | ALPHA-GLUCOSIDASE | + |
| BETA-N-ACETYL-GLUCOSAMINIDASE | + | D-TAGATOSE | − |
| Ala-Phe-Pro ARYLAMIDASE | + | D-TREHALOSE | − |
| CYCLODEXTRIN | − | INULIN | − |
| D-GALACTOSE | − | D-GLUCOSE | − |
| GLYCOGEN | − | D-RIBOSE | − |
| myo-INOSITOL | − | PUTRESCINE assimilation | − |
| METHYL-A-D-GLUCOPYRANOSIDE acidification | − | GROWTH IN 6.5% NaCl | − |
| ELLMAN | − | KANAMYCIN RESISTANCE | + |
| METHYL-D-XYLOSIDE | − | OLEANDOMYCIN RESISTANCE | − |
| ALPHA-MANNOSIDASE | − | ESCULIN hydrolyse | + |
| MALTOTRIOSE | − | TETRAZOLIUM RED | − |
| Glycine ARYLAMIDASE | − | POLYMIXIN_B RESISTANCE | + |
| Antibiotics | Zone of Inhibitions (mm) |
|---|---|
| Streptomycin (10 µg) | 22, S |
| Clindamycin (5 µg) | 16, I |
| Doxycycline–HCl (30 µg) | 14, S |
| Vancomycin (10 µg) | 11, S |
| Gentamycin (10 µg) | 37, S |
| Azithromycin (15 µg) | R |
| Lincomycin (10 µg) | R |
| Chloromphenicol (30 µg) | R |
| Comparative NMR Data | ||||
|---|---|---|---|---|
| a* Published Data | b Isolated Compound | |||
| Position | δC | δH (J in Hz) | δC | δH (J in Hz) |
| 2 | 162.8 | - | 162.4 | - |
| 3 | 106.6 | 6.86 (s) | 107.14 | 6.96 (1H, d, J = 2.4) |
| 4 | 176.4 | - | 176.9 | - |
| 5 | 126.5 | 7.86 (d, J = 8.7) | 127.07 | 7.85 (1H, d, J = 9.2) |
| 6 | 115.1 | 6.91 (dd, J = 2.3) | 115.60 | 6.88 (1H, m) |
| 7 | 161.9 | 163.3 | - | |
| 8 | 102.6 | 6.97 (d, J =2.2) | 103.05 | 6.88 (1H, m) |
| 9 | 157.5 | - | 158 | - |
| 10 | 116.1 | - | 116.67 | - |
| 1′ | 131.3 | - | 131.8 | - |
| 2′ | 126.2 | 8.02 (d, J = 7.9) | 126.68 | 8.01 (1H, m) |
| 3′ | 129.1 | 7.52 (m) | 129.59 | 7.52 (1H, m) |
| 4′ | 131.6 | 7.52 (m) | 132.1 | 7.52 (1H, m) |
| 5′ | 129.1 | 7.52 (m) | 129.59 | 7.52 (1H, m) |
| 6′ | 126.2 | 8.02 (d, J = 7.9) | 126.68 | 8.01 (1H, m) |
| 7-OH | 10.80 (s) | - | 10.92 | |
| Pathogens | MIC (µg/mL) | Standards (µg/mL) |
|---|---|---|
| B. cereus | 3.12 | 0.8 |
| S. aureus | 1.6 | 0.8 |
| S. flexneri | 6.25 | 1.6 |
| C. glabrata | 3.12 | 0.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Math, H.H.; Kumar, R.S.; Chakraborty, B.; Almansour, A.I.; Perumal, K.; Kantli, G.B.; Nayaka, S. Antimicrobial Efficacy of 7-Hydroxyflavone Derived from Amycolatopsis sp. HSN-02 and Its Biocontrol Potential on Cercospora Leaf Spot Disease in Tomato Plants. Antibiotics 2023, 12, 1175. https://doi.org/10.3390/antibiotics12071175
Math HH, Kumar RS, Chakraborty B, Almansour AI, Perumal K, Kantli GB, Nayaka S. Antimicrobial Efficacy of 7-Hydroxyflavone Derived from Amycolatopsis sp. HSN-02 and Its Biocontrol Potential on Cercospora Leaf Spot Disease in Tomato Plants. Antibiotics. 2023; 12(7):1175. https://doi.org/10.3390/antibiotics12071175
Chicago/Turabian StyleMath, Halaswamy Hire, Raju Suresh Kumar, Bidhayak Chakraborty, Abdulrahman I. Almansour, Karthikeyan Perumal, Girish Babu Kantli, and Sreenivasa Nayaka. 2023. "Antimicrobial Efficacy of 7-Hydroxyflavone Derived from Amycolatopsis sp. HSN-02 and Its Biocontrol Potential on Cercospora Leaf Spot Disease in Tomato Plants" Antibiotics 12, no. 7: 1175. https://doi.org/10.3390/antibiotics12071175
APA StyleMath, H. H., Kumar, R. S., Chakraborty, B., Almansour, A. I., Perumal, K., Kantli, G. B., & Nayaka, S. (2023). Antimicrobial Efficacy of 7-Hydroxyflavone Derived from Amycolatopsis sp. HSN-02 and Its Biocontrol Potential on Cercospora Leaf Spot Disease in Tomato Plants. Antibiotics, 12(7), 1175. https://doi.org/10.3390/antibiotics12071175