First Bacteremia Due to Corynebacterium gottingense in an Immunocompromised Child: A Case Report, 16S rDNA-Based Phylogenetic Analyses and Review of the Literature
Abstract
1. Introduction
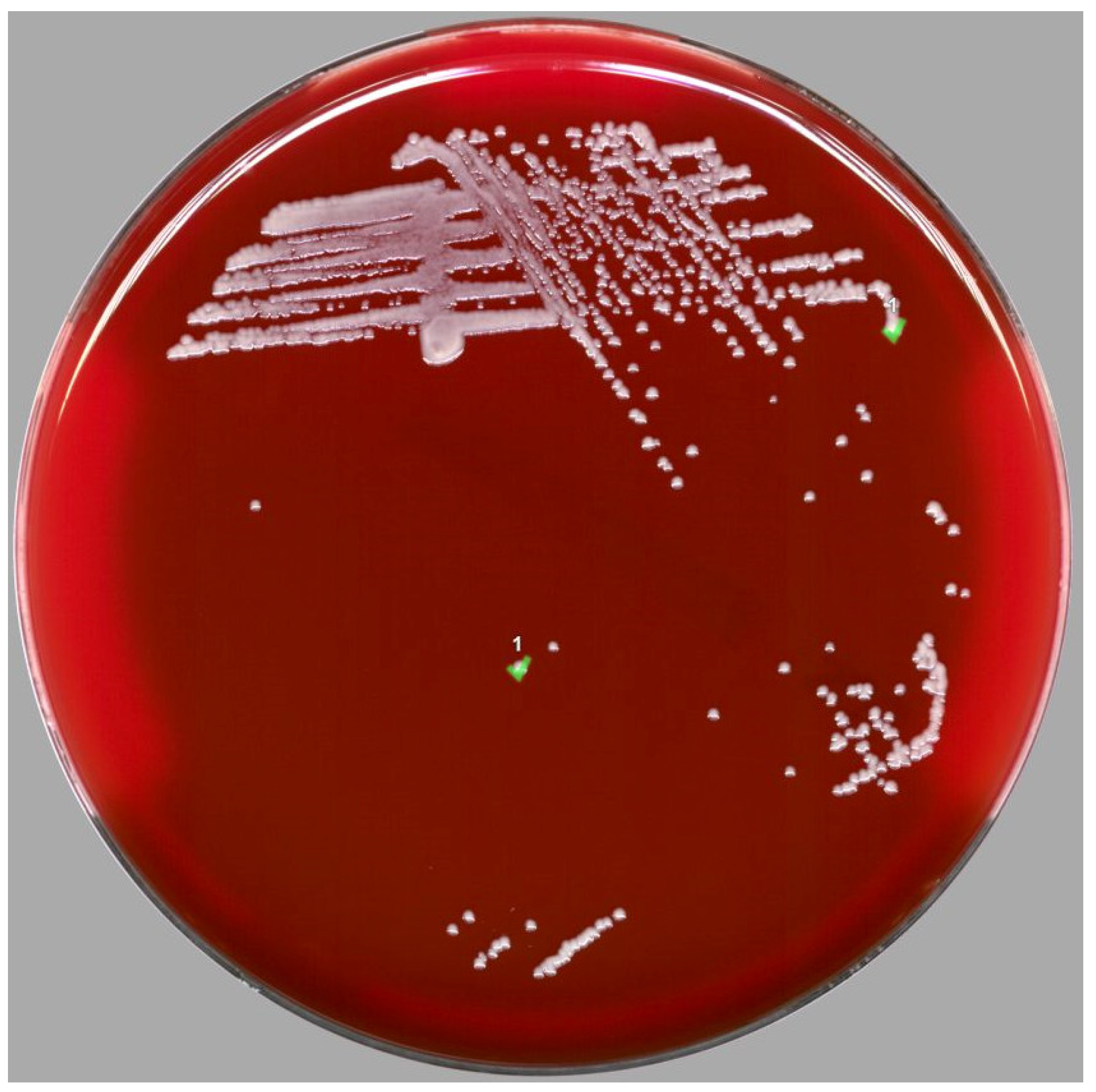
2. Case Report
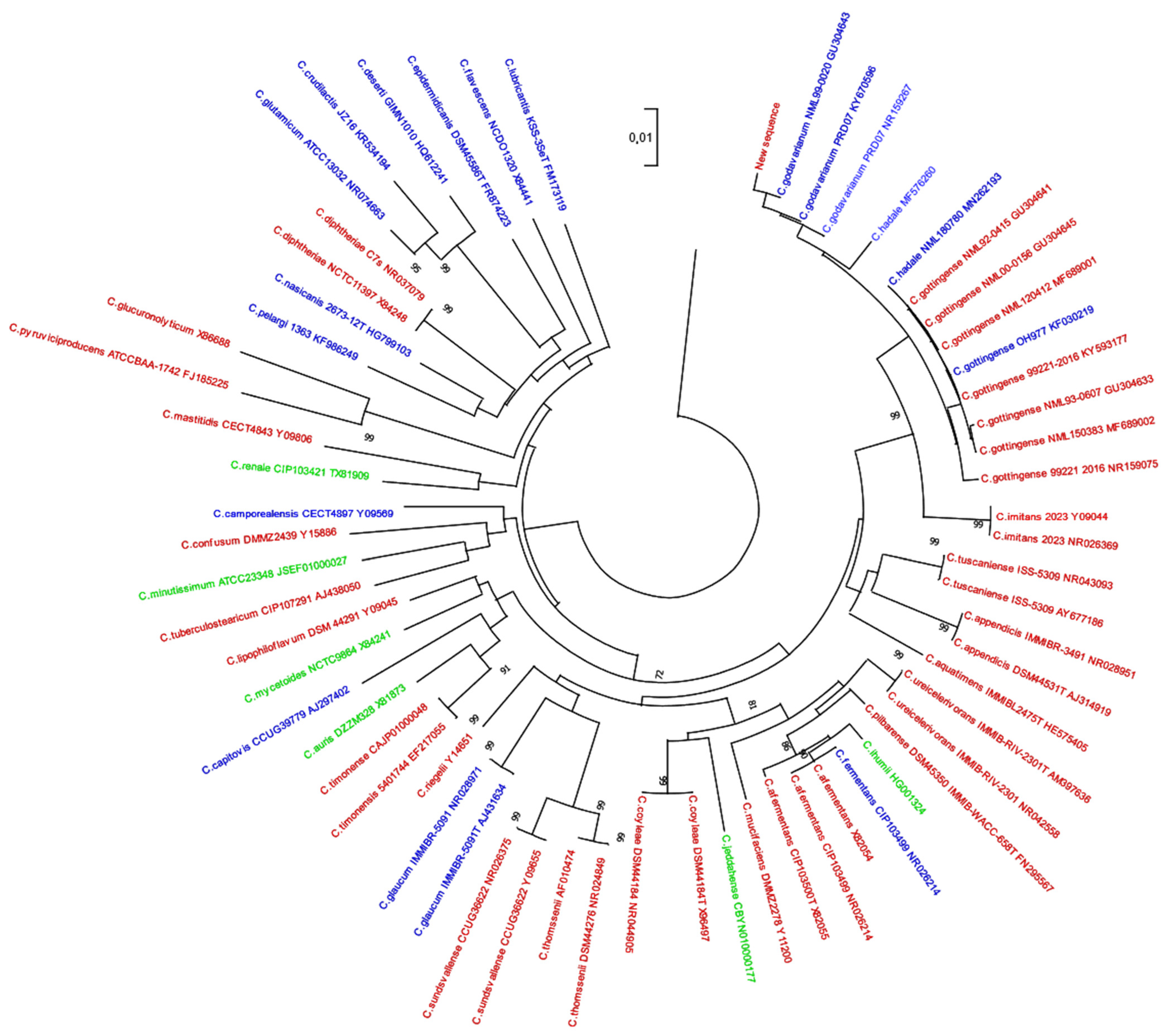
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Atasayar, E.; Zimmermann, O.; Spröer, C.; Schumann, P.; Groß, U. Corynebacterium gottingense sp. Nov., Isolated from a Clinical Patient. Int. J. Syst. Evol. Microbiol. 2017, 67, 4494–4499. [Google Scholar] [CrossRef] [PubMed]
- Bernard, K.A.; Pacheco, A.L.; Burdz, T.; Wiebe, D.; Bernier, A.-M. Corynebacterium Godavarianum Jani et al. 2018 and Corynebacterium Hadale Wei et al. 2018 Are Both Later Heterotypic Synonyms of Corynebacterium Gottingense Atasayar et al. 2017, Proposal of an Emended Description of Corynebacterium Gottingense Atasayar et al. 2017. Int. J. Syst. Evol. Microbiol. 2020, 70, 3534–3540. [Google Scholar] [CrossRef] [PubMed]
- LPSN.Dsmz.De. Available online: https://lpsn.dsmz.de/genus/corynebacterium (accessed on 23 November 2022).
- Tozzo, P.; Delicati, A.; Caenazzo, L. Skin Microbial Changes during Space Flights: A Systematic Review. Life 2022, 12, 1498. [Google Scholar] [CrossRef] [PubMed]
- Bernard, K. The Genus Corynebacterium and Other Medically Relevant Coryneform-like Bacteria. J. Clin. Microbiol. 2012, 50, 3152–3158. [Google Scholar] [CrossRef] [PubMed]
- Jani, K.; Khare, K.; Senik, S.; Karodi, P.; Vemuluri, V.R.; Bandal, J.; Shouche, Y.; Rale, V.; Sharma, A. Corynebacterium godavarianum sp. Nov., Isolated from the Godavari River, India. Int. J. Syst. Evol. Microbiol. 2018, 68, 241–247. [Google Scholar] [CrossRef]
- Wei, Y.; Fang, J.; Xu, Y.; Zhao, W.; Cao, J. Corynebacterium hadale sp. Nov. Isolated from Hadopelagic Water of the New Britain Trench. Int. J. Syst. Evol. Microbiol. 2018, 68, 1474–1478. [Google Scholar] [CrossRef] [PubMed]
- Broutin, L.; Deroche, L.; Michaud, A.; Le Moal, G.; Burucoa, C.; Gayet, L.-E.; Plouzeau, C.; Pichon, M. First Description of Bacteremia Caused by Oscillibacter Valericigenes in a Patient Hospitalized for Leg Amputation. Anaerobe 2020, 64, 102244. [Google Scholar] [CrossRef] [PubMed]
- Cabrol, M.; Rammaert, B.; Plouzeau, C.; Burucoa, C.; Pichon, M. Phylogeny of Catabacter Hongkongensis Strains Responsible for Bacteremia Is Not Associated with Clinical Outcomes or Therapeutic Efficacy. Diseases 2021, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Flandrois, J.-P.; Perrière, G.; Gouy, M. LeBIBIQBPP: A Set of Databases and a Webtool for Automatic Phylogenetic Analysis of Prokaryotic Sequences. BMC Bioinform. 2015, 16, 251. [Google Scholar] [CrossRef] [PubMed]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Kurgan, L. Sequence Similarity Searching. Curr. Protoc. Protein Sci. 2019, 95, e71. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.C.; Efstratiou, A.; Mokrousov, I.; Mutreja, A.; Das, B.; Ramamurthy, T. Diphtheria. Nat. Rev. Dis. Prim. 2019, 5, 81. [Google Scholar] [CrossRef] [PubMed]
- Costales, J.; Alsyouf, M.; Napolitan, P.; Wang, S.; Hu, B. Corynebacterium Urealyticum: Rare Urinary Tract Infection with Serious Complications. Can. J. Urol. 2019, 26, 9680–9682. [Google Scholar] [PubMed]
- Gupta, R.; Popli, T.; Ranchal, P.; Khosla, J.; Aronow, W.S.; Frishman, W.H.; El Khoury, M.Y. Corynebacterium Jeikeium Endocarditis: A Review of the Literature. Cardiol. Rev. 2021, 29, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Yamamuro, R.; Hosokawa, N.; Otsuka, Y.; Osawa, R. Clinical Characteristics of Corynebacterium Bacteremia Caused by Different Species, Japan, 2014–2020. Emerg. Infect. Dis. 2021, 27, 2981–2987. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Kim, S.B.; Choi, S.H.; Kim, S. Comparison of 16S RRNA Gene Based Microbial Profiling Using Five Next-Generation Sequencers and Various Primers. Front. Microbiol. 2021, 12, 715500. [Google Scholar] [CrossRef] [PubMed]
| Classification | C. godavarianum, 2018 [6] | C. hadale, 2018 [7] | C. gottingense, 2017 [1] | Emended Description, 2020 [2] | Present Description |
|---|---|---|---|---|---|
| Phylum | Actinobacteria | ||||
| Family | Corynebacteriaceae | ||||
| Genus | Corynebacterium | ||||
| Reference Strain | NR_159267 | MF576260 | NR_159075 | - | OP897049.1 |
| Isolation | Godavari river in India | Deep sea water in the New Britain trench | Blood of clinical patient with bacteraemia in a German city | - | Blood of clinical patient with bacteraemia in a French city |
| Morphology | |||||
| Rod-shaped | ||||
| No | - | - | - | - |
| Positive | - | - | - | Positive |
| Oxygen tolerance | Aerobic | Anaerobic | Aero-anaerobic | - | Aero-anaerobic |
| Catalase | Positive | - | - | - | Positive |
| Length | 1 µm | 1.3 µm | - | - | - |
| Diameter | 0.5 µm | 0.57 µm | - | - | - |
| Colony color on blood agar | Creamy-whitish and non-translucent | Ivory-white | White cream | - | White cream |
| Hemolysis on agar supplemented with 5% sheep blood | Non haemolytic | - | - | - | Non haemolytic |
| Culture and growth conditions | |||||
| Temperature | 37° (range 25–40 °C) | Optimal 35 °C (range 25–41 °C) | 37° | 35–37° | 35–37° |
| pH | Optimal: 7.0 (range 6–11) | - | - | 6–7 | - |
| NaCl | Optimum 1% (tolerance up to 11%) | Optimal 4% 0–10% | - | 0–4% | - |
| Enzyme production | |||||
| Catalase | + | + | + | + | |
| Oxidase | - | - | - | - | - |
| Gelatinase | - | / | / | - | / |
| Nitrate reductase | - | - | - | - | - |
| H2S | - | / | / | / | / |
| Acid production as the only carbon substrate from | |||||
| Glucose | - | / | + | + | / |
| Ribose | + | / | + | + | / |
| Maltose | + | / | + | + | / |
| Antibiotic Susceptibility Testing (AST) | |||||
| Susceptible | Amikacin, cefixime, cefotaxime, ceftriaxone, chloramphenicol, ciprofloxacin, colistin, co-trimoxazole, furazolidone, gentamicin, levofloxacin, netillin, norfloxacin, rifampicin, streptomycin, tetracycline and vancomycin | - | - | - | Amoxicillin, ciprofloxacin, cotrimoxazole, rifampicin, linezolid, tetracycline and vancomycin |
| Resistance | Amoxicillin, co-amoxiclav, cefpodoxime, and clindamycin | - | - | - | Penicillin G and clindamycin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouguerra, L.; Dupraz, C.; Plouzeau, C.; Michaud, A.; Broutin, L.; Cremniter, J.; Burucoa, C.; Pichon, M. First Bacteremia Due to Corynebacterium gottingense in an Immunocompromised Child: A Case Report, 16S rDNA-Based Phylogenetic Analyses and Review of the Literature. Antibiotics 2023, 12, 528. https://doi.org/10.3390/antibiotics12030528
Bouguerra L, Dupraz C, Plouzeau C, Michaud A, Broutin L, Cremniter J, Burucoa C, Pichon M. First Bacteremia Due to Corynebacterium gottingense in an Immunocompromised Child: A Case Report, 16S rDNA-Based Phylogenetic Analyses and Review of the Literature. Antibiotics. 2023; 12(3):528. https://doi.org/10.3390/antibiotics12030528
Chicago/Turabian StyleBouguerra, Lucas, Chrystelle Dupraz, Chloé Plouzeau, Anthony Michaud, Lauranne Broutin, Julie Cremniter, Christophe Burucoa, and Maxime Pichon. 2023. "First Bacteremia Due to Corynebacterium gottingense in an Immunocompromised Child: A Case Report, 16S rDNA-Based Phylogenetic Analyses and Review of the Literature" Antibiotics 12, no. 3: 528. https://doi.org/10.3390/antibiotics12030528
APA StyleBouguerra, L., Dupraz, C., Plouzeau, C., Michaud, A., Broutin, L., Cremniter, J., Burucoa, C., & Pichon, M. (2023). First Bacteremia Due to Corynebacterium gottingense in an Immunocompromised Child: A Case Report, 16S rDNA-Based Phylogenetic Analyses and Review of the Literature. Antibiotics, 12(3), 528. https://doi.org/10.3390/antibiotics12030528










