Antimicrobial and Antibiofilm Effect of Bacteriocin-Producing Pediococcus inopinatus K35 Isolated from Kimchi against Multidrug-Resistant Pseudomonas aeruginosa
Abstract
1. Introduction
2. Results
2.1. Isolation of Antimicrobial LAB from Kimchi
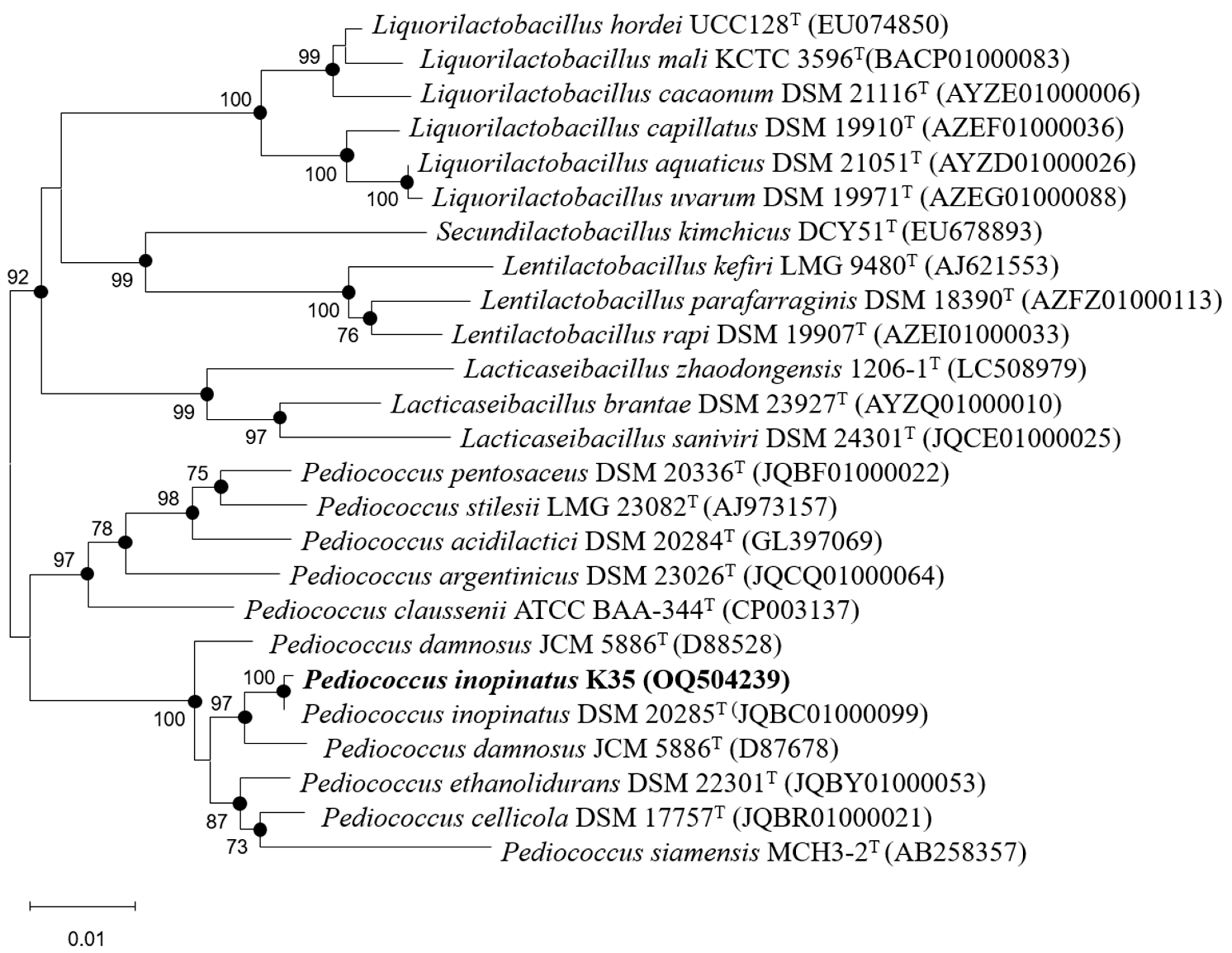
2.2. Identification of Strain K35
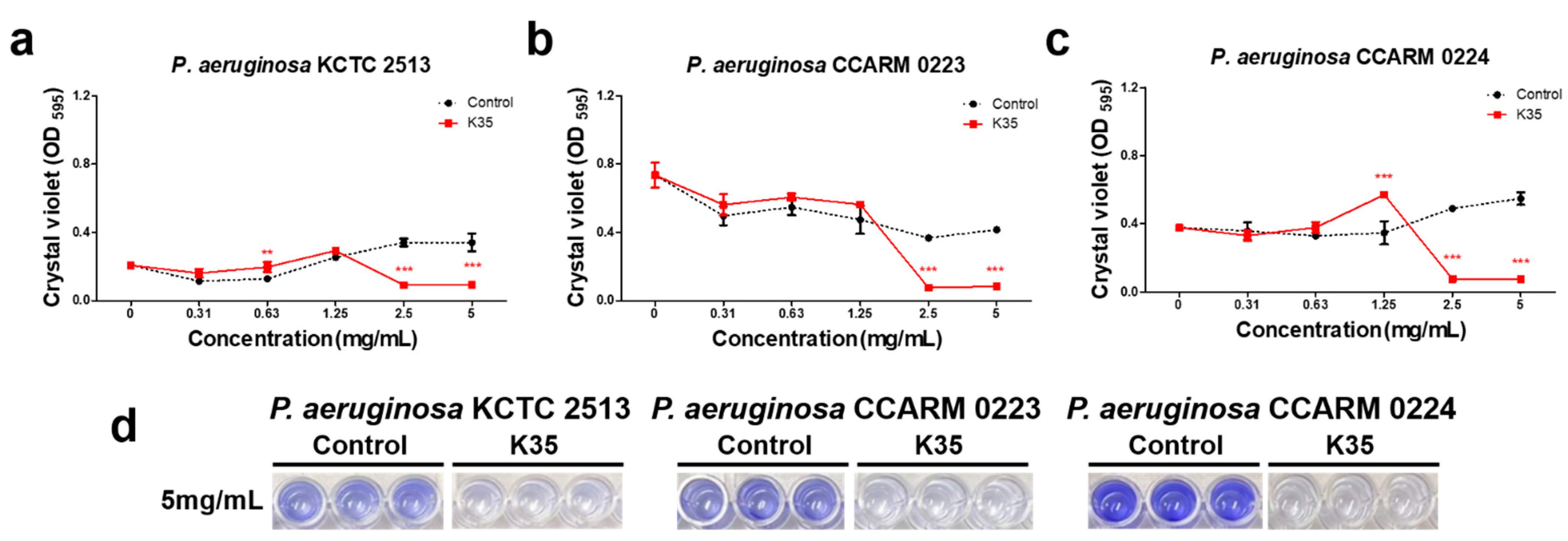
2.3. Biofilm Formation Inhibitory Activity
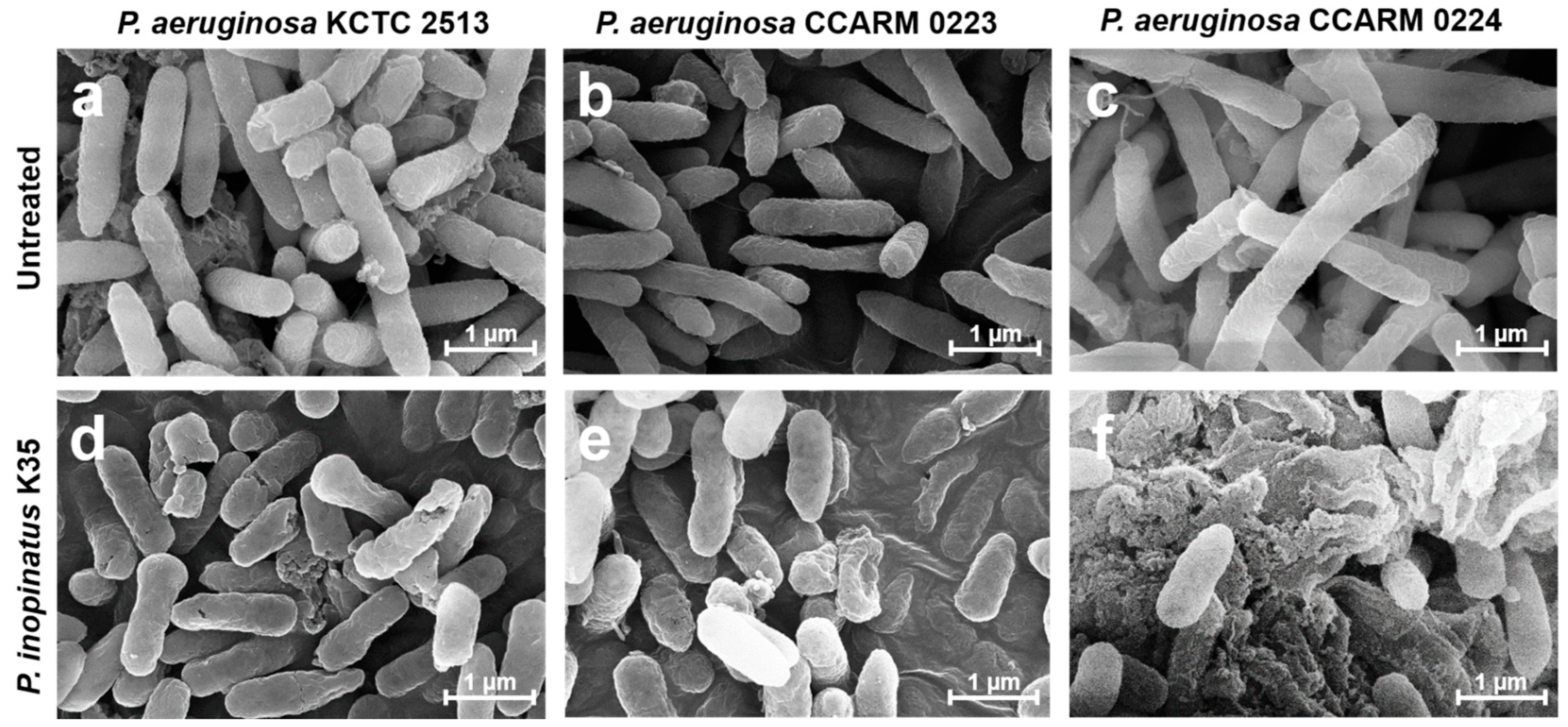
2.4. Observation of the Morphological Changes of Bacterial Cells
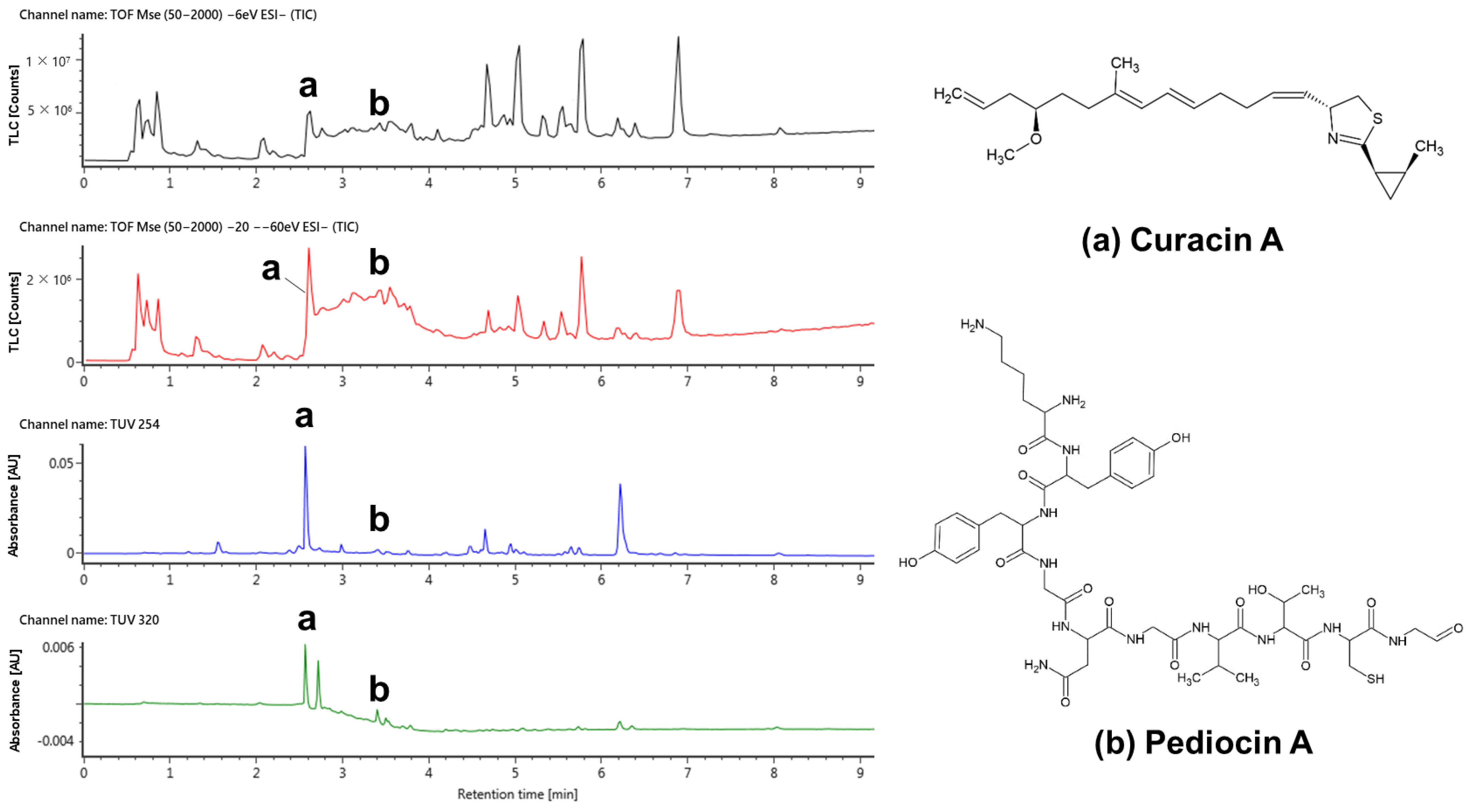
2.5. Detection of Substances with Antimicrobial and Anti-Biofilm-Formation Effects
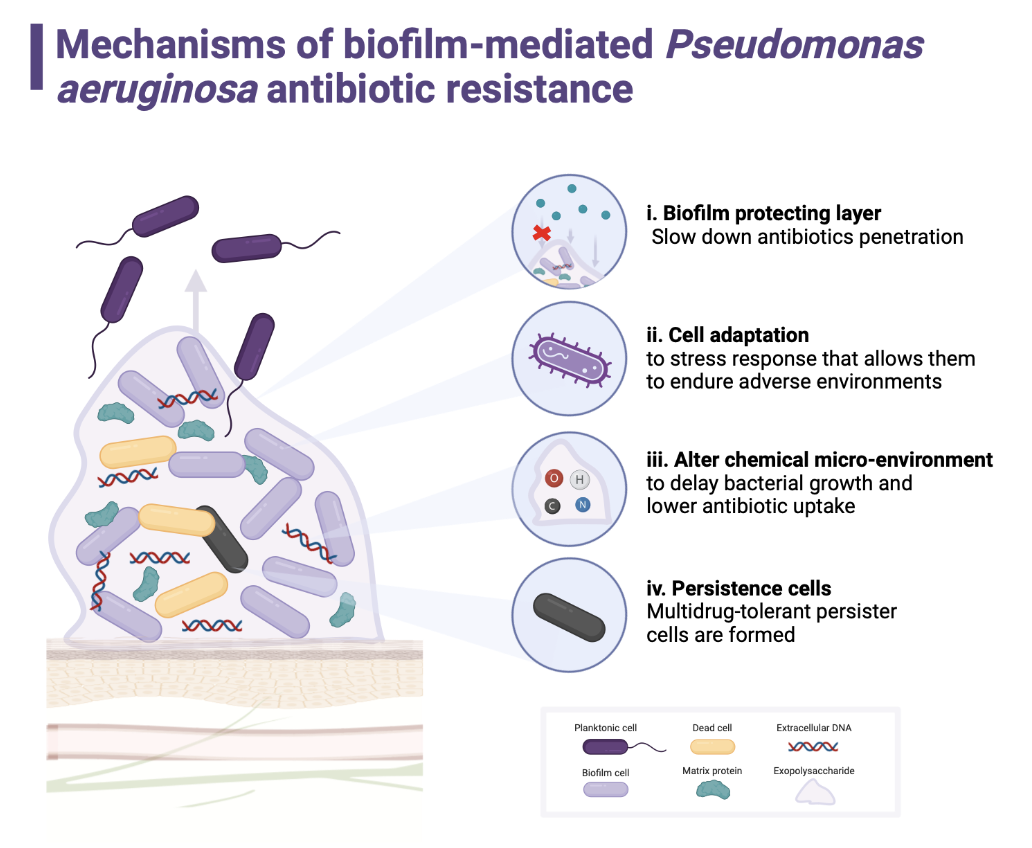
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Isolation and Identification of LAB from Kimchi
5.2. Bacterial Strains and Growth Conditions
5.3. Disc Diffusion Assay
5.4. Broth Microdilution Method
5.5. Biofilm Formation Inhibition Assay
5.6. Scanning Electron Microscopy (SEM) of Bacteria
5.7. UPLC-QTOF-MS Analysis
5.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Strateva, T.; Mitov, I. Contribution of an arsenal of virulence factors to pathogenesis of Pseudomonas aeruginosa infections. Ann. Microbiol. 2011, 61, 717–732. [Google Scholar] [CrossRef]
- Wu, D.C.; Chan, W.W.; Metelitsa, A.I.; Fiorillo, L.; Lin, A.N. Pseudomonas skin infection: Clinical features, epidemiology, and management. Am. J. Clin. Dermatol. 2011, 12, 157–169. [Google Scholar] [CrossRef]
- Breidenstein, E.B.; de la Fuente-Núñez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Henrichfreise, B.; Wiegand, I.; Pfister, W.; Wiedemann, B. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob. Agents Chemother. 2007, 51, 4062–4070. [Google Scholar] [CrossRef]
- Gellatly, S.L.; Hancock, R.E. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef]
- Shokri, D.; Khorasgani, M.R.; Mohkam, M.; Fatemi, S.M.; Ghasemi, Y.; Taheri-Kafrani, A. The inhibition effect of lactobacilli against growth and biofilm formation of Pseudomonas aeruginosa. Probiotics Antimicrob. Proteins 2018, 10, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Marsh, P.D. Dental plaque as a biofilm: The significance of pH in health and caries. Compend. Contin. Educ. Dent. 2009, 30, 76–78, 80, 83. [Google Scholar]
- Taraszkiewicz, A.; Fila, G.; Grinholc, M.; Nakonieczna, J. Innovative strategies to overcome biofilm resistance. BioMed Res. Int. 2013, 2013, 150653. [Google Scholar] [CrossRef]
- Bajaj, B.K.; Claes, I.J.; Lebeer, S. Functional mechanisms of probiotics. J. Microbiol. Biotechnol. Food Sci. 2021, 2021, 321–327. [Google Scholar]
- Vemuri, R.; Shinde, T.; Shastri, M.D.; Perera, A.P.; Tristram, S.; Martoni, C.J.; Gundamaraju, R.; Ahuja, K.D.; Ball, M.; Eri, R. A human origin strain Lactobacillus acidophilus DDS-1 exhibits superior in vitro probiotic efficacy in comparison to plant or dairy origin probiotics. Int. J. Med. Sci. 2018, 15, 840. [Google Scholar] [CrossRef]
- Kumar Bajaj, B.; Andrabi, T.; JJ Claes, I.; Lebeer, S. Bioprospecting for functionally-proficient potential probiotics. Curr. Nutr. Food Sci. 2014, 10, 251–263. [Google Scholar] [CrossRef]
- Alexandre, Y.; Le Berre, R.; Barbier, G.; Le Blay, G. Screening of Lactobacillus spp. for the prevention of Pseudomonas aeruginosa pulmonary infections. BMC Microbiol. 2014, 14, 107. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Ganger, S.; Patil, S. Characterization of Lactobacillus brevis with Potential Probiotic Properties and Biofilm Inhibition against Pseudomonas aeruginosa. Proceedings 2020, 66, 14. [Google Scholar]
- Valdez, J.; Peral, M.; Rachid, M.; Santana, M.; Perdigon, G. Interference of Lactobacillus plantarum with Pseudomonas aeruginosa in vitro and in infected burns: The potential use of probiotics in wound treatment. Clin. Microbiol. Infect. 2005, 11, 472–479. [Google Scholar] [CrossRef]
- Wilson, R.M.; Walker, J.M.; Yin, K. Different Concentrations of Lactobacillus acidophilus cell free filtrate have differing anti-biofilm and immunomodulatory effects. Front. Cell. Infect. Microbiol. 2021, 11, 864. [Google Scholar] [CrossRef]
- Papagianni, M.; Anastasiadou, S. Pediocins: The bacteriocins of Pediococci. Sources, production, properties and applications. Microb. Cell Factories 2009, 8, 3. [Google Scholar] [CrossRef]
- Garvie, E. Genus Pediococcus Claussen 1903, 68. Bergey’s Man. Syst. Bacteriol. 1986, 2, 1075–1079. [Google Scholar]
- Reynolds, D.; Kollef, M. The epidemiology and pathogenesis and treatment of Pseudomonas aeruginosa infections: An update. Drugs 2021, 81, 2117–2131. [Google Scholar] [CrossRef]
- Smith, A.W. Biofilms and antibiotic therapy: Is there a role for combating bacterial resistance by the use of novel drug delivery systems? Adv. Drug Deliv. Rev. 2005, 57, 1539–1550. [Google Scholar] [CrossRef]
- Kang, M.-S.; Lee, Y.-G.; Kim, H.-S.; Chung, H.-S.; Hwang, D.-Y.; Kim, D.-S. Therapeutic Antiallergy Effect of Fermented Soy Curd by Pediococcus inopinatus Y2. J. Life Sci. 2019, 29, 478–483. [Google Scholar]
- Lim, S.-J.; Bae, E.-Y.; Seol, M.-K.; Cho, Y.-j.; Jung, H.-Y.; Kim, B.-O. Change in Lactobacillus brevis GS1022 and Pediococcus inopinatus GS316 in Gajami Sikhae Fermentation. J. Life Sci. 2020, 30, 491–500. [Google Scholar]
- Tateda, K.; Ishii, Y.; Matsumoto, T.; Kobayashi, T.; Miyazaki, S.; Yamaguchi, K. Potential of macrolide antibiotics to inhibit protein synthesis of Pseudomonas aeruginosa: Suppression of virulence factors and stress response. J. Infect. Chemother. 2000, 6, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Benbrook, D.M.; Miller, R.V. Effects of norfloxacin on DNA metabolism in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1986, 29, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ye, C.; Guo, L.; Chen, C.; Kong, X.; Chen, Y.; Shu, L.; Wang, P.; Yu, X.; Fang, J. Assessment of the UV/Chlorine Process in the Disinfection of Pseudomonas aeruginosa: Efficiency and Mechanism. Environ. Sci. Technol. 2021, 55, 9221–9230. [Google Scholar] [CrossRef] [PubMed]
- Khorshidian, N.; Khanniri, E.; Mohammadi, M.; Mortazavian, A.M.; Yousefi, M. Antibacterial activity of pediocin and pediocin-producing bacteria against Listeria monocytogenes in meat products. Front. Microbiol. 2021, 12, 709959. [Google Scholar] [CrossRef] [PubMed]
- Piva, A.; Headon, D.R. Pediocin A, a bacteriocin produced by Pediococcus pentosaceus FBB61. Microbiology 1994, 140, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Rath, C.M.; Scaglione, J.B.; Kittendorf, J.D.; Sherman, D.H. NRPS/PKS hybrid enzymes and their natural products. Chem. Mol. Sci. Chem. Eng. 2010, 1, 453–492. [Google Scholar]
- Vellai, T.; Vida, G. The origin of eukaryotes: The difference between prokaryotic and eukaryotic cells. Proc. R. Soc. London. Ser. B Biol. Sci. 1999, 266, 1571–1577. [Google Scholar] [CrossRef]
- Mills, S.; Ross, R.P.; Hill, C. Bacteriocins and bacteriophage; a narrow-minded approach to food and gut microbiology. FEMS Microbiol. Rev. 2017, 41, S129–S153. [Google Scholar] [CrossRef]
- Fitch, W.M. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst. Biol. 1971, 20, 406–416. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Abdelbasset, M.; Djamila, K. Antimicrobial activity of autochthonous lactic acid bacteria isolated from Algerian traditional fermented milk “Raïb”. Afr. J. Biotechnol. 2008, 7, 2908–2914. [Google Scholar]
- Zaidan, M.; Noor Rain, A.; Badrul, A.; Adlin, A.; Norazah, A.; Zakiah, I. In vitro screening of five local medicinal plants for antibacterial activity using disc diffusion method. Trop. Biomed. 2005, 22, 165–170. [Google Scholar]
- Palepou, M.; Johnson, A.; Cookson, B.; Beattie, H.; Charlett, A.; Woodford, N. Evaluation of disc diffusion and Etest for determining the susceptibility of Staphylococcus aureus to mupirocin. J. Antimicrob. Chemother. 1998, 42, 577–583. [Google Scholar] [CrossRef] [PubMed]
| No. | Source | LAB Strain Name | Inhibition | Citation |
|---|---|---|---|---|
| 1 | Oral cavity | Limosilactobacillus fermentum ES.A.2 | 95% | [12] |
| 2 | Limosilactobacillus fermentum ES.F.115 | 93% | ||
| 3 | Limosilactobacillus fermentum ES.A.6a | 88% | ||
| 4 | Limosilactobacillus fermentum ES.A.1a | 88% | ||
| 5 | Milk and yogurt | Limosilactobacillus fermentum L1 | 100% | [6] |
| 6 | Limosilactobacillus fermentum L2 | 100% | ||
| 7 | Milk, yogurt, buttermilk, idli batter | Levilactobacillus brevis SII | 52.63% | [13] |
| 8 | Sauerkraut | Lactiplantibacillus plantarum ATCC 10241 | 100% | [14] |
| 9 | Human | Lactobacillus acidophilus ATCC 4356T | 99.99% | [15] |
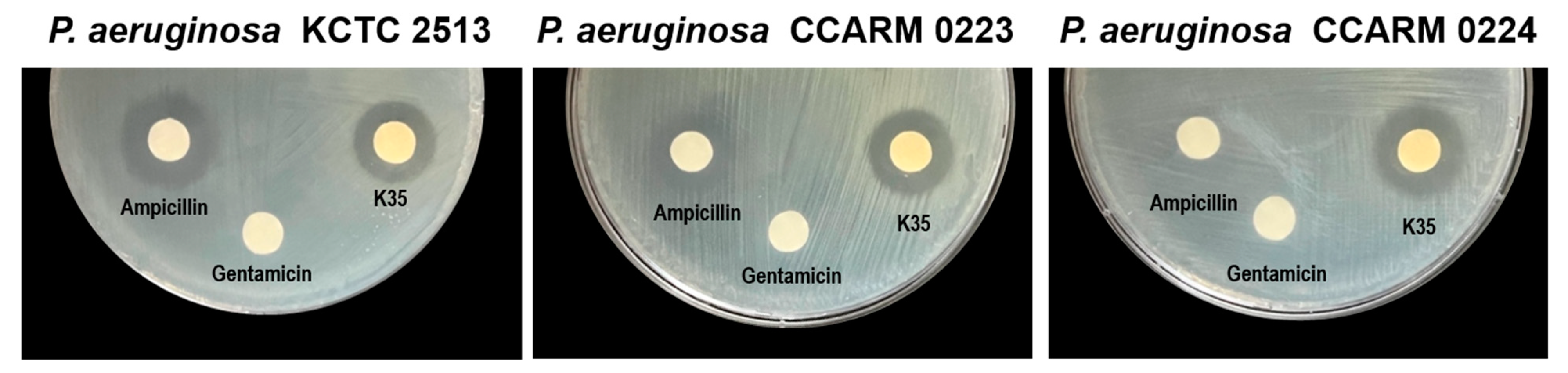
| Strain | Zone of Inhibition (mm) | |||
|---|---|---|---|---|
| P. aeruginosa KCTC 2513 | P. aeruginosa CCARM 0223 | P. aeruginosa CCARM 0224 | ||
| LAB supernatant (100 µL/disc) | ||||
| P. inopinatus K35 | 17.5 ± 0.87 | 17.33 ± 1.26 | 18.5 ± 0.50 | |
| Antibiotic | ||||
| Ampicillin (5 mg/disc) | 20.17 ± 1.04 | 16.67 ± 0.58 | ND | |
| Gentamicin (2 µg/disc) | ND * | ND | ND | |
| Strain | MIC (mg/mL) | MBC (mg/mL) |
|---|---|---|
| P. aeruginosa KCTC 2513 | ≥2.5 | 5 |
| P. aeruginosa CCARM 0223 | ≥2.5 | 5 |
| P. aeruginosa CCARM 0224 | ≥2.5 | 5 |
| Characteristics | K35 | DSM 20285T |
|---|---|---|
| Isolation source | Kimchi | Brewery yeast |
| Growth at | ||
| 25 °C | + | + |
| 30 °C | + | + |
| 37 °C | w | w |
| 45 °C | w | w |
| Acid produced from: | ||
| D-galactose | + | + |
| D-glucose | + | + |
| D-fructose | + | + |
| D-mannose | + | + |
| Methyl-α-D-mannopyranoside | + | − |
| N-acetylglucosamine | + | + |
| Amygdalin | w | + |
| Esculin ferric citrate | + | + |
| Salicin | + | + |
| D-cellobiose | + | + |
| D-maltose | w | w |
| D-trehalose | + | + |
| Gentiobiose | + | + |
| D-turanose | + | − |
| No. | Compound | Observed RT (min) | Formula | Theoretical Mass (m/z) | Observed Mass (m/z) | Mass Error (mDa) | Adducts |
|---|---|---|---|---|---|---|---|
| 1 | Curacin A | 2.61 | C23H35NOS | 372.2367 | 372.2344 | −2.2 | -H |
| 2 | Pediocin A | 3.37 | C46H68N12O14S | 1043.4626 | 1043.4577 | −4.9 | -H |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, E.-J.; Kim, A.-J. Antimicrobial and Antibiofilm Effect of Bacteriocin-Producing Pediococcus inopinatus K35 Isolated from Kimchi against Multidrug-Resistant Pseudomonas aeruginosa. Antibiotics 2023, 12, 676. https://doi.org/10.3390/antibiotics12040676
Yi E-J, Kim A-J. Antimicrobial and Antibiofilm Effect of Bacteriocin-Producing Pediococcus inopinatus K35 Isolated from Kimchi against Multidrug-Resistant Pseudomonas aeruginosa. Antibiotics. 2023; 12(4):676. https://doi.org/10.3390/antibiotics12040676
Chicago/Turabian StyleYi, Eun-Ji, and Ae-Jung Kim. 2023. "Antimicrobial and Antibiofilm Effect of Bacteriocin-Producing Pediococcus inopinatus K35 Isolated from Kimchi against Multidrug-Resistant Pseudomonas aeruginosa" Antibiotics 12, no. 4: 676. https://doi.org/10.3390/antibiotics12040676
APA StyleYi, E.-J., & Kim, A.-J. (2023). Antimicrobial and Antibiofilm Effect of Bacteriocin-Producing Pediococcus inopinatus K35 Isolated from Kimchi against Multidrug-Resistant Pseudomonas aeruginosa. Antibiotics, 12(4), 676. https://doi.org/10.3390/antibiotics12040676





