Abstract
Pseudomonas aeruginosa is a significant pathogen identified with healthcare-associated infections. The present study evaluates the role of biofilm and efflux pump activities in influencing high-level resistance in virulent P. aeruginosa strains in clinical infection. Phenotypic resistance in biotyped Pseudomonas aeruginosa (n = 147) from diagnosed disease conditions was classified based on multiple antibiotic resistance (MAR) indices and analysed with logistic regression for risk factors. Efflux pump activity, biofilm formation, and virulence factors were analysed for optimal association in Pseudomonas infection using receiver operation characteristics (ROC). Age-specificity (OR [CI] = 0.986 [0.946–1.027]), gender (OR [CI] = 1.44 [0.211–9.827]) and infection sources (OR [CI] = 0.860 [0.438–1.688]) were risk variables for multidrug resistance (MDR)-P. aeruginosa infection (p < 0.05). Biofilm formers caused 48.2% and 18.5% otorrhea and wound infections (95% CI = 0.820–1.032; p = 0.001) respectively and more than 30% multidrug resistance (MDR) strains demonstrated high-level efflux pump activity (95% CI = 0.762–1.016; p = 0.001), protease (95% CI = 0.112–0.480; p = 0.003), lipase (95% CI = 0.143–0.523; p = 0.001), and hemolysin (95% CI = 1.109–1.780; p = 0.001). Resistance relatedness of more than 80% and 60% to cell wall biosynthesis inhibitors (ceftazidime, ceffproxil, augumentin, ampicillin) and, DNA translational and transcriptional inhibitors (gentamicin, ciprofloxacin, ofloxacin, nitrofurantoin) were observed (p < 0.05). Strong efflux correlation (r = 0.85, p = 0.034) with MDR strains, with high predictive performances in efflux pump activity (ROC-AUC 0.78), biofilm formation (ROC-AUC 0.520), and virulence hierarchical-clustering. Combine activities of the expressed efflux pump and biofilm formation in MDR-P. aeruginosa pose risk to clinical management and infection control.
1. Introduction
Pseudomonas aeruginosa is one of the significant pathogens identified with increasing healthcare-associated infections (HAIs) [1,2]. It is a ubiquitous and non-fastidious organism that grows in humid or wet environments. This opportunistic pathogen is commonly found among immune-compromised patients, cases of underlying diseases (diabetes), traumatized or invasive surgical procedures, sepsis, bloodstream, otorrhea, catheterized or indwelling devices, skin, and soft tissue infections (mostly related to burns, and pressure ulcer) [3,4,5]. Frontline antibiotics are usually selected to treat Pseudomonas infection to prevent growing resistance. Antibiotic selective pressures facilitate intrinsic resistance mechanisms to the drug of choice, making therapeutic applications difficult, expensive, and exacerbating morbidity [6].
P. aeruginosa biofilm functionally creates favourable medium that prevents accessibility of antibiotics to infection sites and promotes cell-surface interactions for tissue degradation. Biofilm formation in P. aeruginosa infection further enhances in vivo colonization, adaptation and persistence, through aggregation of exopolysaccharides (EPS) and biomolecules (lipids, proteins, carbohydrates) induced by poly-N-acetylglucosamine [7]. The activities of the multi-drug efflux system and low outer membrane permeability are important intrinsic components which are characterized with broad substrate specificity and genetic domains mediating unrelated multi-drug resistance (MDR) in P. aeruginosa [8]. MexAB-OprM is one of the major efflux pumps belonging to the resistance-nodulation-division (RND) which plays significant roles in intrinsic and acquired resistance to cell wall biosynthesis inhibitors (such as β-lactam classes), DNA and protein synthesis inhibitors (mostly fluoroquinolones and aminoglycosides respectively) [9,10]. Persistence expression of MexAB-OprM efflux pump of the RND superfamily constituting an inner membrane (MexB), periplasmic membrane fusion protein (MexA) and a channel-forming outer membrane protein (OprM) [10], are preferential and active extruding mechanisms mediating poor permeability, facilitating high-level P. aeruginosa resistance and increase virulence factor productions.
Increase efflux activity and virulence factors usually intensify disease morbidity, with activation of hemolysin that cause tissue damage, dysfunctional cellular immune responses, and red cell destruction [11,12]. Involvement of extracellular secretions of protease and lipases (lipolytic enzymes) in biofilm formers aids the functional capacity of the pathogen for invasion of host cells, and subversion of host defenses leading to tissue damage [13]. The ability of the virulence factor to degrade mucosal lipids and phospholipids on the epithelial surface alters mucosal integrity and promotes constant invasion [14]. Combine activities of virulent proteins promote the formation of membrane blebs and lipid rafts, aiding P. aeruginosa intracellular survival and cytosol lipid degradation leading to severe tissue pathology [15]. Several reports of antibiotic resistance among the clinical P. aeruginosa isolates are associated with biofilm and efflux pump activities [16,17,18,19,20,21]. However, there is paucity of relevant information on the assessment and synergy of efflux pump, biofilm formation, and virulence factors activities with the antibiotic resistance profile of healthcare-associated P. aeruginosa and possible implications on antibiotic stewardship in Nigerian hospital settings. The association of biofilm formation, efflux pump activities, and virulence proteins from P. aeruginosa strains resulting in high-level multi-drug resistance in clinical infection is yet to be defined. The study aims to evaluate the functional role of biofilm-forming capacity and efflux pump activities of virulent MDR-P. aeruginosa isolates from clinical infections and association of these factors with pathogen tissue tropism.
2. Results
2.1. MDR Pseudomonas aeruginosa Resistance Pattern
Of the 147 P. aeruginosa strains collected over the 6-month period, 27 were multi-drug resistance (MDR), showing significant resistance of more than 80% to cell wall biosynthesis inhibitors (ceftazidime, augmentin, ampicillin) and over 60% resistance to DNA translational and transcriptional inhibitors (ofloxacin, ciprofloxacin, nitrofurantoin); (p < 0.05, Figure 1A). To identify antibiotic resistance relatedness of the MDR P. aeruginosa, clustergram was built with strains from clinical samples. The heatmap revealed a comparative and related resistance pattern to cefprozil, ofloxacin and gentamicin (Figure 1B). Overall low median susceptibility rate of less than 10% was shown by the MDR P. aeruginosa obtained from all the clinical samples (Figure 1C).
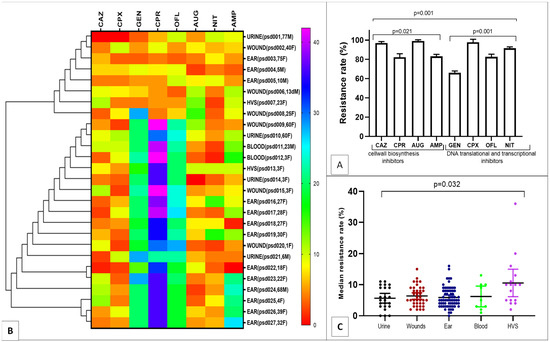
Figure 1.
Resistance pattern and Clustergram of the MDR strains ((A), Strain resistance to antibiotic classes; (B), Resistance relatedness; (C), Median resistance rates).
2.2. Risk Factors for MDR Pseudomonas aeruginosa Infection
The univariate analysis of the strain collections from the subjects presenting various systemic infections is shown in Table 1. Age-specificity of the subjects showed a significant risk for MDR P. aeruginosa infection (OR = 0.986, CI = 0.946–1.027, p = 0.024) with the highest prevalence rate among children ages 0–12 years (47.6%) and adult ages 24–50 (25.7%). Higher rate was recorded among female (20.8%) compared to male (OR = 1.440, CI = 0.211–9.827, p = 0.001). Considerable high risk for otorrhea (37.0%), wounds (21.4%) and bloodstream (18.2%) infections were noted (OR = 0.860, CI = 0.438–1.688, p = 0.002), but no MDR P. aeruginosa strain was isolated from other infection sites (including the eyes, throat, lungs and endocervical).

Table 1.
Univariate analysis of risk factor for MDR-P. aeruginosa infections.
2.3. Phenotypic Virulence Factor Expression
Of the MDR-P. aeruginosa strains (Table S2), significant rates of 48.2% and 18.5% (95% CI = 0.820–1.032; p = 0.001) were biofilm producers causing otorrhea and wound infections respectively. Strains found in otorrhea infection expressed significant efflux pump activity (44.4%; 95% CI = 0.762–1.016; p = 0.001), protease (18.5%; 95% CI = 0.112–0.480; p = 0.003), lipase (40.7%; 95% CI = 0.143–0.523; p = 0.001), and hemolysin (33.3%; 95% CI = 1.109–1.780; p = 0.001) compared to strains from other infections (Table 2).

Table 2.
Detection rate of virulence factors produced by the MDR Pseudomonas aeruginosa.
2.4. Correlation and Prediction Performance of EP Activity and Expressed Biofilm
Assessment of correlation coefficients of biofilm, efflux pump activity and virulence factors with MDR P. aeruginosa was shown in Figure 2A,B. A strong correlation of the efflux activity (r = 0.85, p = 0.034) with the MDR strain mostly from the ear infection, was recorded. Higher proportion of MDR strains overproducing efflux and biofilm clustered together (cluster a) compared to less number recorded in cluster b (Figure 2A). In clusters c and d, less proportion of strains (producing protease, lipase and hemolysin) from ear, wound, urine, vaginal and bloodstream infections further confirm low correlation as shown in Figure 2B. Figure 2C showed the Receiver Operating Characteristic (ROC) curves for predicting efflux and biofilm production with MAR indices of MDR P. aeruginosa. High biofilm ROC-AUC 0.78 (95% CI: 0.580–0.915) highlights a stronger predictive performance compare to efflux ROC-AUC 0.520 (95% CI: 0.321–0.715), showing a significant phenotypic adaptive resistance mechanism (Figure 2C). Weak MARI correlation with protease, lipase and hemolysin was noted based on the strain frequency from the clinical samples and biofilm production (Table 3). Hierarchical-clustering heat-map provided three clusters of MDR P. aeruginosa compared to age, sex and efflux proteins (MexA and MexB) with the degree of biofilm, hemolysin and lipase production (Figure 3). Four main clusters defined the degree of strain resistance relatedness. Cluster b included strains with high MexA and MexB encoded genes, hemolytic and lipase productions, and a mild MARI level, while strains in cluster c expressed high-level MARI and biofilm production, mostly among females aged 0 to 40 years.
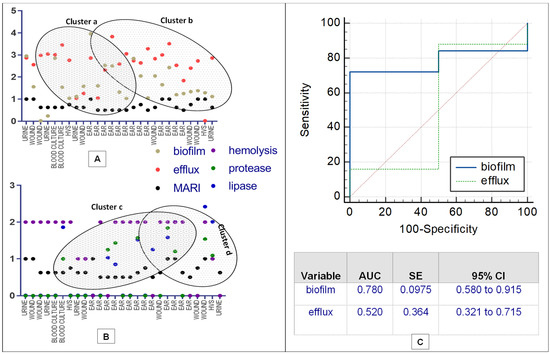
Figure 2.
Correlation matrix of phenotypic antibiotic resistance ((A,B), Association of antibiotic resistance, biofilm formation, efflux activity, protease, lipase and hemolysin production; (C), ROC curves).

Table 3.
Analysis of strain efflux, biofilm, lipase, protease and hemolysis production with MARI.
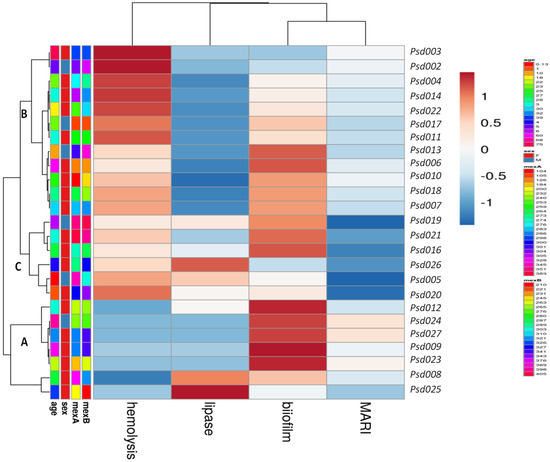
Figure 3.
Heatmap of hierarchical clustering of MDR-Psa resistance and virulence profiles in comparison with the age, sex, MexA and MexB efflux proteins (ranging between 158–395 bp), MARI and expressed virulence factors. Rows clustered by UPGMA method using Euclidean distances to define clusters.
3. Discussion
In the current study, age–specificity is a considerable risk factor for MDR P. aeruginosa infection rate showing a significant prevalence among children. Low immunity due to poor nutrition and maternal care, persistent nosocomial infection during admission, and consistent use of medical devices (urinary catheters, intravascular and endotracheal tubes etc), facilitate strain colonization and high infection rate [22,23]. The declining demography of the admitted adult group in these healthcare settings with the poor socio-economic condition, prolonged hospital stay, and antibiotic misuse are common factors that persistently influence the infection spread [24]. Self-prescription and consumption of counter drugs are common practices that enhance MDR and continuous treatment failure. The high rate of recorded MDR P. aeruginosa among female patients presents a different Pseudomonas infection phenomenon attributed to hormonal changes during the menstrual cycle, influencing behavioral and physiological changes that alter the immune responses [25]. Increased poor clinical outcomes in several gynecological conditions and urinary cases are potential risks for Pseudomonas infection [26]. Though ear infection (otorrhea) is observed to be frequent among infants and young children [1], poor clinical management by quacks, unregulated antibiotic prescription, and unhygienic perforation of the ear lobe usually facilitate the middle ear infection. Observed Pseudomonas wound infections are exacerbated by expired or ineffective topical antibiotics, contaminated or poorly sterilized dressing gauze. These further pose a significant challenge to clinical management, causing prolonged hospital stay and increasing morbidity [9].
Furthermore, the produced biofilm matrix plays structural function of inhibiting neutrophil elastase (a bactericidal enzyme) from the host to enable persistence infectivity and virulence [21]. The ability to produce protease once attached or colonized with biofilm intensifies necrotization of epithelia mucosa and functional invasive mechanism. As P. aeruginosa protease degrades basement membranes and extracellular matrix components, it cleaves the laminin, fibronectin, proteoglycans, and various collagens, causing direct tissue invasion with extensive ulceration leading to the down-regulation of cascade path of humoral immune responses [27]. During invasion, P. aeruginosa lipase (an extracellular lipolytic enzymes), interacts with inflammatory mediators released from human platelets, neutrophilic and basophilic granulocytes, and monocytes to enhance massive tissue damage and bloodstream infection [28]. Hemolysin production further suggests impending disease severity by forming cellular pore and necrotic cell death. As host cell lysis increases, there is increase in erythrocytes depletion, cytotoxicity, host cell plasma membrane fluidity, bilayer structure deformities resulting to immune evasion, physiological imbalance and septic shock mostly in severe conditions [29]. These virulence factors contribute to several stages of tissue damage that involve adherence, colonization and invasion leading to local and systemic dissemination of biofilm lipopolysaccharide, polysaccharide slime (alginate), hemolysin, and proteases [29].
Clustering of highly related MDR P. aeruginosa obtained from wounds, urinary tract and ear infections reveal active circulation and re-dissemination of strains with potential to exchange resistant genetic elements (plasmids, transposons, integrons and prophages), thereby facilitating horizontal gene transfer. This results to relatively high recombination that could give rise to multi-clonal MDR population [30]. Dissemination of these MDR strains portrays an imminent health risk that could escalate to an outbreak of multiple extra-intestinal infections but the inclusion of genomic-wide surveillance centered on the community antimicrobial stewardship is highly important to curb the spread. Low ciprofloxacin and ceftazidime susceptibility presented possible intrinsic resistance to the available drug of choice [31]. Hospital and community Pseudomonas aeruginosa infection control could suffer major setbacks due to high-level resistance to cell wall biosynthesis inhibitors (particularly augmentin, ceftazidime and ampicillin) which are most prescribed antibiotics. The recorded antibiotic resistance of more than 60% to translational and transcriptional inhibitors poses a serious challenge to clinical management with a remarkable capacity to develop an extend-spectrum resistance to other classes of antibiotics. Low outer membrane permeability, high efflux activity and producted inactivating enzymes are possible intrinsic or acquired resistance gained through horizontal gene transfer or mutational changes [32], limiting antibiotic efficacy. The formation of biofilm is one of the P. aeruginosa adaptive resistance mechanisms, acting as a diffusion barrier to limit antibiotic access to the bacterial cells making the strains capable of surviving antibiotic attacks [7]. Hospital and community antibiotic stewardship needs to be strengthened with proper information on combining these antibiotic classes and regulation of drug prescription, particularly in local outlets.
Very low clinical MDR P. aeruginosa susceptibility corroborate the previous report of increasing MDR in similar clinical settings in northern and southern Nigerian with more prevalence rate of hospital-acquired Pseudomonas infection [18,19,20,33], which is evident in different infection sources. Newer clinical approaches are needed to curtail the increasing resistance by considering the innovative integrated system in prescription and therapeutic formulation, with combined synergistic mechanism of action.
Over-expression of a well-developed efflux system embedded across the outer membrane (OM) permeability barrier continuously enables active expulsion of antibacterial compounds, improving the cell persistence and multiplication during infection [34]. Reports have shown that P. aeruginosa OM carries several substrate-specific porins that limit the intake of certain compounds and nutrients, further enhance expression of MexAB-OprM efflux, which provides resistance to varieties of antibiotics (chloramphenicol, β-lactams, quinolones, macrolides, tetracycline and novobiocin) [35]. The abundance of porin on the OM constitutes high permeability properties that usually influence the active expulsion of accumulated antibiotics [36]. The weak association of the resistance pattern of the isolates with produced biofilm and virulence factors indicates the interdependent strategies of P. aeruginosa to intensify infection while maintaining high-level resistance. The interplay of virulence factor activity and biofilm formation demonstrates a dominant influence that increases infection burden, therapeutic failure, morbidity and poor quality of life [36]. The observed association of the biofilm-forming ability, virulence factor, and efflux activities provides insight into the mechanism of P. aeruginosa-host adaptation in severe infection. Further genomic studies would be needed to ascertain the pathways that facilitate the MDR-P. aeruginosa pathology. Clustering efflux pump and biofilm-producing P. aeruginosa obtained from different infection sources with related multi-antibiotic resistance indices reveals a remarkable MDR strains relatedness that could trigger an hospital and community-acquired P. aeruginosa infection which may be difficult to treat.
The predictive performance of the biofilm matrix indicates phenotypic adaptations and protective capacity provided by the matrix for cellular efflux pump and persistent induction of modulators (phosphonate degradation, lipid biosynthesis, and polyamine biosynthesis) that contribute to persistent resistance [37]. Functional mediation of biofilm enhances genetic regulatory pathways involving multidrug efflux activities and possible alteration of drug targets [38]. These findings are helpful in understanding biofilm-mediated resistance in P. aeruginosa infection pathology and important gene targets to be considered for new drug discovery. Comparative analysis of hierarchical clustering of MDR strains characterized with a combination of efflux genes, virulence factors, and biofilm further suggests a major impending public health challenge that calls for routine surveillance, monitoring, and definitive strategic interventions. The study provides a clue to the emerging clusters of virulent MDR P. aeruginosa that could be regarded as high risk for public health.
4. Materials and Methods
4.1. Bacterial Strain Collections
One hundred and forty-seven P. aeruginosa isolates from diagnosed disease conditions were obtained from clinical samples of patients attending two major healthcare facilities; Federal Medical Centre, Abeokuta and General Hospital, Ota; in southwest Nigeria between September 2020 and March 2021 with ethical approval from Covenant Health Research Committees, Covenant University, Ota, Nigeria (CHREC/055/2020). All the isolates were preserved in semi-solid Brain Heart Infusion (BHI) (Oxoid, Basingstoke, UK) supplemented with glycerol and re-characterized for confirmation following standard biochemical methods previously described [8].
4.2. Antimicrobial Susceptibility
The susceptibility profile of commonly prescribed antibiotics for Pseudomonas infections was determined using Kirby-Bauer disc diffusion in accordance with CLSI recommendations and guidelines [39]. Overnight culture from cetrimide agar was sub-cultured on BHI and further incubated for 24 h at 37 °C. Bacterial suspension of 0.5 MacFarland turbidity was spread on BHI using sterile swab stick. Antibiotic discs including ceffproxil (CPR, 10 µg), ofloxacin (OFL, 30 µg), amoxicillin-clavulanic acid (AUG, 5/30 µg), nitrofurantoin (NIT, 30 µg), ciprofloxacin (CPX, 30 µg), ceftazidime (CAZ, 30 µg), gentamicin (GEN, 10 µg), and ampicillin (AMP, 30 µg) were gently placed on the plate and incubated at 37 °C for 24 h. Phenotypic resistance was interpreted according to the CLSI guidelines (2018) [39,40]. Isolates showing resistance to at least one agent in more than three classes of the antibiotic group were classified as multi-drug resistant P. aeruginosa (MDR P. aeruginosa) according to Magiorakos et al. [41]. Multiple antibiotic resistance index (MARI) ranging between 0 and 1 was determined by dividing the total number of detected resistance to antimicrobials for each isolate by the total number of tested antimicrobials [42].
4.3. Biofilm Assay
The formation of biofilm was assayed in a microtitre plate as described by Mathur et al. [43]. Briefly, 200 µL of 0.5 McFarland inoculum were placed in 96-well microtiter plate along with negative control. After incubation at 37 °C for 24 h, broth suspension was discarded and rinsed three times with sterile water to remove the planktonic bacteria cell. To air-dried plate, 200 µL of 1% (v/v) crystal violet solution was added and further incubated at room temperature for 1 h. Following two washing to remove the stain, 200 µL glacial acetic was added to dissolve the attached stain. The optical density (OD) was measured using UV Microplate reader [44].
4.4. Phenotypic Detection of Strain Efflux Pump (EP) Activity
MDR P. aeruginosa strains were selected for EP activity and biofilm detection. The EP activity was assayed with the Ethidium Bromide (EtBr) which is a substrate of efflux pumps using agar cartwheel method described by Ugwuanyi, et al. [21]. Adjusted 0.5 MacFarland turbid broth suspension was streaked on Mueller–Hinton agar plates containing 0 mg/L, 0.5 mg/L, 1 mg/L, 1.5 mg/L, and 2 mg/L concentrations of EtBr and incubated for 24 h at 37 °C. The plates were examined after incubation for fluorescence under UV transilluminator, and isolates that did not fluoresce were identified to express active EP activity and were scored according to the concentration of EtBr. In contrast, bacteria that fluoresced at a minimum concentration of EtBr were recorded as not possessing active efflux pumps.
4.5. Phenotypic Detection of Virulence Factors
Hemolysin production was demonstrated as described by Edberg et al. [45]. Overnight single colony was sub-cultured on 5% defibrinated sheep blood agar overlaid on the Nutrient agar base (Oxoid, UK) and incubated at 37 °C for 72 h. A clear halo zone indicating lysis of the red blood cells around the colony indicates haemolysin production [46]. A phenotypic assay for protease production was performed according to Suganthi et al. [47]. Briefly, broth of 0.5MacFarland was dropped on Skim milk agar supplemented with 1% casein and allowed to be adsorbed and incubated at 37 °C for 24 h. Casein hydrolysis was positive, indicating a clear zone around the inoculum spot. A loopful of 18 h bacteria colonies was streaked onto tributyrin agar plate, incubated at 37 °C for 24 h and then observed for a zone of hydrolysis around the colony. The zone of hydrolysis produced by the strain was observed as an indication for lipase production [48].
4.6. Efflux Gene Genotyping
Genomic DNA was isolated from pure broth culture using a DNA extraction kit (Zymo, Irvine, CA, USA) according to the manufacturer’s instruction and evaluated for quality and purity with Nanospectrophotometer. Encoded efflux pump genes (mexA, mexB and OprM), which belong to the RND pump, was assayed with PCR [49]. In a total reaction volume of 25 µL, 1 µL of DNA template, 12 µL 5X FIREPOL Solis Biodyne (Tartu, Estonia) master mix, 1 µL each of reverse and forward primers (Table S1) and 10 µL deionized water (Sigma-Aldrich, St. Louis, MO, USA). DNA template obtained from P. aeruginosa ATCC 27853 was used as positive control and nuclease-free water as negative control. The PCR assays were performed in Bio-Rad MJ thermal cycler (T100cycler, Bio-Rad, Hemel Hempstead, UK) with initiation of 95 °C for 5 min; 30 cycles of denaturation at 95 °C for 60 s, annealing (listed in Table 1) for 30 s, extension 72 °C for 45 s and final elongation at 72 °C for 10 min. Obtained amplicons were electrophoresed in 1.5% agarose gel at 100 V for 60 min and visualized under a trans-illuminator.
4.7. Data Analysis
Among the patients, risk factors for P. aeruginosa infection were analysed with univariate logistic regression to determine the significance of dependable variables taking the odd ratio at 95% confidence intervals (CIs) using SPSS v22. Level of virulence factors production from MDR P. aeruginosa were expressed as percentages and further analysed with unpaired t-tests (two-tailed, unequal variance) to determine the differences between the variables and p-values less than 0.05 were considered statistically significant. The significance of the resistance rate of the selected translational and transcriptional inhibitors was determined with ANOVA in GraphPad Prism 8.0. Median resistance of MDR P. aeruginosa obtained from various clinical sample sources was evaluated with ANOVA. The correlation matrix of phenotypic antibiotic resistance with biofilm formation, efflux activity, protease, lipase, and hemolysin productions was calculated using the Pearson method. The possible association of these variables was illustrated in graphical presentations. To predict the intensity of antibiotic resistance of MDR P. aeruginosa with biofilm and efflux pump activity, receiver operating characteristics (ROC) curves were calculated, taking the significance of optimal predictions given by the area under the curve (AUC) [50], using the statistical program MedCalc Statistical Sofware (Ostend, Belgium).
5. Conclusions
The activities of the produced virulence proteins, efflux pump and biofilm, contribute significantly to high antibiotic resistance and persistent chronic infection with possible transmission and exchange of genetic materials among the MDR P. aeruginosa population. Observed association and hierarchical clustering of virulent MDR strains reveal high capacity for effective adaptability, survival, and infectivity. Periodic community-wide intervention, monitoring of local drug prescriptions, and appropriate clinical management of the Pseudomonas infections are needed to mitigate the impending spread of virulent MDR P. aeruginosa.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics12030626/s1, Table S1: Primers used for amplification of isolated P. aeruginosa; Table S2: Antimicrobial susceptibility pattern of the isolated multidrug resistant Pseudomonas aeruginosa strains.
Author Contributions
Conceptualization, P.A.A., S.O.R. and C.A.O.; Data curation, O.W.G., O.E.A., S.T.P., K.S.A., J.O.P. and G.I.O.; Formal analysis, P.A.A., O.W.G., H.U.O., O.E.A., A.I.A., F.O.O. and G.I.O.; Funding acquisition, O.W.G., H.U.O., S.T.P., A.I.A., K.S.A., J.O.P., S.O.R. and F.O.O.; Investigation, P.A.A., O.W.G., H.U.O., J.O.P. and G.I.O.; Methodology, O.E.A., K.S.A., F.O.O. and C.A.O.; Project administration, P.A.A., S.T.P., A.I.A., J.O.P., S.O.R., F.O.O., C.A.O. and G.I.O.; Resources, O.W.G., H.U.O., O.E.A., S.T.P., K.S.A., S.O.R., F.O.O. and C.A.O.; Software, P.A.A. and A.I.A.; Visualization, G.I.O. and K.S.A.; Writing—original draft, P.A.A. All authors have read and agreed to the published version of the manuscript.
Funding
Publication of this article was supported by the Covenant University Centre for Research, Innovation and Development (CUCRID), Covenant University, Ota, Nigeria. The afore-mentioned support agents had no role in study design, data collection, and analysis, interpretation of data, the decision to publish, or preparation of the manuscript.
Institutional Review Board Statement
Informed consent of the patients was obtained during sample collection and ethical permission for the study was approved by the Covenant Health Research Committees, Covenant University, Ota, Nigeria (CHREC/055/2020). All methods were carried out in accordance with relevant guidelines and regulations as approved by the Ethics Committee and with appropriate and published references.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All data generated during the study are presented in this paper.
Acknowledgments
Authors kindly appreciate the staff of the Department of Medical Microbiology, Federal Medical Centre, Abeokuta and General Hospital, Ota, Nigeria, for the collection and storage of the bacterial isolates. We thank the Biotechnology Research Cluster Group, Covenant University, Ota Nigeria for provision of genomic facilities.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Reynolds, D.; Kollef, M. The Epidemiology and Pathogenesis and Treatment of Pseudomonas aeruginosa Infections: An Update. Drugs 2021, 81, 2117–2131. [Google Scholar] [CrossRef] [PubMed]
- Litwin, A.; Rojek, S.; Gozdzik, W.; Duszynska, W. Pseudomonas aeruginosa device associated–healthcare associated infections and its multidrug resistance at intensive care unit of University Hospital: Polish, 8.5-year, prospective, single-centre study. BMC Infect. Dis. 2021, 21, 180. [Google Scholar] [CrossRef] [PubMed]
- Olasehinde, O.; Lamikanra, A. Pattern of Esbls in Uro-Pathogens Obtained from a Nigerian Tertiary Hospital. Niger. J. Pharm. Res. 2021, 16, 139–147. [Google Scholar] [CrossRef]
- Araos, R.; D’Agata, E. Pseudomonas aeruginosa and Other Pseudomonas Species. In Mandell, Douglas, and Benett’s Principles and Practice of Infectious Diseases; Churchill Livingstone Elsevier: Philadelphia, PA, USA, 2019; pp. 2686–2699. [Google Scholar]
- Herrera, S.; Bodro, M.; Soriano, A. Predictors of multidrug resistant Pseudomonas aeruginosa involvement in bloodstream infections. Curr. Opin. Infect. Dis. 2021, 34, 686–692. [Google Scholar] [CrossRef]
- Langendonk, R.F.; Neill, D.R.; Fothergill, J.L. The building blocks of antimicrobial resistance in Pseudomonas aeruginosa: Implications for current resistance-breaking therapies. Front. Cell. Infect. Microbiol. 2021, 11, 307. [Google Scholar] [CrossRef]
- Brindhadevi, K.; LewisOscar, F.; Mylonakis, E.; Shanmugam, S.; Verma, T.N.; Pugazhendhi, A. Biofilm and Quorum sensing mediated pathogenicity in Pseudomonas aeruginosa. Process Biochem. 2020, 96, 49–57. [Google Scholar] [CrossRef]
- Zahedani, S.S.; Tahmasebi, H.; Jahantigh, M. Coexistence of Virulence Factors and Efflux Pump Genes in Clinical Isolates of Pseudomonas aeruginosa: Analysis of Biofilm-Forming Strains from Iran. Int. J. Microbiol. 2021, 2021, 5557361. [Google Scholar] [CrossRef]
- Rahbar, M.; Hamidi-Farahani, R.; Asgari, A.; Esmailkhani, A.; Soleiman-Meigooni, S. Expression of RND efflux pumps mediated antibiotic resistance in Pseudomonas aeruginosa clinical strains. Microb. Pathog. 2021, 153, 104789. [Google Scholar]
- Mohanty, S.; Baliyarsingh, B.; Nayak, S.K. Antimicrobial Resistance in Pseudomonas aeruginosa: A Concise. Antimicrob. Resist. A One Health Perspect. 2021, 49, 177–192. [Google Scholar]
- Palaniappan, B.; Solomon, A.P. Targeting AgrA quorum sensing regulator by bumetanide attenuates virulence in Staphylococcus aureus–A drug repurposing approach. Life Sci. 2021, 273, 119306. [Google Scholar] [CrossRef]
- Ebersole, J.L.; Dawson, D., III; Emecen-Huja, P.; Nagarajan, R.; Howard, K.; Grady, M.E.; Gonzalez, O.A. The periodontal war: Microbes and immunity. Periodontology 2017, 75, 52–115. [Google Scholar] [CrossRef] [PubMed]
- Allen, P.E.; Martinez, J.J. Modulation of host lipid pathways by pathogenic intracellular bacteria. Pathogens 2020, 9, 614. [Google Scholar] [CrossRef] [PubMed]
- Angus, A.A.; Lee, A.A.; Augustin, D.K.; Lee, E.J.; Evans, D.J.; Fleiszig, S.M. Pseudomonas aeruginosa induces membrane blebs in epithelial cells, which are utilized as a niche for intracellular replication and motility. Infect. Immun. 2008, 76, 1992–2001. [Google Scholar] [CrossRef] [PubMed]
- Cianciotto, N.P.; White, R.C. Expanding role of type II secretion in bacterial pathogenesis and beyond. Infect. Immun. 2017, 85, e00014-17. [Google Scholar] [CrossRef] [PubMed]
- Olayinka, A.T.; Onile, B.A.; Olayinka, B.O. Prevalence of multi-drug resistant (MDR) Pseudomonas aeruginosa isolates in surgical units of Ahmadu Bello University Teaching Hospital, Zaria, Nigeria: An indication for effective control measures. Ann. Afr. Med. 2004, 3, 13–16. [Google Scholar]
- Omoregie, R.; Erebor, J.O.; Ahonkhai, I.; Isibor, J.O.; Ogefere, H.O. Observed changes in the prevalence of uropathogens in Benin City, Nigeria. N. Z. J. Med. Lab. Sci. 2008, 62, 29–31. [Google Scholar]
- Agwu, E.; Lhongbe, J.; Inyang, N. Prevalence of Quinolone-susceptible Pseudomonas aeruginosa and Staphylococcus aureus in Delayed-healing DFU’s in Ekpoma, Nigeria. Int. Wound J. 2010, 22, 100–105. [Google Scholar]
- Garba, I.; Lusa, Y.H.; Bawa, E.; Tijjani, M.B.; Aliyu, M.S.; Zango, U.U.; Raji, M.I.O. Antibiotics susceptibility pattern of Pseudomonas aeruginosa isolated from wounds in patients attending Ahmadu Bello University Teaching Hospital, Zaria, Nigeria. Niger. J. Basic Appl. Sci. 2012, 20, 32–34. [Google Scholar]
- Zubair, K.O.; Iregbu, K.C. Resistance pattern and detection of metallo-beta-lactamase genes in clinical isolates of Pseudomonas aeruginosa in a central Nigeria tertiary hospital. Niger. J. Clin. Pract. 2018, 21, 176–182. [Google Scholar]
- Ugwuanyi, F.C.; Ajayi, A.; Ojo, D.A.; Adeleye, A.I.; Smith, S.I. Evaluation of efflux pump activity and biofilm formation in multidrug resistant clinical isolates of Pseudomonas aeruginosa isolated from a Federal Medical Center in Nigeria. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 11. [Google Scholar] [CrossRef]
- George, C.R.R.; Jeffery, H.E.; Lahra, M.M. Infection of Mother and Baby. Keeling’s Fetal Neonatal Pathol. 2022, 1, 207–245. [Google Scholar]
- Alshaikh, B.N.; Reyes Loredo, A.; Knauff, M.; Momin, S.; Moossavi, S. The Role of Dietary Fats in the Development and Prevention of Necrotizing Enterocolitis. Nutrients 2022, 14, 145. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, K.; Khan, M.H. Antimicrobial Mechanisms and Mode of Actions of Nanoemulsion Against Drug-Resistant ESKAPE Pathogens. In Handbook of Research on Nanoemulsion Applications in Agriculture, Food, Health, and Biomedical Sciences; IGI Global: Hershey, PA, USA, 2022; pp. 142–168. [Google Scholar]
- da Cruz, D.G.; de Magalhães, R.F.; Padilha, G.A.; da Silva, M.C.; Braga, C.L.; Silva, A.R.; Silva, P.L. Impact of positive biphasic pressure during low and high inspiratory efforts in Pseudomonas aeruginosa-induced pneumonia. PLoS ONE 2021, 16, e0246891. [Google Scholar] [CrossRef] [PubMed]
- Gudiol, C.; Durà-Miralles, X.; Aguilar-Company, J.; Hernández-Jiménez, P.; Martínez-Cutillas, M.; Fernandez-Avilés, F.; Carratalà, J. Co-infections and superinfections complicating COVID-19 in cancer patients: A multicentre, international study. J. Infect. 2021, 83, 306–313. [Google Scholar] [CrossRef]
- Johnson, D.I. Beck. In Bacterial Pathogens and Their Virulence Factors; Springer: Cham, Switzerland, 2018; pp. 363–379. [Google Scholar]
- Mobarak-Qamsari, E.; Kasra-Kermanshahi, R.; Moosavi-Nejad, Z. Isolation and identification of a novel, lipase-producing bacterium, Pseudomnas aeruginosa KM110. Iran. J. Microbiol. 2011, 3, 92–98. [Google Scholar]
- Ali, N.M.; Rehman, S.; Mazhar, S.A.; Liaqat, I.; Mazhar, B. Pseudomonas aeruginosa-Associated Acute and Chronic Pulmonary Infections. In Pathogenic Bacteria; IntechOpen: London, UK, 2020. [Google Scholar]
- Winter, M.; Buckling, A.; Harms, K.; Johnsen, P.J.; Vos, M. Antimicrobial resistance acquisition via natural transformation: Context is everything. Curr. Opin. Microbiol. 2021, 64, 133–138. [Google Scholar] [CrossRef]
- Brzozowski, M.; Krukowska, Ż.; Galant, K.; Jursa-Kulesza, J.; Kosik-Bogacka, D. Genotypic characterisation and antimicrobial resistance of Pseudomonas aeruginosa strains isolated from patients of different hospitals and medical centres in Poland. BMC Infect. Dis. 2020, 20, 693. [Google Scholar] [CrossRef]
- Azam, M.W.; Khan, A.U. Updates on the pathogenicity status of Pseudomonas aeruginosa. Drug Discov. Today 2019, 24, 350–359. [Google Scholar] [CrossRef]
- Ohore, H.U.; Akinduti, P.A.; Ahuekwe, E.F.; Ajayi, A.S.; Olasehinde, G.I. Molecular Detection of ESBLs, TEM, SHV, and CTX-M in Clinical Pseudomonas aeruginosa Isolates in Ogun State. Bioenergy Biochem. Process. Technol. 2022, 127–136. [Google Scholar]
- Ferrand, A.; Vergalli, J.; Davin-Regli, A. An intertwined network of regulation controls membrane permeability Including drug influx and efflux in Enterobacteriaceae. Microorganisms 2020, 8, 833. [Google Scholar] [CrossRef]
- Ashwath, P.; Sannejal, A.D. A quest to the therapeutic arsenal: Novel strategies to combat multidrug-resistant bacteria. Curr. Gene Ther. 2022, 22, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Ayepola, O.O.; Olasupo, N.A.; Egwari, L.O.; Schaumburg, F. Characterization of Panton–Valentine leukocidin-positive Staphylococcus aureus from skin and soft tissue infections and wounds in Nigeria: A cross-sectional study. F1000Research 2018, 7, 30345027. [Google Scholar] [CrossRef] [PubMed]
- da Cruz Nizer, W.S.; Inkovskiy, V.; Versey, Z.; Strempel, N.; Cassol, E.; Overhage, J. Oxidative Stress Response in Pseudomonas aeruginosa. Pathogens 2021, 10, 1187. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Wang-Kan, X.; Neuberger, A.; van Veen, H.W.; Pos, K.M.; Piddock, L.J.; Luisi, B.F. Multidrug efflux pumps: Structure, function and regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, Sixteenth Informational Supplement; Document M100-S20; CLSI: Wayne, PA, USA, 2018. [Google Scholar]
- EUCAST Clinical Breakpoints and Dosing. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 5 August 2021).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Paterson, D.L. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Sadat, A.; El-Sherbiny, H.; Zakaria, A.; Ramadan, H.; Awad, A. Prevalence, antibiogram and virulence characterization of Vibrio isolates from fish and shellfish in Egypt: A possible zoonotic hazard to humans. J. Appl. Microbiol. 2020, 131, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Mathur, T.; Singhal, S.; Khan, S.; Upadhyay, D.J.; Fatma, T.; Rathan, A. Detection of biofilm formation among the clinical isolates of Staphylococci: An evaluation of three different screening methods. Indian J. Med. Microbiol. 2006, 24, 25–29. [Google Scholar] [CrossRef]
- Akinduti, A.P.; Osiyemi, J.A.; Banjo, T.T.; Ejilude, O.; El-Ashker, M.; Adeyemi, A.G.; Isibor, P.O. Clonal diversity and spatial dissemination of multi-antibiotics resistant Staphylococcus aureus pathotypes in Southwest Nigeria. PLoS ONE 2021, 16, e0247013. [Google Scholar] [CrossRef]
- Edberg, S.C.; Gallo, P.; Kontnick, C. Analysis of the virulence characteristics of bacteria isolated from bottled, water cooler, and tap water. Microb. Ecol. Health Dis. 1996, 9, 67–77. [Google Scholar] [CrossRef]
- Lopes, E.S.; Cardoso, W.M.; Nishi, D.M.; Horn, R.V.; Albuquerque, A.H.; Lima, S.V.G.; Beleza, A.J.F.; Gaio, F.C.; Carmo, C.; Pascoal Filho, M.N.; et al. Sero-group identification, phenotypic detection of hemolysis and extended spectrum beta-lactamases of Escherichia coli isolated from psittacine of illegal wildlife trade in Fortaleza, Brazil. Arq. Bras. Med. Vet. Zootec. 2018, 70, 823–829. [Google Scholar] [CrossRef]
- Suganthi, C.; Mageswari, A.; Karthikeyan, S.; Anbalagan, M.; Sivakumar, A.; Gothandam, K.M. Screening and optimization of protease production from a halotolerant Bacillus licheniformis isolated from saltern sediments. J. Genet. Eng. Biotechnol. 2013, 11, 47–52. [Google Scholar] [CrossRef]
- Banoth, L.; Devarapalli, K.; Paul, I.; Thete, K.N.; Pawar, S.V.; Banerjee, U.C. Screening, isolation and selection of a potent lipase producing microorganism and its use in the kinetic resolution of drug intermediates. J. Indian Chem. Soc. 2021, 98, 100143. [Google Scholar] [CrossRef]
- Su, F.; Wang, J. Berberine inhibits the MexXY-OprM efflux pump to reverse imipenem resistance in a clinical carbapenem-resistant Pseudomonas aeruginosa isolate in a planktonic state. Exp. Ther. Med. 2018, 15, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Gupta, P.; Kumar, R.; Bhardwaj, A. dPABBs: A novel in silico approach for predicting and designing anti-biofilm peptides. Sci. Rep. 2016, 6, 21839. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).