New Antimicrobial Resistance Strategies: An Adaptive Resistance Network Conferring Reduced Glycopeptide Susceptibility in VISA
Abstract
1. Introduction
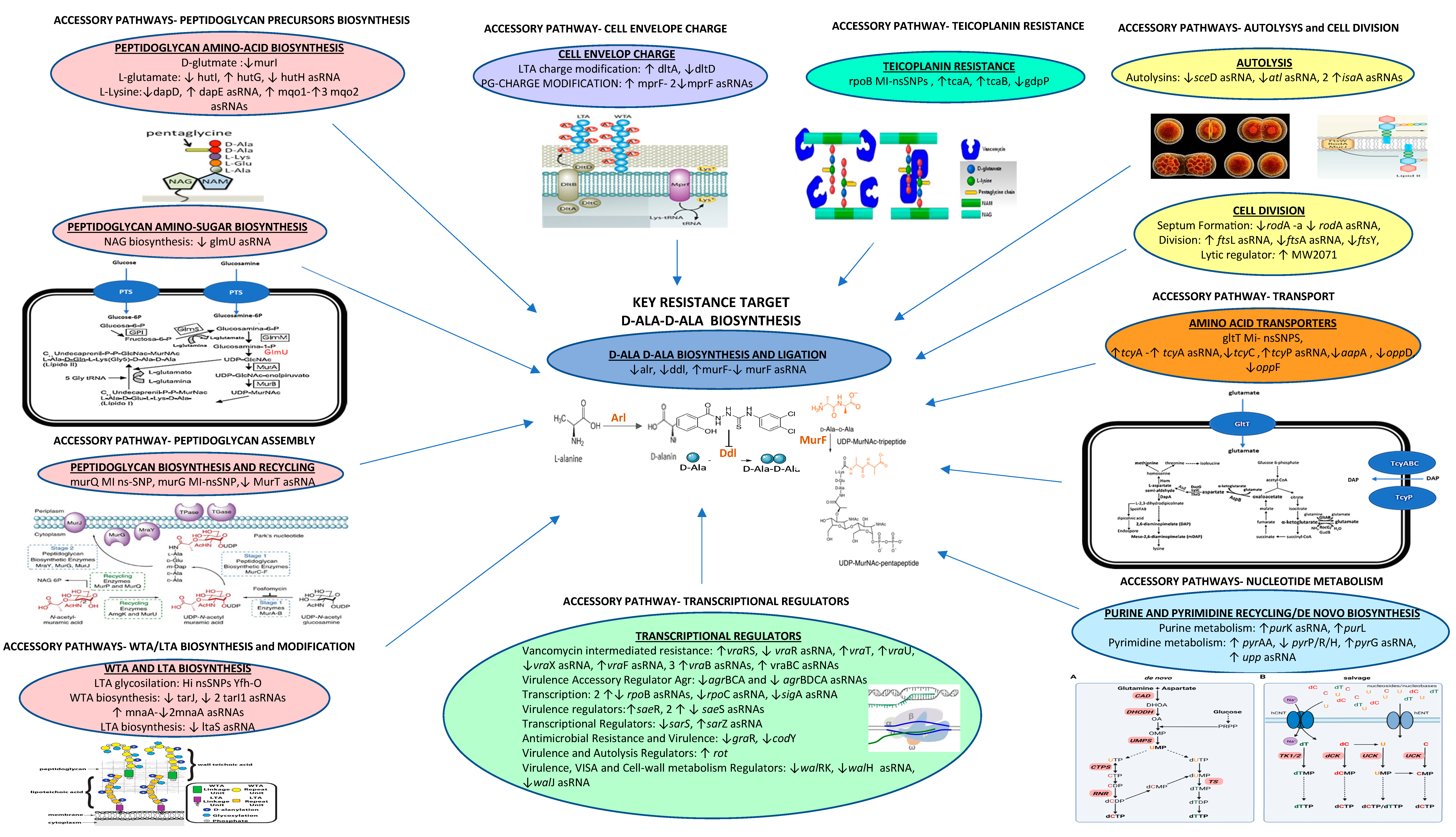
2. Results
2.1. VISA-Related nsSNPs
2.2. Comparative Transcriptomics
2.3. Comparative Analysis of nsSNPs in Intergenic Genomic Regions
2.4. Validation of the Transcriptional Trends
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Strains
5.2. Whole-Genome Sequencing
5.3. De Novo Genome Assembly
5.4. Gene Annotation
5.5. Single Nucleotide Variants (SNVs)
5.6. Predicted Effects of Whole-Genome Single Nucleotide Polymorphisms (wgSNPs)
5.7. Phylogeny and Genomic Epidemiology
5.8. RNA-Seq
5.8.1. RNA-Seq Bacterial Cultures
5.8.2. RNA-Seq Libraries
5.8.3. RNA Extraction
5.8.4. Preparation of the Tru-Seq Library
5.8.5. Preparation of the Short-Insert Library
5.8.6. Post-Processing of the Tru-Seq Library’s Raw Reads
5.8.7. Post-Processing of the Short-Insert Library’s Raw Reads
5.8.8. Analysis of the Tru-Seq and Short-Insert Reads
5.9. DAVID Enrichment Analysis
5.10. Real-Time qPCR Validation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Howden, B.P.; Davies, J.K.; Johnson, P.D.R.; Stinear, T.P.; Grayson, M.L. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: Resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 2010, 23, 99–139. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Ma, X.; Sato, K.; Okuma, K.; Tenover, F.C.; Mamizuka, E.M.; Gemmell, C.G.; Kim, M.N.; Ploy, M.C.; El Solh, N.; et al. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 2003, 41, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Iwamoto, A.; Lian, J.Q.; Neoh, H.M.; Maruyama, T.; Horikawa, Y.; Hiramatsu, K. Novel mechanism of antibiotic resistance originating in vancomycin-Intermediate Staphylococcus aureus. Antimicrob. Agents. Chemother. 2006, 50, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.M.; Filipe, S.R.; Tomasz, A.; Pinho, M.G. Fluorescence ratio imaging microscopy shows decreased access of vancomycin to cell wall synthetic sites in vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents. Chemother. 2007, 51, 3627–3633. [Google Scholar] [CrossRef]
- Gardete, S.; Kim, C.; Hartmann, B.M.; Mwangi, M.; Roux, C.M.; Dunman, P.M.; Chambers, H.F.; Tomasz, A. Genetic pathway in acquisition and loss of vancomycin resistance in a methicillin resistant Staphylococcus aureus (MRSA) strain of clonal type USA300. PLOS Pathog. 2012, 8, e1002505. [Google Scholar] [CrossRef]
- Boyle-Vavra, S.; Berke, S.K.; Lee, J.C.; Daum, R.S. Reversion of the glycopeptide resistance phenotype in Staphylococcus aureus clinical isolates. Antimicrob. Agents. Chemother. 2000, 44, 272–277. [Google Scholar] [CrossRef]
- Boyle-Vavra, S.; Carey, R.B.; Daum, R.S. Development of vancomycin and lysostaphin resistance in a methicillin-resistant Staphylococcus aureus isolate. J. Antimicrob. Chemother. 2001, 48, 617–625. [Google Scholar] [CrossRef]
- Daum, R.S.; Gupta, S.; Sabbagh, R.; Milewski, W.M. Characterization of Staphylococcus aureus isolates with decreased susceptibility to vancomycin and teicoplanin: Isolation and purification of a constitutively produced protein associated with decreased susceptibility. J. Infect. Dis. 1992, 166, 1066–1072. [Google Scholar] [CrossRef]
- Hanaki, H.; Kuwahara-Arai, K.; Boyle-Vavra, S.; Daum, R.; Labischinski, H.; Hiramatsu, K. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 1998, 42, 199–209. [Google Scholar] [CrossRef]
- Moreira, B.; Boyle-Vavra, S.; Daum, R.S. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1997, 41, 1788–1793. [Google Scholar] [CrossRef]
- Howden, B.P.; Johnson, P.D.; Ward, P.B.; Stinear, T.P.; Davies, J.K. Isolates with low-level vancomycin resistance associated with persistent methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 2006, 50, 3039–3047. [Google Scholar] [CrossRef]
- Boyle-Vavra, S.; Challapalli, M.; Daum, R.S. Resistance to autolysis in vancomycin-selected Staphylococcus aureus isolates precedes vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 2003, 47, 2036–2039. [Google Scholar] [CrossRef] [PubMed]
- Boyle-Vavra, S.; Labischinski, H.; Ebert, C.C.; Ehlert, K.; Daum, R.S. A spectrum of changes occurs in peptidoglycan composition of glycopeptide-intermediate clinical Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 2001, 45, 280–287. [Google Scholar] [CrossRef]
- Renzoni, A.; Barras, C.; François, P.; Charbonnier, Y.; Huggler, E.; Garzoni, C.; Kelley, W.L.; Majcherczyk, P.; Schrenzel, J.; Lew, D.P.; et al. Transcriptomic and functional analysis of an autolysis-deficient, teicoplanin-resistant derivative of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 3048–3061. [Google Scholar] [CrossRef]
- Scherl, A.; François, P.; Charbonnier, Y.; Deshusses, J.M.; Koessler, T.; Huyghe, A.; Bento, M.; Stahl-Zeng, J.; Fischer, A.; Masselot, A.; et al. Exploring glycopeptide-resistance in Staphylococcus aureus: A combined proteomics and transcriptomics ap-proach for the identification of resistance-related markers. BMC Genom. 2006, 7, 296. [Google Scholar] [CrossRef] [PubMed]
- Vaudaux, P.; Francois, P.; Berger-Bächi, B.; Lew, D.P. In vivo emergence of subpopulations expressing teicoplanin or vancomycin resistance phenotypes in a glycopeptide-susceptible, methicillin-resistant strain of Staphylococcus aureus. J. Antimicrob. Chemother. 2001, 47, 163–170. [Google Scholar] [CrossRef]
- Koehl, J.L.; Muthaiyan, A.; Jayaswal, R.K.; Ehlert, K.; Labischinski, H.; Wilkinson, B.J. Cell wall composition and decreased autolytic activity and lysostaphin susceptibility of glycopeptide-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 2004, 48, 3749–3757. [Google Scholar] [CrossRef]
- McCallum, N.; Karauzum, H.; Getzmann, R.; Bischoff, M.; Majcherczyk, P.; Berger-Bächi, B.; Landmann, R. In vivo survival of teicoplanin-resistant Staphylococcus aureus and fitness cost of teicoplanin resistance. Antimicrob. Agents Chemother. 2006, 50, 2352–2360. [Google Scholar] [CrossRef] [PubMed]
- Muthaiyan, A.; Jayaswal, R.K.; Wilkinson, B.J. Intact mutS in laboratory-derived and clinical glycopeptide-intermediate Staphylococcus aureus strains. Antimicrob. Agents Chemother. 2004, 48, 623–625. [Google Scholar] [CrossRef]
- Pfeltz, R.F.; Singh, V.K.; Schmidt, J.L.; Batten, M.A.; Baranyk, C.S.; Nadakavukaren, M.J.; Jayaswal, R.K.; Wilkinson, B.J. Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds. Antimicrob. Agents Chemother. 2000, 44, 294–303. [Google Scholar] [CrossRef]
- Sakoulas, G.; Eliopoulos, G.M.; Moellering, R.C., Jr.; Wennersten, C.; Venkataraman, L.; Novick, R.P.; Gold, H.S. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 2002, 46, 1492–1502. [Google Scholar] [CrossRef] [PubMed]
- Mwangi, M.M.; Wu, S.W.; Zhou, Y.; Sieradzki, K.; de Lencastre, H.; Richardson, P.; Bruce, D.; Rubin, E.; Myers, E.; Siggia, E.D.; et al. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 2007, 104, 9451–9456. [Google Scholar] [CrossRef] [PubMed]
- Sieradzki, K.; Roberts, R.B.; Haber, S.W.; Tomasz, A. The Development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 1999, 340, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Roch, M.; Clair, P.; Renzoni, A.; Reverdy, M.E.; Dauwalder, O.; Bes, M.; Martra, A.; Freydière, A.M.; Laurent, F.; Reix, P.; et al. Exposure of Staphylococcus aureus to subinhibitory concentrations of β-Lactam antibiotics induces heteroge-neous vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 2014, 58, 530614. [Google Scholar] [CrossRef]
- Haaber, J.; Friberg, C.; McCreary, M.; Lin, R.; Cohen, S.N.; Ingmer, H. Reversible antibiotic tolerance induced in Staphylococcus aureus by concurrent drug exposure. mBio 2015, 6, e02268-14. [Google Scholar] [CrossRef]
- Cafiso, V.; Stracquadanio, S.; Lo Verde, F.; De Guidi, I.; Zega, A.; Pigola, G.; Stefani, S. Genomic and Long-Term Transcriptomic Imprints Related to the Daptomycin Mechanism of Action Occurring in Daptomycin- and Methicillin-Resistant Staphylococcus aureus Under Daptomycin Exposure. Front. Microbiol. 2020, 11, 1893. [Google Scholar] [CrossRef]
- Mishra, N.N.; McKinnell, J.; Yeaman, M.R.; Rubio, A.; Nast, C.C.; Chen, L.; Kreiswirth, B.N.; Bayer, A.S. In vitro cross-resistance to daptomycin and host defense cationic antimicrobial peptides in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 2011, 55, 4012–4018. [Google Scholar] [CrossRef]
- Steinkraus, G.; White, R.; Friedrich, L. Vancomycin MIC creep in non-vancomycin-intermediate Staphylococcus aureus (VISA), vancomycin-susceptible clinical methicillin-resistant S. aureus (MRSA) blood isolates from 2001–05. J. Antimicrob. Chemother. 2007, 60, 788–794. [Google Scholar] [CrossRef]
- Howe, R.A.; Monk, A.; Wootton, M.; Walsh, T.R.; Enright, M.C. Vancomycin susceptibility within methicillin-resistant Staphylococcus aureus lineages. Emerg. Infect. Dis. 2004, 10, 855–857. [Google Scholar] [CrossRef]
- Meehl, M.; Herbert, S.; Götz, F.; Cheung, A. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2007, 51, 2679–2689. [Google Scholar] [CrossRef]
- McEvoy, C.R.E.; Tsuji, B.; Gao, W.; Seemann, T.; Porter, J.L.; Doig, K.; Ngo, D.; Howden, B.P.; Stinear, T.P. Decreased vancomycin susceptibility in Staphylococcus aureus caused by IS256 tempering of walKR expression. Antimicrob. Agents Chemother. 2013, 57, 3240–3249. [Google Scholar] [CrossRef] [PubMed]
- Herbert, S.; Bera, A.; Nerz, C.; Kraus, D.; Peschel, A.; Goerke, C.; Meehl, M.; Cheung, A.; Götz, F. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog. 2007, 3, e102. [Google Scholar] [CrossRef]
- Howden, B.P.; Smith, D.J.; Mansell, A.; Johnson, P.D.; Ward, P.B.; Stinear, T.P.; Davies, J.K. Different bacterial gene expression patterns and attenuated host immune responses are associated with the evolution of low-level vancomycin resistance during persistent methicillin-resistant Staphylococcus aureus bacteraemia. BMC Microbiol. 2008, 8, 39. [Google Scholar] [CrossRef]
- Nishi, H.; Komatsuzawa, H.; Fujiwara, T.; McCallum, N.; Sugai, M. Reduced content of lysyl-phosphatidylglycerol in the cytoplasmic membrane affects susceptibility to moenomycin, as well as vancomycin, gentamicin, and antimicrobial peptides, in Staphylococcus aureus. Antimicrob. Agents Chemother. 2004, 48, 4800–4807. [Google Scholar] [CrossRef]
- Peschel, A.; Otto, M.; Jack, R.W.; Kalbacher, H.; Jung, G.; Götz, F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 1999, 274, 8405–8410. [Google Scholar] [CrossRef] [PubMed]
- Ruzin, A.; Severin, A.; Moghazeh, S.L.; Etienne, J.; Bradford, P.A.; Projan, S.J.; Shlaes, D.M. Inactivation of mprF affects vancomycin susceptibility in Staphylococcus aureus. Biochim. Biophys. Acta 2003, 1621, 117–121. [Google Scholar] [CrossRef]
- McAleese, F.; Wu, S.W.; Sieradzki, K.; Dunman, P.; Murphy, E.; Projan, S.; Tomasz, A. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S. aureus-type resistance to vancomycin. J. Bacteriol. 2006, 188, 1120–1133. [Google Scholar] [CrossRef] [PubMed]
- Utaida, S.; Dunman, P.M.; Macapagal, D.; Murphy, E.; Projan, S.J.; Singh, V.K.; Jayaswal, R.K.; Wilkinson, B.J. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 2003, 149, 2719–2732. [Google Scholar] [CrossRef] [PubMed]
- Gardete, S.; Wu, S.; Gill, S.; Tomasz, A. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 3424–3434. [Google Scholar] [CrossRef] [PubMed]
- Howden, B.P.; Stinear, T.P.; Allen, D.L.; Johnson, P.D.; Ward, P.B.; Davies, J.K. Genomic analysis reveals a point mutation in the two-component sensor gene graS that leads to intermediate vancomycin resistance in clinical Staphylococcus aureus. Antimicrob. Agents Chemother. 2008, 52, 3755–3762. [Google Scholar] [CrossRef]
- Kuroda, M.; Kuroda, H.; Oshima, T.; Takeuchi, F.; Mori, H.; Hiramatsu, K. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 2003, 49, 80721. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Isii, T.; Fukuda, M.; Ochiai, T.; Neoh, H.M.; Camargo, I.L.; Watanabe, Y.; Shoji, M.; Hishinuma, T.; Hiramatsu, K. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 2010, 54, 5222–5233. [Google Scholar] [CrossRef]
- Matsuo, M.; Hishinuma, T.; Katayama, Y.; Cui, L.; Kapi, M.; Hiramatsu, K. Mutation of RNA polymerase beta subunit (rpoB) promotes hVISA to-VISA phenotypic conversion of strain Mu3. Antimicrob. Agents Chemother. 2011, 55, 4188–4195. [Google Scholar] [CrossRef] [PubMed]
- Capone, A.; Cafiso, V.; Campanile, F.; Parisi, G.; Mariani, B.; Petrosillo, N.; Stefani, S. In Vivo development of daptomycin resistance in vancomycin-susceptible methicillin-resistant Staphylococcus aureus severe infections previously treated with glycopeptides. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Salemi, R.; Zega, A.; Aguglia, E.; Lo Verde, F.; Pigola, G.; Stefani, S.; Cafiso, V. Balancing the Virulence and Antimicrobial Resistance in VISA DAP-R CA-MRSA Superbug. Antibiotics 2022, 11, 1159. [Google Scholar] [CrossRef] [PubMed]
- Saberi, F.; Kamali, M.; Najafi, A.; Yazdanparast, A.; Moghaddam, M.M. Natural antisense RNAs as mRNA regulatory elements in bacteria: A review on function and applications. Cell. Mol. Biol. Lett. 2016, 21, 6. [Google Scholar] [CrossRef]
- Peschel, A.; Vuong, C.; Otto, M.; Götz, F. The D-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob. Agents Chemother. 2000, 44, 2845–2847. [Google Scholar] [CrossRef]
- Paige, M.; Fox, M.W.C.; Gordon, L.A. Lack of Relationship between Purine Biosynthesis and Vancomycin Resistance in Staphylococcus aureus: A Cautionary Tale for Microarray Interpretation. Antimicrob. Agents Chemother. 2007, 51, 1274–1280. [Google Scholar]
- Gillen, A.L.; Conrad, J.; Cargill, M. The Genesis and Emergence of Community-Associated Methicillin-Resistant Staphylococcus aureus (CA-MRSA): An Example of Evolution in Action? Answ. Res. J. 2015, 8, 391–401. [Google Scholar]
- Cafiso, V.; Bertuccio, T.; Spina, D.; Purrello, S.; Campanile, F.; Di Pietro, C.; Purrello, M.; Stefani, S. Modulating activity of vancomycin and daptomycin on the expression of autolysis cell-wall turnover and membrane charge genes in hVISA and VISA strains. PLoS ONE. 2012, 7, e29573. [Google Scholar] [CrossRef]
- Griffiths, J.M.; O’Neill, A.J. Loss of function of the gdpP protein leads to joint β-lactam/glycopeptide tolerance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012, 56, 579–581. [Google Scholar] [CrossRef]
- Cafiso, V.; Bertuccio, T.; Purrello, S.; Campanile, F.; Mammina, C.; Sartor, A.; Raglio, A.; Stefani, S. dltA overexpression: A strain-independent keystone of daptomycin resistance in methicillin-resistant Staphylococcus aureus. Int. J. Antimicrob. Agents 2014, 43, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Almeida, S.T.; Paulo, A.C.; Babo, J.; Borralho, J.; Figueiredo, C.; Gonçalves, B.; Lança, J.; Louro, M.; Morais, H.; Queiroz, J.; et al. Absence of methicillin-resistant Staphylococcus aureus colonization among immunocompetent healthy adults: Insights from a longitudinal study. PLoS ONE 2021, 16, e0253739. [Google Scholar] [CrossRef] [PubMed]
- EUCAST. Available online: https://www.eucast.org/ecast_news/news_sileview/?tx_ttnews%5Btt_news%5D=459cHash=160a5b91371e598957e10178fb3aa143 (accessed on 1 July 2022).
- Cafiso, V.; Lo Verde, F.; Zega, A.; Pigola, G.; Rostagno, R.; Borrè, S.; Stefani, S. Genomic Characterization of a New Biofilm Forming and Adhesive ST398 Human-Adapted MSSA Lineage Causing Septic Knee Arthritis Following Surgical Reconstruction. Microorganisms 2021, 9, 305. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, l.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Bartels, M.D.; Petersen, A.; Worning, P.; Nielsen, J.B.; Larner-Svensson, H.; Johansen, H.K.; Andersen, L.P.; Jarløv, J.O.; Boye, K.; Larsen, A.R.; et al. Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2014, 52, 4305–4308. [Google Scholar] [CrossRef]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef]
- Carattoli, A.; Hasman, H. PlasmidFinder and in silico pMLST: Identification and typing of plasmid replicons in whole-genome sequencing (WGS). In Horizontal Gene Transfer; Humana: New York, NY, USA, 2020; pp. 285–294. [Google Scholar]
- Johansson, M.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A fast phage search tool. Nucleic Acids. Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef]
- Van Der Mee-Marquet, N.; Corvaglia, A.R.; Valentin, A.S.; Hernandez, D.; Bertrand, X.; Girard, M.; Kluytmansf, J.; Donnio, P.Y.; Francois, P. Analysis of prophages harboured by the human-adapted subpopulation of Staphylococcus aureus CC398. Infect. Genet. Evol. 2013, 18, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Ramadhan, A.A.; Hegedus, E. Survivability of vancomycin resistant enterococci and fitness cost of vancomycin resistance acquisition. J. Clin. Pathol. 2005, 58, 744–746. [Google Scholar] [CrossRef]
- Cafiso, V.; Stracquadanio, S.; Lo Verde, F.; Dovere, V.; Zega, A.; Pigola, G.; Aranda, J.; Stefani, S. COLR Acinetobacter baumannii sRNA Signatures: Computational Comparative Identification and Biological Targets. Front. Microbiol. 2020, 17, 3075. [Google Scholar] [CrossRef] [PubMed]
| Gene | Function | Product | nsSNPs AA Changes |
|---|---|---|---|
| Peptidoglycan Assembly | |||
| MW0165 | N-acetyl muramic acid recycling | N-acetylmuramic acid 6-phosphate etherase MurQ | MI-nsSNPs Ile156Asn |
| MW1307 | Lipid II PG substrate for biosynthesis of peptidoglycan | UDP-NAG-NAM-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase MurG | MI-nsSNPs Ile121Asn |
| Biosynthesis of Peptidoglycan AA | |||
| MW2286 | Intermediate metabolic compound of lysine biosynthesis | Malate:quinone oxidoreductase 1 Mqo1 | MI-nsSNPs Val280Glu |
| MW1283 | L-lysine biosynthesis via the DAP pathway | Dihydrodipicolinate synthase DapA | MI-nsSNPs Ala101Thr |
| Biosynthesis of lipotheicoic acid | |||
| MW1125 | LTA glycosylation | YfhO membrane protein | HI-nsSNPs Gly75 * |
| Charge of the Cell Envelope | |||
| MW1247 | Charge of the cell envelope | Phosphatidylglycerol lysyl-transferase MprF | MI-nsSNPs Thr345Ala Leu538Phe |
| Regulators of Vancomycin-Intermediate Resistance | |||
| MW1826 | Methicillin resistance and activation of vraSR TCRS | VraT | MI-nsSNPs Ala59Glu |
| Glycopeptide-Β-Lactams Resistant-Related Genes | |||
| MW0014 | β-lactams and/or glycopeptide cross-resistance | GdpP | MI-nsSNPs Ile186Met |
| Metabolic Substrate Transporters | |||
| MW2304 | Proton/sodium-glutamate symport | GltT | MI-nsSNPs Val232Glu |
| CA-MRSA MW2 Locus Tag | sRNA | RefGen Position (nt) | sRNA Size (bp) | Library | RPKM 1R | RPKM 1S | |
|---|---|---|---|---|---|---|---|
| MW1303 | Predicted antisense small RNA | 1423626 | 1423663 | 37 | SI | 969 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguglia, E.; Chines, E.; Stefani, S.; Cafiso, V. New Antimicrobial Resistance Strategies: An Adaptive Resistance Network Conferring Reduced Glycopeptide Susceptibility in VISA. Antibiotics 2023, 12, 783. https://doi.org/10.3390/antibiotics12040783
Aguglia E, Chines E, Stefani S, Cafiso V. New Antimicrobial Resistance Strategies: An Adaptive Resistance Network Conferring Reduced Glycopeptide Susceptibility in VISA. Antibiotics. 2023; 12(4):783. https://doi.org/10.3390/antibiotics12040783
Chicago/Turabian StyleAguglia, Elvira, Eleonora Chines, Stefania Stefani, and Viviana Cafiso. 2023. "New Antimicrobial Resistance Strategies: An Adaptive Resistance Network Conferring Reduced Glycopeptide Susceptibility in VISA" Antibiotics 12, no. 4: 783. https://doi.org/10.3390/antibiotics12040783
APA StyleAguglia, E., Chines, E., Stefani, S., & Cafiso, V. (2023). New Antimicrobial Resistance Strategies: An Adaptive Resistance Network Conferring Reduced Glycopeptide Susceptibility in VISA. Antibiotics, 12(4), 783. https://doi.org/10.3390/antibiotics12040783






