Relevance of Pharmacogenomics to the Safe Use of Antimicrobials
Abstract
1. Introduction
2. Adverse Drug Reactions Involving HLA Gene Associations
3. Other Common Adverse Drug Reactions That Appear Independent of HLA Genotype
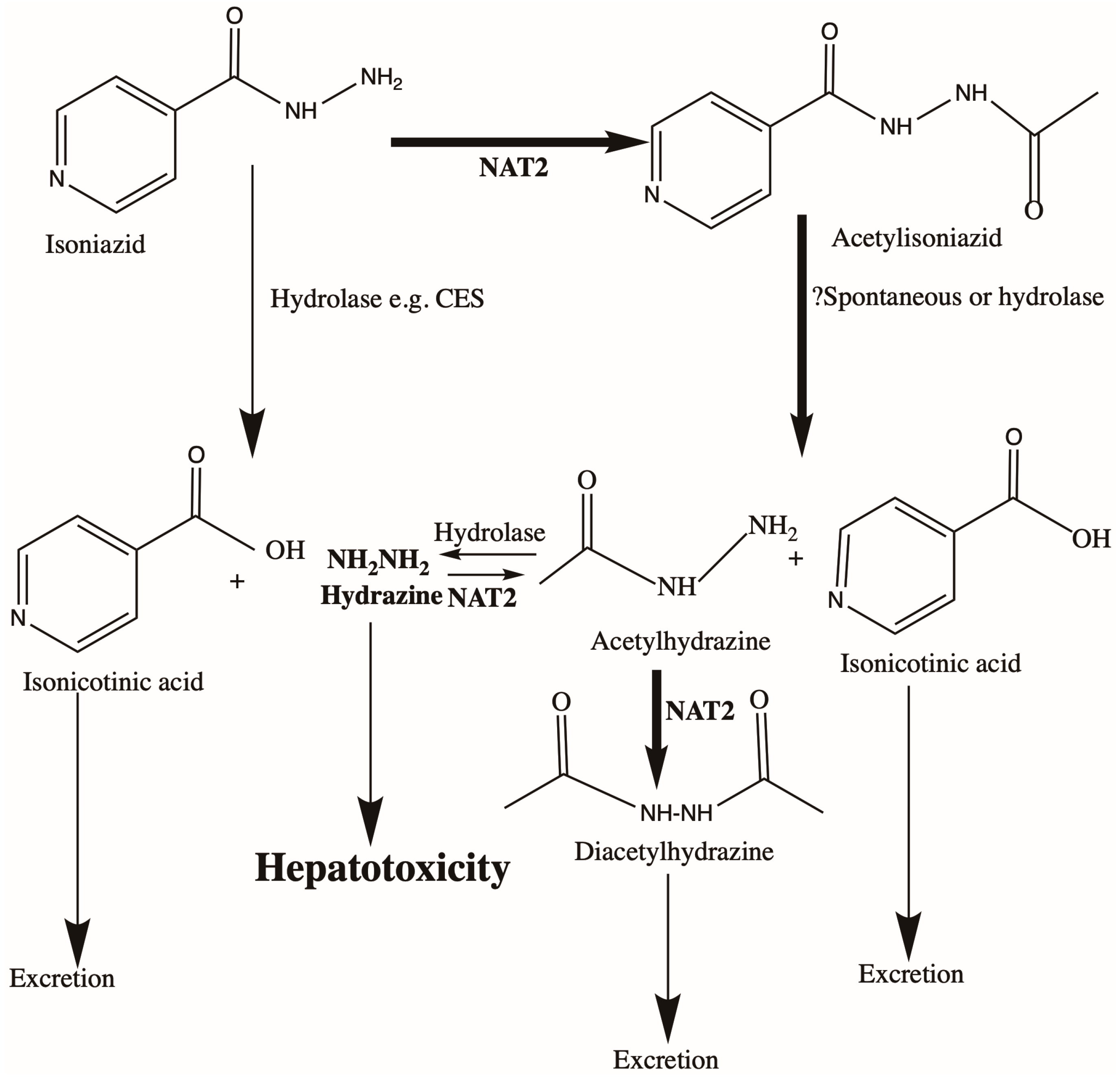
3.1. Anti-TB Drug Treatment and Liver Toxicity
3.2. Aminoglycosides and Deafness
3.3. Fluoroquinolone Toxicity
4. Voriconazole and CYP2C19
5. Polygenic Risk Scores
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- van der Wouden, C.H.; Guchelaar, H.J.; Swen, J.J. Precision Medicine Using Pharmacogenomic Panel-Testing: Current Status and Future Perspectives. Clin. Lab. Med. 2022, 42, 587–602. [Google Scholar] [CrossRef]
- Cusato, J.; Allegra, S.; Nicolò, A.; Calcagno, A.; D’Avolio, A. Precision medicine for HIV: Where are we? Pharmacogenomics 2018, 19, 145–165. [Google Scholar] [CrossRef]
- Brunton, L.; Knollmann, B. Goodman and Gilman’s the Pharmacological Basis of Therapeutics, 14th ed.; McGraw-Hill Education: New York, NY, USA, 2022. [Google Scholar]
- Blumenthal, K.G.; Peter, J.G.; Trubiano, J.A.; Phillips, E.J. Antibiotic allergy. Lancet 2019, 393, 183–198. [Google Scholar] [CrossRef]
- Uetrecht, J.; Naisbitt, D.J. Idiosyncratic adverse drug reactions: Current concepts. Pharmacol. Rev. 2013, 65, 779–808. [Google Scholar] [CrossRef]
- Daly, A.K. Genetics of drug-induced liver injury: Current knowledge and future prospects. Clin. Transl. Sci. 2022, 16, 37–42. [Google Scholar] [CrossRef]
- Lucena, M.I.; Molokhia, M.; Shen, Y.; Urban, T.J.; Aithal, G.P.; Andrade, R.J.; Day, C.P.; Ruiz-Cabello, F.; Donaldson, P.T.; Stephens, C.; et al. Susceptibility to Amoxicillin-Clavulanate-Induced Liver Injury Is Influenced by Multiple HLA Class I and II Alleles. Gastroenterology 2011, 141, 338–347. [Google Scholar] [CrossRef]
- Nicoletti, P.; Aithal, G.P.; Bjornsson, E.S.; Andrade, R.J.; Sawle, A.; Arrese, M.; Barnhart, H.X.; Bondon-Guitton, E.; Hayashi, P.H.; Bessone, F.; et al. Association of Liver Injury From Specific Drugs, or Groups of Drugs, With Polymorphisms in HLA and Other Genes in a Genome-Wide Association Study. Gastroenterology 2017, 152, 1078–1089. [Google Scholar] [CrossRef]
- Zhang, F.R.; Liu, H.; Irwanto, A.; Fu, X.A.; Li, Y.; Yu, G.Q.; Yu, Y.X.; Chen, M.F.; Low, H.Q.; Li, J.H.; et al. HLA-B*13:01 and the dapsone hypersensitivity syndrome. N. Engl. J. Med. 2013, 369, 1620–1628. [Google Scholar] [CrossRef]
- Li, Y.J.; Phillips, E.J.; Dellinger, A.; Nicoletti, P.; Schutte, R.; Li, D.; Ostrov, D.A.; Fontana, R.J.; Watkins, P.B.; Stolz, A.; et al. Human Leukocyte Antigen B*14:01 and B*35:01 Are Associated With Trimethoprim-Sulfamethoxazole Induced Liver Injury. Hepatology 2021, 73, 268–281. [Google Scholar] [CrossRef]
- Nicoletti, P.; Dellinger, A.; Li, Y.J.; Barnhart, H.X.; Chalasani, N.; Fontana, R.J.; Odin, J.A.; Serrano, J.; Stolz, A.; Etheridge, A.S.; et al. Identification of Reduced ERAP2 Expression and a Novel HLA Allele as Components of a Risk Score for Susceptibility to Liver Injury Due to Amoxicillin-Clavulanate. Gastroenterology 2022, in press. [Google Scholar] [CrossRef]
- Urban, T.J.; Nicoletti, P.; Chalasani, N.; Serrano, J.; Stolz, A.; Daly, A.K.; Aithal, G.P.; Dillon, J.; Navarro, V.; Odin, J.; et al. Minocycline hepatotoxicity: Clinical characterization and identification of HLA-B∗35:02 as a risk factor. J. Hepatol. 2017, 67, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Krebs, K.; Bovijn, J.; Zheng, N.; Lepamets, M.; Censin, J.C.; Jürgenson, T.; Särg, D.; Abner, E.; Laisk, T.; Luo, Y.; et al. Genome-wide Study Identifies Association between HLA-B(∗)55:01 and Self-Reported Penicillin Allergy. Am. J. Hum. Genet. 2020, 107, 612–621. [Google Scholar] [CrossRef]
- Daly, A.K.; Donaldson, P.T.; Bhatnagar, P.; Shen, Y.; Pe’er, I.; Floratos, A.; Daly, M.J.; Goldstein, D.B.; John, S.; Nelson, M.R.; et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat. Genet. 2009, 41, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Petros, Z.; Kishikawa, J.; Makonnen, E.; Yimer, G.; Habtewold, A.; Aklillu, E. HLA-B*57 Allele Is Associated with Concomitant Anti-tuberculosis and Antiretroviral Drugs Induced Liver Toxicity in Ethiopians. Front. Pharmacol. 2017, 8, 90. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, P.; Aithal, G.P.; Chamberlain, T.C.; Coulthard, S.; Alshabeeb, M.; Grove, J.I.; Andrade, R.J.; Bjornsson, E.; Dillon, J.F.; Hallberg, P.; et al. Drug-Induced Liver Injury due to Flucloxacillin: Relevance of Multiple Human Leukocyte Antigen Alleles. Clin. Pharmacol. Ther. 2019, 106, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, P.; Carr, D.F.; Barrett, S.; McEvoy, L.; Friedmann, P.S.; Shear, N.H.; Nelson, M.R.; Chiriac, A.M.; Blanca-López, N.; Cornejo-García, J.A.; et al. Beta-lactam-induced immediate hypersensitivity reactions: A genome-wide association study of a deeply phenotyped cohort. J. Allergy Clin. Immunol. 2021, 147, 1830–1837.e1815. [Google Scholar] [CrossRef] [PubMed]
- Chalasani, N.; Li, Y.J.; Dellinger, A.; Navarro, V.; Bonkovsky, H.; Fontana, R.J.; Gu, J.; Barnhart, H.; Phillips, E.; Lammert, C.; et al. Clinical features, outcomes, and HLA risk factors associated with nitrofurantoin-induced liver injury. J. Hepatol. 2023, 78, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; Oussalah, A.; Chery, C.; Guéant-Rodriguez, R.M.; Gaeta, F.; Cornejo-Garcia, J.A.; Rouyer, P.; Josse, T.; Mayorga, C.; Torres, M.J.; et al. Next-generation sequencing and genotype association studies reveal the association of HLA-DRB3*02:02 with delayed hypersensitivity to penicillins. Allergy 2022, 77, 1827–1834. [Google Scholar] [CrossRef]
- Jee, A.; Sernoskie, S.C.; Uetrecht, J. Idiosyncratic Drug-Induced Liver Injury: Mechanistic and Clinical Challenges. Int. J. Mol. Sci. 2021, 22, 2954. [Google Scholar] [CrossRef]
- Manson, L.E.N.; van den Hout, W.B.; Guchelaar, H.J. Genotyping for HLA Risk Alleles to Prevent Drug Hypersensitivity Reactions: Impact Analysis. Pharmaceuticals 2021, 15, 4. [Google Scholar] [CrossRef]
- Alffenaar, J.W.C.; Stocker, S.L.; Forsman, L.D.; Garcia-Prats, A.; Heysell, S.K.; Aarnoutse, R.E.; Akkerman, O.W.; Aleksa, A.; van Altena, R.; de Oñata, W.A.; et al. Clinical standards for the dosing and management of TB drugs. Int. J. Tuberc. Lung Dis. 2022, 26, 483–499. [Google Scholar] [CrossRef] [PubMed]
- Saukkonen, J.J.; Cohn, D.L.; Jasmer, R.M.; Schenker, S.; Jereb, J.A.; Nolan, C.M.; Peloquin, C.A.; Gordin, F.M.; Nunes, D.; Strader, D.B.; et al. An official ATS statement: Hepatotoxicity of antituberculosis therapy. Am. J. Respir. Crit. Care Med. 2006, 174, 935–952. [Google Scholar] [CrossRef]
- Ramappa, V.; Aithal, G.P. Hepatotoxicity Related to Anti-tuberculosis Drugs: Mechanisms and Management. J. Clin. Exp. Hepatol. 2013, 3, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Garibaldi, R.A.; Drusin, R.E.; Ferebee, S.H.; Gregg, M.B. Isoniazid-associated hepatitis. Report of an outbreak. Am. Rev. Respir. disease 1972, 106, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Senousy, B.E.; Belal, S.I.; Draganov, P.V. Hepatotoxic effects of therapies for tuberculosis. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 543–556. [Google Scholar] [CrossRef] [PubMed]
- Hein, D.W.; Millner, L.M. Arylamine N-acetyltransferase acetylation polymorphisms: Paradigm for pharmacogenomic-guided therapy- a focused review. Expert Opin. Drug Metab. Toxicol. 2021, 17, 9–21. [Google Scholar] [CrossRef]
- Richardson, M.; Kirkham, J.; Dwan, K.; Sloan, D.J.; Davies, G.; Jorgensen, A.L. NAT2 variants and toxicity related to anti-tuberculosis agents: A systematic review and meta-analysis. Int. J. Tuberc. Lung Dis. 2019, 23, 293–305. [Google Scholar] [CrossRef]
- Suvichapanich, S.; Wattanapokayakit, S.; Mushiroda, T.; Yanai, H.; Chuchottawon, C.; Kantima, T.; Nedsuwan, S.; Suwankesawong, W.; Sonsupap, C.; Pannarunothai, R.; et al. Genomewide Association Study Confirming the Association of NAT2 with Susceptibility to Antituberculosis Drug-Induced Liver Injury in Thai Patients. Antimicrob. Agents Chemother. 2019, 63, e02692-18. [Google Scholar] [CrossRef]
- Nicoletti, P.; Devarbhavi, H.; Goel, A.; Venkatesan, R.; Eapen, C.E.; Grove, J.I.; Zafer, S.; Bjornsson, E.; Lucena, M.I.; Andrade, R.J.; et al. Genetic Risk Factors in Drug-Induced Liver Injury Due to Isoniazid-Containing Antituberculosis Drug Regimens. Clin. Pharmacol. Ther. 2021, 109, 1125–1135. [Google Scholar] [CrossRef]
- Fukunaga, K.; Kato, K.; Okusaka, T.; Saito, T.; Ikeda, M.; Yoshida, T.; Zembutsu, H.; Iwata, N.; Mushiroda, T. Functional Characterization of the Effects of N-acetyltransferase 2 Alleles on N-acetylation of Eight Drugs and Worldwide Distribution of Substrate-Specific Diversity. Front. Genet. 2021, 12, 652704. [Google Scholar] [CrossRef]
- Richards, V.E.; Chau, B.; White, M.R.; McQueen, C.A. Hepatic gene expression and lipid homeostasis in C57BL/6 mice exposed to hydrazine or acetylhydrazine. Toxicol. Sci. 2004, 82, 318–332. [Google Scholar] [CrossRef]
- Azuma, J.; Ohno, M.; Kubota, R.; Yokota, S.; Nagai, T.; Tsuyuguchi, K.; Okuda, Y.; Takashima, T.; Kamimura, S.; Fujio, Y.; et al. NAT2 genotype guided regimen reduces isoniazid-induced liver injury and early treatment failure in the 6-month four-drug standard treatment of tuberculosis: A randomized controlled trial for pharmacogenetics-based therapy. Eur. J. Clin. Pharmacol. 2013, 69, 1091–1101. [Google Scholar] [CrossRef]
- Steyger, P.S. Mechanisms of Aminoglycoside- and Cisplatin-Induced Ototoxicity. Am. J. Audiol. 2021, 30, 887–900. [Google Scholar] [CrossRef]
- Hutchin, T.P.; Cortopassi, G.A. Mitochondrial defects and hearing loss. Cell Mol. Life Sci. 2000, 57, 1927–1937. [Google Scholar] [CrossRef]
- McDermott, J.H.; Mahaveer, A.; James, R.A.; Booth, N.; Turner, M.; Harvey, K.E.; Miele, G.; Beaman, G.M.; Stoddard, D.C.; Tricker, K.; et al. Rapid Point-of-Care Genotyping to Avoid Aminoglycoside-Induced Ototoxicity in Neonatal Intensive Care. JAMA Pediatr. 2022, 176, 486–492. [Google Scholar] [CrossRef]
- Medicines and Healthcare Products Regulatory Agency. Aminoglycosides (Gentamicin, Amikacin, Tobramycin, and Neomycin): Increased Risk of Deafness in Patients with Mitochondrial Mutations; Medicines and Healthcare Products Regulatory Agency: London, UK, 2021; pp. 19–21.
- McDermott, J.H.; Wolf, J.; Hoshitsuki, K.; Huddart, R.; Caudle, K.E.; Whirl-Carrillo, M.; Steyger, P.S.; Smith, R.J.H.; Cody, N.; Rodriguez-Antona, C.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline for the Use of Aminoglycosides Based on MT-RNR1 Genotype. Clin. Pharmacol. Ther. 2022, 111, 366–372. [Google Scholar] [CrossRef]
- Kros, C.J.; Steyger, P.S. Aminoglycoside- and Cisplatin-Induced Ototoxicity: Mechanisms and Otoprotective Strategies. Cold Spring Harb. Perspect. Med. 2019, 9, a033548. [Google Scholar] [CrossRef]
- Kuula, L.S.M.; Backman, J.T.; Blom, M.L. Healthcare costs and mortality associated with serious fluoroquinolone-related adverse reactions. Pharmacol. Res. Perspect. 2022, 10, e00931. [Google Scholar] [CrossRef]
- Bove, C.; Baldock, R.A.; Champigneulle, O.; Martin, L.; Bennett, C.L. Fluoroquinolones: Old drugs, putative new toxicities. Expert Opin. Drug Saf. 2022, 21, 1365–1378. [Google Scholar] [CrossRef]
- Walsh, T.J.; Anaissie, E.J.; Denning, D.W.; Herbrecht, R.; Kontoyiannis, D.P.; Marr, K.A.; Morrison, V.A.; Segal, B.H.; Steinbach, W.J.; Stevens, D.A.; et al. Treatment of aspergillosis: Clinical practice guidelines of the Infectious Diseases Society of America. Clin. Infect. Dis. 2008, 46, 327–360. [Google Scholar] [CrossRef]
- Botton, M.R.; Whirl-Carrillo, M.; Del Tredici, A.L.; Sangkuhl, K.; Cavallari, L.H.; Agúndez, J.A.G.; Duconge, J.; Lee, M.T.M.; Woodahl, E.L.; Claudio-Campos, K.; et al. PharmVar GeneFocus: CYP2C19. Clin. Pharmacol. Ther. 2021, 109, 352–366. [Google Scholar] [CrossRef]
- Trifilio, S.; Pennick, G.; Pi, J.; Zook, J.; Golf, M.; Kaniecki, K.; Singhal, S.; Williams, S.; Winter, J.; Tallman, M.; et al. Monitoring plasma voriconazole levels may be necessary to avoid subtherapeutic levels in hematopoietic stem cell transplant recipients. Cancer 2007, 109, 1532–1535. [Google Scholar] [CrossRef]
- Moriyama, B.; Obeng, A.O.; Barbarino, J.; Penzak, S.R.; Henning, S.A.; Scott, S.A.; Agúndez, J.; Wingard, J.R.; McLeod, H.L.; Klein, T.E.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP2C19 and Voriconazole Therapy. Clin. Pharmacol. Ther. 2017, 102, 45–51. [Google Scholar] [CrossRef]
- Evans, D.M.; Visscher, P.M.; Wray, N.R. Harnessing the information contained within genome-wide association studies to improve individual prediction of complex disease risk. Hum. Mol. Genet. 2009, 18, 3525–3531. [Google Scholar] [CrossRef]
- Cross, B.; Turner, R.; Pirmohamed, M. Polygenic risk scores: An overview from bench to bedside for personalised medicine. Front. Genet. 2022, 13, 1000667. [Google Scholar] [CrossRef]
- Koido, M.; Kawakami, E.; Fukumura, J.; Noguchi, Y.; Ohori, M.; Nio, Y.; Nicoletti, P.; Aithal, G.P.; Daly, A.K.; Watkins, P.B.; et al. Polygenic architecture informs potential vulnerability to drug-induced liver injury. Nat. Med. 2020, 26, 1541–1548. [Google Scholar] [CrossRef]
- Cirulli, E.T.; Nicoletti, P.; Abramson, K.; Andrade, R.J.; Bjornsson, E.S.; Chalasani, N.; Fontana, R.J.; Hallberg, P.; Li, Y.J.; Lucena, M.I.; et al. A Missense Variant in PTPN22 is a Risk Factor for Drug-induced Liver Injury. Gastroenterology 2019, 156, 1707–1716.e1702. [Google Scholar] [CrossRef]
- Baudhuin, L.M.; Train, L.J.; Goodman, S.G.; Lane, G.E.; Lennon, R.J.; Mathew, V.; Murthy, V.; Nazif, T.M.; So, D.Y.F.; Sweeney, J.P.; et al. Point of care CYP2C19 genotyping after percutaneous coronary intervention. Pharm. J. 2022, 22, 303–307. [Google Scholar] [CrossRef]
- Ji, X.; Ning, B.; Liu, J.; Roberts, R.; Lesko, L.; Tong, W.; Liu, Z.; Shi, T. Towards population-specific pharmacogenomics in the era of next-generation sequencing. Drug Discov. Today 2021, 26, 1776–1783. [Google Scholar] [CrossRef]
| HLA Allele | Type of Idiosyncratic Adverse Reaction | Drug | Odds Ratio (Allelic) | p-Value | Reference |
|---|---|---|---|---|---|
| A*02:01 | Liver injury | Amoxicillin-clavulanate | 2.3 (1.8–2.9) | 1.8 × 10−10 | [7] |
| A*33:01 | Liver injury | Terbinafine | 40.5 (12.5–131.4) | 6.7 × 10−10 | [8] |
| B*13:01 | Hypersensitivity syndrome (particularly involving skin, with possible liver involvement) | Dapsone | 21.7 (10.4–45.1) | 2.0 × 10−16 | [9] |
| B*14:01-C*08:02 | Liver injury | Trimethoprim-sulfamethoxazole | 8.7 (3.2–19.5) | 2.3 × 10−4 | [10] |
| B*15:18 | Liver injury | Amoxicillin-clavulanate | 3.1 (6.08–1.58) | 0.001 | [11,12] |
| B*35:02 | Liver injury | Minocycline | 29.6 (7.8–89.8) | 2.57 × 10−8 | [12] |
| B*55:01 | Early and delayed allergic reactions (mainly involving skin) | Penicillins | 1.4 (1.3–1.5) | 2.0 × 10−31 | [13] |
| B*57:01 | Liver injury | Flucloxacillin | 36.6 (26.1–51.3) | 2.6 × 10−97 | [14] |
| B*57:02 and B*57:03 | Liver injury | Anti-HIV and anti-TB comb | 30.1 (3.4–263.1) | 0.002 | [15] |
| B*57:03 | Liver injury | Flucloxacillin | 19.8 (3.37–116.1) | 0.001 | [16] |
| DRB1*10:01 | Immediate hypersensitivity reaction | Penicillins | 2.9 (2.0–4.4) | 5.4 × 10−7 | [17] |
| DRB1*11:04 | Liver injury | Nitrofurantoin | 4.29 (2.18–7.66) | 1.2 × 10−4 | [18] |
| DRB1*15:01 | Liver injury | Amoxicillin-clavulanate | 2.8 (2.1–3.8) | 3.5 × 10−11 | [7] |
| DRB3*02:02 | Delayed hypersensitivity reaction | Penicillins | 8.9 (3.4–23.3) | p < 0.0001 | [19] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daly, A.K. Relevance of Pharmacogenomics to the Safe Use of Antimicrobials. Antibiotics 2023, 12, 425. https://doi.org/10.3390/antibiotics12030425
Daly AK. Relevance of Pharmacogenomics to the Safe Use of Antimicrobials. Antibiotics. 2023; 12(3):425. https://doi.org/10.3390/antibiotics12030425
Chicago/Turabian StyleDaly, Ann K. 2023. "Relevance of Pharmacogenomics to the Safe Use of Antimicrobials" Antibiotics 12, no. 3: 425. https://doi.org/10.3390/antibiotics12030425
APA StyleDaly, A. K. (2023). Relevance of Pharmacogenomics to the Safe Use of Antimicrobials. Antibiotics, 12(3), 425. https://doi.org/10.3390/antibiotics12030425






