Abstract
Background. Quinolones are commonly used for reducing the duration of diarrhea, infection severity, and limiting further transmission of disease related to Vibrio cholerae, but V. cholerae susceptibility to quinolone decreases over time. In addition to mutations in the quinolone-resistance determining regions (QRDRs), the presence of qnr and other acquired genes also contributes to quinolone resistance. Results. We determined the prevalence of quinolone resistance related genes among V. cholerae O139 strains isolated in China. We determined that eight strains carried qnrVC, which encodes a pentapeptide repeat protein of the Qnr subfamily, the members of which protect topoisomerases from quinolone action. Four qnrVC alleles were detected: qnrVC1, qnrVC5, qnrVC12, and qnrVC9. However, the strains carrying qnrVC1, qnrVC5, and qnrVC12 were ciprofloxacin (CIP)-sensitive. Contrastingly, the strain carrying qnrVC9 demonstrated high CIP resistance. qnrVC9 was carried by a small plasmid, which was conjugative and contributed to the high CIP resistance to the receptor V. cholerae strain. The same plasmid was also detected in V. vulnificus. The qnrVC1, qnrVC5, and qnrVC12 were cloned into expression plasmids and conferred CIP resistance on the host V. cholerae O139 strain. Conclusions. Our results revealed the contribution of quinolone resistance mediated by the qnrVC9 carried on the small plasmid and its active horizontal transfer among Vibrio species. The results also suggested the different effects of qnrVC alleles in different V. cholerae strains, which is possibly due to differences in sequences of qnrVC alleles and even the genetic characteristics of the host strains.
1. Introduction
Cholera is an acute gastrointestinal tract disease caused by Vibrio cholerae, of which two serogroups (O1 and O139) demonstrate epidemic potential. To date, seven major cholera pandemics have occurred since the 19th century, the most recent of which originated in Indonesia in the 1960s and is still ongoing [1]. Major cholera epidemics that have occurred periodically in recent years were those in Zimbabwe (2008), Haiti (2010), Sierra Leone (2012), Mexico (2013), and South Sudan and Ghana (2014) [1]. Despite the availability of effective prevention and treatment methods, cholera persistence remains a major public health problem worldwide mainly due to socioeconomic factors such as unsatisfactory hygienic conditions. The World Health Organization (WHO) estimated that 3–5 million cholera cases occur globally every year, with 100,000–120,000 deaths [2]. Since the first 1993 outbreak caused by the V. cholerae O139 strains in Xinjiang, China [3], limited foodborne outbreaks associated with food poisoning and sporadic cases were frequently reported and expanded in various regions, especially southeastern China [4]. The organisms resided and survived within environmental reservoirs during inter-epidemic periods and the threat of a cholera epidemic is ever-present.
Antibiotic therapy reduces the duration of diarrhea and infection severity, and limits further transmission of disease related to V. cholerae [5]. Currently, azithromycin and ciprofloxacin (CIP) are commonly used for treating patients with cholera [6]. CIP belongs to the quinolones, a family of broad-spectrum, systemic antibacterial agents that act by inhibiting DNA gyrase (gyrA and gyrB) and DNA topoisomerase IV (parC and parE), which are required for bacterial mRNA synthesis and DNA replication [7]. Due to excessive antibiotic use and misuse in humans, agriculture and aquaculture systems, antibiotic resistance emerged and evolved in bacteria, including Vibrio species, during the past few decades [8]. Bacteria often combine more than one mechanism to increase drug resistance acquisition efficacy. In addition to mutations in the quinolone-resistance determining regions (QRDRs) and overexpression of efflux pumps that reduce intracellular concentrations of the drug [9], Qnr proteins and the inactivation of drug by quinolone-modifying enzyme (AAC (6′) Ib-Cr) [10] also contribute to quinolone resistance.
In a previous survey, we examined the V. cholerae O139 strains recovered in 1993–2009 in China and detected strains that were increasingly resistant to nalidixic acid (NAL), which is the first member of the quinolones, and decreasingly sensitive to CIP [4]. Subsequent analysis revealed that the accumulation of mutations in the DNA gyrase and topoisomerase IV genes contributed to fluoroquinolone resistance [11]. However, it is unclear whether quinolone resistance related genes contribute to quinolone resistance in V. cholerae. Therefore, we investigated the prevalence of quinolone resistance related genes in V. cholerae serogroup O139 strains and estimated whether they could mediate quinolone resistance to their host strains.
2. Results
2.1. Prevalence of Quinolone Resistance Related Genes
In a previous study, we conducted a comprehensive investigation of the antibiotic resistance of V. cholerae O139 strains isolated in China from 1993 to 2009. The multidrug resistance in O139 isolates increased suddenly and became common after 1998. Different resistance profiles were observed in the V. cholerae O139 isolates from different years, while V. cholerae O1 strains isolated in the same period were much less resistant to these antibiotics and no obvious multidrug resistance patterns were detected. We further found decreased susceptibility was exhibited to CIP in V. cholera O139 strains [4]. Between 2001 and 2009, the median MIC of CIP was 0.5 μg/mL, without a change over this interval. These values were 16.7-fold and 4.2-fold higher than the median MICs of CIP for isolates of V. cholerae in 1993–1997 (0.03 μg/mL) and 1998–2000 (0.12 μg/mL), respectively. The seven strains resistant to CIP, which carried these accumulated mutations in QRDRs, were isolated in 1998, 2001 and 2003, respectively [11]. In 2006, there was one isolated strain named VC1699 possessing no mutation in QRDR of parE but resistant to CIP.
To determine whether quinolone resistance related genes contributed to quinolone resistance in V. cholera, we performed PCR screening for the prevalence of these genes in V. cholerae O139 strains. qnrVC genes were detected from eight V. cholerae O139 strains: VC422, VC1435, VC707, VC454, VC319, VC515, VC1692, and VC1699. Furthermore, QRDR genes were also detected from these strains and sequenced to identify existing mutations. Theses strains contained a series of mutations such as the meaningful mutations GyrA S83I and ParC S85L and nonsense mutations such as GyrA A171S.
MEGA 7.0 alignment of the qnrVC PCR amplicon revealed a series of qnrVC alleles. The qnrVC allele amplified from VC422, VC515, and VC707 was identical to qnrVC1; that from VC454, VC319, and VC1692 was identical to qnrVC5; and that from VC1699 was identical to qnrVC9. Lastly, the qnrVC allele from VC1435 differed from qnrVC4 by two single amino acids (Met18Leu and Gly167Glu) and was designated qnrVC12 (GenBank accession no. OQ442957). The QnrVC12 protein sequence was found to have 99% identity with QnrVC4. Table 1 lists the main characteristics of these eight strains. Phylogenetic analysis showed the QnrVC12, QnrVC4, QnrVC7, QnrVC9, and qnrVC5 belonged to the same group, whereas QnrVC10, QnrVC1, QnrVC3, QnrVC6, and QnrVC8 formed another group (Figure 1).

Table 1.
Characteristics of V. cholera O139 strains containing qnrVC alleles.
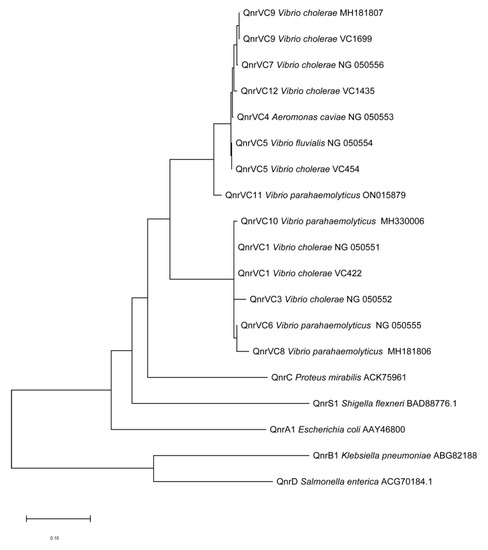
Figure 1.
Neighbor-joining tree of Qnr protein sequences. Phylogenetic analysis of Qnr protein sequences was performed using Neighbor-Joining method by MEGA 7.0 after multiple alignment of the data via CLUSTAL 2.1. The sequences of other Qnr were obtained from GenBank and compared with QnrVC9 of VC1699, QnrVC12 of VC1435, Qnr5 of VC454 and Qnr1 of VC422.
2.2. Characterization of Plasmids Containing qnrVC9
Among the eight strains containing qnrVC genes, we extracted a plasmid termed pVC1699 from VC1699, which was isolated from a patient in Jiangxi in 2006. Sequencing of pVC1699 determined that it was 7081-bp long with a GC content of 42.85%. The plasmid sequence was submitted to GenBank under the accession number OP821998. The ORF search revealed that pVC1699 carried qnrVC9 genes related to quinolone resistance. pVC1699 also carried genes for antitoxin (parD1), plasmid stabilization system protein (parE), integrase 1 (int1), and dihydrofolate reductase type 1 (dhfrI). BLAST revealed that pVC1699 had low identity with plasmids harbored qnrVC5, including pBD146 from V. fluvialis (GenBank accession no. EU574928.1), plasmids v110 from V. parahaemolyticus (GenBank accession no. KC540630.1), and pVN84 from V. cholerae O1 (GenBank accession no. AB200915.1) (Figure 2). However, pVC1699 had high identity to 99% with a plasmid termed 307 from V. vulnificus (GenBank accession no. MZ325519) (Figure 2).
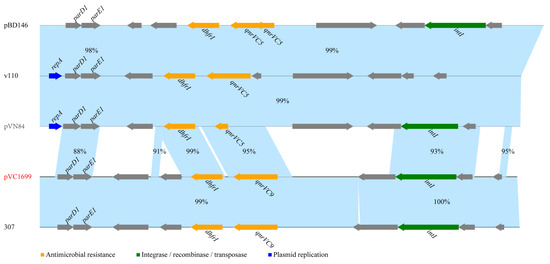
Figure 2.
Comparing plasmid pVC1699 with sequences of four plasmids of Vibrios obtained from GenBank.
2.3. Effect of qnrVC on Quinolone Resistance
Except VC1699, the qnrVC-containing strains were CIP-sensitive (Table 1). A series of plasmids were constructed to determine the effect of different qnrVC alleles on quinolone resistance (Table 2). First, when plasmid pVC1699 was translated into VC401, the CIP MIC of the constructed strain increased from 0.375 µg/mL to 4/6 µg/mL, a 10.7/16.0-fold increase. There was a slight difference among the CIP MIC for the same VC401 strains harboring pBAD24-qnrVC1, -qnrVC5, -qnrVC12, and -qnrVC9. Expressing qnr9 genes from VC1699 through vector pBAD24 in VC401, the CIP MIC was 3/4 µg/mL, and inducing the expression of the qnrVC1, qnrVC5, and qnrVC12 genes from the quinolone-sensitive strains through pBAD24 increased the CIP MIC to 1.5–2 μg/mL. Furthermore, expressing qnrVC9 genes through vector pBAD24 in N16961, a reference V. cholera O1 strain without QRDR mutations, the CIP MIC was increased from 0.015 to 0.125 μg/mL, an 8.3-fold increase. Moreover, expressing qnrVC9 genes through vector pBAD24 in VC1891, a V. cholera O139 stain harboring nonsense A171S mutation in QRDR, the CIP MIC was increased from 0.03 to 0.125/0.25 μg/mL, a 4.2/8.3-fold increase.

Table 2.
Characteristics of Cloned V. cholera strains.
As qnr expression required induction by arabinose, and the CIP MICs of stains containing pBAD24-qnrVC9 were lower than that of stains harboring plasmid pVC1699, the mRNA expression level of qnrVC9 was also analyzed by RT-PCR. As shown in Figure 3, the mRNA expression level of qnrVC9 in pBAD24 in N16961, VC1891, and VC401 were higher than that in pVC1699 in VC401.
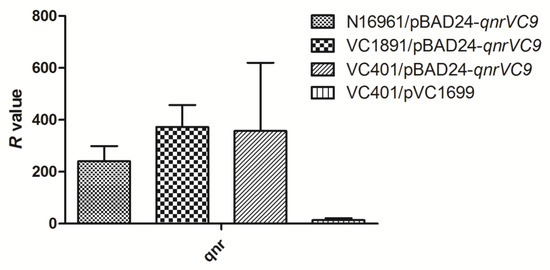
Figure 3.
Comparisons of qnrVC gene expression in N16961/pBAD24-qnrVC9, VC1891/pBAD24-qnrVC9, VC401/pBAD24-qnrVC9, and VC401/pVC1699 cultured by Mueller–Hinton agar containing 0.1% (w/v) arabinose. The relative expression values (R) were measured by RT-PCR. All assays were performed in triplicate, and the means and standard deviations were shown.
3. Discussion
Qnr, encoding by qnr gene, was a pentapeptide repeat protein of 218 amino acids which protect DNA gyrase from quinolone action [12]. qnr gene was first discovered in a plasmid from Klebsiella pneumoniae in 1998 [13], and increasingly being reported worldwide from bacteria isolated from both clinical and aquatic environments [14]. To date, several qnr gene families have been identified, mainly containing qnrA, qnrB, qnrC, qnrD, qnrE, qnrS, and qnrVC [14,15,16,17,18]. These qnr genes generally differ in sequence by 35% or more from each other. Allelic variants differing by 10% or less have also been described in almost every qnr family [9]. Since the qnrVC1 gene was first described in a class 1 integron from a V. cholerae O1 strain recovered in 1998 [15], twelve qnrVC alleles (qnrVC1–12) have been reported, mainly in Vibrionaceae to date [16,19,20,21,22,23], as well as in Enterobacterales in Brazilian coastal waters [24]. qnrB, qnrC, qnrD, and qnrA were harbored by 20-35%, aac(6’)-Ib-cr was harbored by 15% of the CIP unsusceptible P. aeruginosa isolates [25]. The predominant resistant genes were aac(6’)-Ib-cr (48.9%) and qnrD (25.6%) in fluoroquinolone resistant E. coli isolates [26]. In our survey, qnrVC genes were more prevalent in V. cholerae as compared to other quinolone resistance related genes. qnrVC was harbored by 2.4% (8/340) of V. cholera stains, and aac(6’)-Ib-cr was not found in them. In our study, qnrVC1 was discovered in 2002, then qnrVC12 in 2003, qnrVC5 in 2004, and the qnrVC9 was discovered in 2006, demonstrating the undergone successive mutations over time.
The most common mechanism of high-level quinolone resistance was due to accumulation of mutations within QRDRs for both Gram-negative and Gram-positive organisms [27]. Usually, bacteria carrying qnr genes only presented low-level quinolone resistance by blocking CIP inhibition of topoisomerase IV or DNA gyrase [28]. For example, the CIP MIC of V. cholerae O1 carrying qnrVC1 was 0.25 µg/mL [15], the CIP MIC of V. cholerae O1 strain N16961 carrying qnrVC9 was 0.125 µg/mL in our study. Furthermore, Qnr enhanced the frequency of spontaneous mutations in DNA gyrase and topoisomerase IV genes when facing antibiotic stress [13]. The qnr-bearing strains contained a series of spontaneous mutations in the QRDR genes, but only VC1699 contained mutations S83I in GyrA and S85L in ParC, which typically caused the CIP MIC to increase up to 0.25–2 µg/mL [11]. When combined with QRDR gene mutations and with an efflux pump, a high-level fluoroquinolone resistance phenotype easily emerged and resulted in treatment failure [23]. The CIP MIC of V. cholerae O1 carrying qnrVC3 and QRDR gene mutations was 0.5–0.75 µg/mL [29]. The CIP MIC of V. fluvialis carrying qnrVC3 and QRDR gene mutations was 2.5 µg/mL [12]. In our study, qnrVC9 resided on plasmid pVC1699 combined with QRDR gene mutations could caused higher-level CIP resistance. It was also reported qnrB, qnrS combined with QRDR gene mutations could cause higher-level CIP resistance in Enterobacteriaceae [30].
The drug resistance genes often resided on mobile genetic elements for easy dissemination of drug resistance to other organisms. The mobile genetic elements, especially plasmids, played a major role in the dissemination of antimicrobial resistance [31]. qnrVC alleles were usually associated with class 1 integron, SXT or plasmid [22,23]. Interestingly, qnr genes are found in a variety of Gram-positive organisms but are chromosomal and not plasmid-mediated [9]. qnrVC9 was first reported residing in the chromosome of a V. cholera isolate obtained from a pork sample in China [19] and we determined that Int1 captured qnrVC9 and inserted it in a plasmid. The qnrVC9 on the plasmid rather than on the chromosome would transfer more easily to other bacteria, indicating that qnrVC9 has the same mobility and dispersion through different hosts as other plasmid-located qnrVC alleles.
Due to the extensive usage of antimicrobials, the presence and accumulation of quinolones in waters has been widely reported [32]. The aquatic environment played a highly important role in the transfer and maintenance of bacterial genes encoding for antibiotic resistance [33,34] due to the antimicrobials selective pressure. qnr genes carried by Gram-negative bacteria were extensively detected in bacteria isolated from aquatic environments [14]. pVC1699, from a V. cholera O139 strain isolated in 2006 in Jiangxi Province, was entirely identical to plasmid 307 isolated from V. vulnificus, whose sequence was submitted in 2021 from Shanghai, China. It appeared that pVC1699 could transmit between V. vulnificus and V. cholerae O139, and this plasmid was maintained in Vibrios for years. It was also reported other qnr-harboring plasmids could transfer within different hosts under antibiotic induced stress [31]. Therefore, more attention should be paid to this highly efficient plasmid pVC1699 in the capture and dissemination of qnrVC9 among Vibrio species.
Except for strain VC1699, the quinolone-sensitive strains also carried qnrVC genes. In these strains, qnrVC might increase the frequency of spontaneous mutations in DNA gyrase and topoisomerase IV genes under antibiotic stress [13]. In our study, the CIP MIC increased by varying degrees when qnrVC1, qnrVC5, and qnrVC12 gene expression was induced through pBAD24, which suggested that qnrVC1, qnrVC5, and qnrVC12 also can contribute to low-level quinolone resistance in V. cholera. This phenomenon was also described in other study, pCN60-qnrVC1 vector elevated the MIC of CIP from 0.003 to 0.25 µg/mL in E. coli, but seemed to have no effect in P. aeruginosa [35]. This different effect was possibly due to the different efficiency of the promoter or the protein in different strains [35].
The three-dimensional structure of Qnr proteins has been determined by X-ray crystallography [36]; essential amino acids were found in the pentapeptide repeat module and in the larger of two loops, where deletion of only a single amino acid compromises activity [37]. The amino acid change in QnrVC7 also could increase or decrease the CIP MIC: the CIP MIC of E. coli harboring pCR2.1-qnrVC7 was 0.06 µg/mL, E. coli harboring pCR2.1-qnrVC7-T152A was 0.25 µg/mL, and E. coli harboring pCR2.1-qnrVC7 -A82T was 0.015 µg/mL [38]. It was supposed that the difference in sequences of qnrVC alleles resulted in the CIP MIC of VC401 strains carrying pBAD24-qnrVC1, -5, -10, -9 were slightly different from each other. In other reports, different qnrVC alleles also showed different effect on susceptibility to CIP. For example, E. coli harboring pET-qnrVC5 vector had a more than 64.2-fold increase CIP MIC from <0.000243 to 0.0156 µg/mL [12]; E. coli harboring pGEMT-qnrVC1 vector had a 41.7-fold increase CIP MIC from 0.003 to 0.125 µg/mL [35]; E. coli harboring pCR2.1–qnrVC6 vector had a more than 5-fold increase CIP MIC from <0.05 to 0.25 µg/mL [16]. However, despite the qnrVC gene over-expression in VC401/pBAD24-qnrVC9, its CIP MIC was still slightly lower than that of VC401/pVC1699. This might reflect a complex interplay of many known and yet unknown genetic factors. This similar phenomenon was also reported in other study; qnrVC1 seemed to have no effect when harboring by another P. aeruginosa. The CIP MIC of P. aeruginosa (pCN60qnrVC1) was only 0.19 µg/mL, compared with >32 µg/mL for the native strain P. aeruginosa Pa25 [35].
4. Materials and Methods
4.1. Bacterial Strains
A total of 340 V. cholerae O139 strains recovered from patients or the environment in 1993–2009 were randomly selected from the V. cholerae strain bank at the Chinese Center for Disease Control and Prevention (Beijing, China) and were used in our previous study [4], including 290 toxigenic (ctxAB gene positive) strains and 50 non-toxingenic (ctxAB genes negative) strains. These strains, including 237 isolated from patients; 8 isolated from healthy carrier; 13 isolated from fish, bullfrog, or soft-shelled turtle; 45 from surface swabs of patient-related places, or river or sewage water samples; and 37 lacking the related information from the isolated province. A typical wild-type V. cholerae O139 strain, VC401, harbored ctxAB gene was chosen as the recipient strain for electroporation experiments. VC401 had a CIP minimum inhibitory concentration (MIC) of 0.375 µg/mL due to the prevalent double mutations of S83I in GyrA and S85L in parC.
4.2. Antibiotic Susceptibility Testing
Antibiotic susceptibility tests were performed using the agar dilution method according to the current guidelines of the Clinical and Laboratory Standards Institute (CLSI). The precise CIP MIC of the cloning strains was determined with intensive serial dilutions: 0.007, 0.015, 0.03, 0.06, 0.125, 0.25, 0.375, 0.5, 1, 1.5, 2, 3, 4, 6, 8 µg/mL. The control for the antibiotic susceptibility tests was Escherichia coli ATCC 25922.
4.3. PCR and DNA Sequencing
The quinolone resistance related genes (qnr [13], aac(6′)-Ib-cr [10], and qepA, which encoded an efflux pump [39]) were amplified by PCR using previously described primers (Table 3) [29,32]. The PCR primers for detecting ct, int1, sxt, gyrA, gyrB, parC, and parE were described in our previous publication [4]. Furthermore, we amplified and sequenced the open reading frame (ORF) of qnr genes from target strains using the primers qnrF/qnrR or 2qnrF/2qnrR (Table 3). Total chromosomal DNA from V. cholerae O139 was prepared with a DNA purification kit (Tiangen Biotech, Beijing, China). PCR amplification was performed with 2× Taq PCR MasterMix (Tiangen Biotech, China). PCR amplicons were sequenced commercially (Shanghai Sangon Biotech, China) and aligned using MEGA 7.0 software. A phylogenetic tree was constructed using Neighbor-Joining method in MEGA 7.0 software after multiple sequence alignments via CLUSTAL 2.1.

Table 3.
Primers used in this study.
4.4. Plasmid Extraction, Electroporation and Sequencing
Plasmids from all qnr-positive V. cholerae strains were extracted using a QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA, USA). The plasmids were sequenced by primer walking and analyzed by National Center for Biotechnology Information (NCBI) BLAST. The possible ORFs were predicted using ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/, accessed on 1 November 2022). Plasmid transformation into V. cholerae was performed by electroporation with selection on Luria–Bertani (LB) agar containing CIP (1 μg/mL). PCR screening was performed for the QRDRs mutations of the VC401/pVC1699 strains to rule out the possibility of any point mutations.
4.5. Cloning and Expression of qnrVC
The qnrVC genes from V. cholerae wild-type strains were cloned into expression vector pBAD24 using the primers qnrEcorI and qnrHindIII for VC454, VC1435, and VC1699 and primers 2qnrF/2qnrR for VC422 and VC707, and then transformed into E. coli TOP10. The constructed plasmids, which were pBAD24-qnrVC1, -qnrVC5, -qnrVC12, and -qnrVC9, were validated by sequencing. These plasmids were transformed into V. cholerae VC401 by electroporation with selection on LB agar containing ampicillin (100 μg/mL). PCR screening was performed for the QRDRs mutations of the VC401/pBAD24-qnrVC strains to rule out the possibility of any point mutations. As qnr expression required induction by arabinose, VC401 harboring pBAD24-qnrVC was first cultured at 37 °C on LB agar containing 0.1% (w/v) arabinose, and then the CIP MICs of the donor and recipient strains were compared by antibiotic resistance testing using Mueller–Hinton agar also containing 0.1% (w/v) arabinose. All experiments were replicated three times.
RT-PCR was mainly carried out to confirm the expression of qnrVC9 gene in the native isolate VC1699 and the series of transformants. Total RNA was isolated with the RNAprep Pure Cell/Bacteria Kit (Tiangen Biotech, China). The RNA preparations were subsequently treated with DNaseI (TaKaRa, Kusatsu, Japan) to remove the genomic DNA contamination. Reverse transcription was carried on by PrimeScript RT reagent kit (TaKaRa, Japan). Analysis of gene expression levels using TB Green Premix Ex Taq II (TaKaRa, Japan) was carried out on Applied Biosystems 7300 Real-Time PCR System (Thermo Scientific, Waltham, MA, USA) following the protocol recommended by the manufacturer. The qnrF2/qnrR2 primers were used for qnr RT-PCR, and recA was used as an internal control with primers recAF/recAR. The relative expression values (R) were calculated using the equation R = 2(-Delta Delta C(T)) (where CT is the fractional threshold cycle) [40].
5. Conclusions
In summary, we determined the prevalence of quinolone resistance related genes among V. cholerae O139 strains isolated in China. Four qnrVC alleles were detected, but only the strain carrying qnrVC9 had high quinolone resistance. qnrVC9 was carried by a small plasmid, which was conjugative and contributed high CIP resistance to the receptor V. cholerae strain. The same plasmid was also detected in V. vulnificus. Our findings expand our knowledge on the transmission of quinolone resistance among Vibrios.
Author Contributions
W.-L.L., B.K. and J.-R.S. conceived, designed the experiments and the revision of the manuscript. Y.-Y.Z. and L.Y. performed the experiments. Y.-Y.Z., L.-Y.M. and X.L. analyzed the data. B.K. and J.-R.S. contributed reagents/materials/analysis tools. Y.-Y.Z. and L.-Y.M. wrote the paper. B.K. and Y.-Y.Z. acquired funding. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Science and Technology Major Project (2018ZX10714002), National Key Research and Development Program of China (2020YFE0205700) and Capital’s Funds for Health Improvement and Research (2016–4-1101).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Acknowledgments
The authors acknowledge the genes annotation and analysis provided by Haiyin Wang, Beijing Bioinfo Biotechnology Co., Ltd. and Pengcheng Du, Beijing Qitan Biotechnology Co., Ltd.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lippi, D.; Gotuzzo, E.; Caini, S. Cholera. Microbiol. Spectr. 2016, 4, 1–6. [Google Scholar] [CrossRef]
- Ali, M.; Lopez, A.L.; You, Y.A.; Kim, Y.E.; Sah, B.; Maskery, B.; Clemens, J. The global burden of cholera. Bull. World Health Organ. 2012, 90, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Qu, M.; Xu, J.; Ding, Y.; Wang, R.; Liu, P.; Kan, B.; Qi, G.; Liu, Y.; Gao, S. Molecular epidemiology of Vibrio cholerae O139 in China: Polymorphism of ribotypes and CTX elements. J. Clin. Microbiol. 2003, 41, 2306–2310. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Zhou, Y.; Wang, R.; Lou, J.; Zhang, L.; Li, J.; Bi, Z.; Kan, B. Multiple antibiotic resistance of Vibrio cholerae serogroup O139 in China from 1993 to 2009. PLoS ONE 2012, 7, e38633. [Google Scholar] [CrossRef]
- Dutta, D.; Kaushik, A.; Kumar, D.; Bag, S. Foodborne Pathogenic Vibrios: Antimicrobial Resistance. Front. Microbiol. 2021, 12, 638331. [Google Scholar] [CrossRef] [PubMed]
- Piarroux, R.; Rebaudet, S. Cholera. Rev. Prat. 2017, 67, 1117–1121. [Google Scholar]
- Pham, T.D.M.; Ziora, Z.M.; Blaskovich, M.A.T. Quinolone antibiotics. Med. Chem. Comm. 2019, 10, 1719–1739. [Google Scholar] [CrossRef]
- Cabello, F.C. Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and for the environment. Environ. Microbiol. 2006, 8, 1137–1144. [Google Scholar] [CrossRef]
- Hooper, D.C.; Jacoby, G.A. Mechanisms of drug resistance: Quinolone resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 12–31. [Google Scholar] [CrossRef]
- Robicsek, A.; Strahilevitz, J.; Jacoby, G.A.; Macielag, M.; Abbanat, D.; Park, C.H.; Bush, K.; Hooper, D.C. Fluoroquinolone-modifying enzyme: A new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 2006, 12, 83–88. [Google Scholar] [CrossRef]
- Zhou, Y.; Yu, L.; Li, J.; Zhang, L.; Tong, Y.; Kan, B. Accumulation of mutations in DNA gyrase and topoisomerase IV genes contributes to fluoroquinolone resistance in Vibrio cholerae O139 strains. Int. J. Antimicrob. Agents 2013, 42, 72–75. [Google Scholar] [CrossRef] [PubMed]
- Vinothkumar, K.; Kumar, G.N.; Bhardwaj, A.K. Characterization of Vibrio fluvialis qnrVC5 Gene in Native and Heterologous Hosts: Synergy of qnrVC5 with other Determinants in Conferring Quinolone Resistance. Front. Microbiol. 2016, 7, 146. [Google Scholar] [CrossRef]
- Martinez-Martinez, L.; Pascual, A.; Jacoby, G.A. Quinolone resistance from a transferable plasmid. Lancet 1998, 351, 797–799. [Google Scholar] [CrossRef] [PubMed]
- Miranda, C.D.; Concha, C.; Godoy, F.A.; Lee, M.R. Aquatic Environments as Hotspots of Transferable Low-Level Quinolone Resistance and Their Potential Contribution to High-Level Quinolone Resistance. Antibiotics 2022, 11, 1487. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, E.L.; Dos Santos Freitas, F.; Vieira, V.V.; Vicente, A.C. New qnr gene cassettes associated with superintegron repeats in Vibrio cholerae O1. Emerg. Infect. Dis. 2008, 14, 1129–1131. [Google Scholar] [CrossRef]
- Kim, H.B.; Park, C.H.; Kim, C.J.; Kim, E.C.; Jacoby, G.A.; Hooper, D.C. Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrob. Agents Chemother. 2009, 53, 639–645. [Google Scholar] [CrossRef]
- Cavaco, L.M.; Hasman, H.; Xia, S.; Aarestrup, F.M. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob. Agents Chemother. 2009, 53, 603–608. [Google Scholar] [CrossRef]
- Albornoz, E.; Tijet, N.; De Belder, D.; Gomez, S.; Martino, F.; Corso, A.; Melano, R.G.; Petroni, A. qnrE1, a Member of a New Family of Plasmid-Located Quinolone Resistance Genes, Originated from the Chromosome of Enterobacter Species. Antimicrob. Agents Chemother. 2017, 61, e02555-16. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, Z.; Chan, E.W.; Dong, N.; Xia, X.; Chen, S. Molecular Characterization of qnrVCGenes Their Novel Alleles in Vibrio spp Isolated from Food Products in China. Antimicrob. Agents Chemother. 2018, 62, e00529-18. [Google Scholar] [CrossRef]
- Liu, M.; Wong, M.H.; Chen, S. Molecular characterisation of a multidrug resistance conjugative plasmid from Vibrio parahaemolyticus. Int. J. Antimicrob. Agents 2013, 42, 575–579. [Google Scholar] [CrossRef]
- Rajpara, N.; Patel, A.; Tiwari, N.; Bahuguna, J.; Antony, A.; Choudhury, I.; Ghosh, A.; Jain, R.; Ghosh, A.; Bhardwaj, A.K. Mechanism of drug resistance in a clinical isolate of Vibrio fluvialis: Involvement of multiple plasmids and integrons. Int. J. Antimicrob. Agents 2009, 34, 220–225. [Google Scholar] [CrossRef]
- Pons, M.J.; Gomes, C.; Ruiz, J. QnrVC, a new transferable Qnr-like family. Enferm. Infecc. Y Microbiol. Clin. 2013, 31, 191–192. [Google Scholar] [CrossRef]
- Fonseca, E.L.; Vicente, A.C.P. Epidemiology of qnrVC alleles and emergence out of the Vibrionaceae family. J. Med. Microbiol. 2013, 62, 1628–1630. [Google Scholar] [CrossRef]
- Kraychete, G.B.; Botelho, L.A.B.; Monteiro-Dias, P.V.; de Araujo, W.J.; Oliveira, C.J.B.; Carvalho-Assef, A.P.D.; Albano, R.M.; Picao, R.C.; Bonelli, R.R. qnrVC occurs in different genetic contexts in Klebsiella and Enterobacter strains isolated from Brazilian coastal waters. J. Glob. Antimicrob. Resist. 2022, 31, 38–44. [Google Scholar] [CrossRef]
- Nabilou, M.; Babaeekhou, L.; Ghane, M. Fluoroquinolone resistance contributing mechanisms and genotypes of ciprofloxacin- unsusceptible Pseudomonas aeruginosa strains in Iran: Emergence of isolates carrying qnr/aac(6)-Ib genes. International microbiology. Off. J. Span. Soc. Microbiol. 2022, 25, 405–415. [Google Scholar]
- Deku, J.G.; Duedu, K.O.; Ativi, E.; Kpene, G.E.; Feglo, P.K. Burden of Fluoroquinolone Resistance in Clinical Isolates of Escherichia coli at the Ho Teaching Hospital, Ghana. Ethiop. J. Health Sci. 2022, 32, 93–102. [Google Scholar] [PubMed]
- Redgrave, L.S.; Sutton, S.B.; Webber, M.A.; Piddock, L.J. Fluoroquinolone resistance: Mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014, 22, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Tran, J.H.; Jacoby, G.A.; Hooper, D.C. Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob. Agents Chemother. 2005, 49, 3050–3052. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.B.; Wang, M.; Ahmed, S.; Park, C.H.; LaRocque, R.C.; Faruque, A.S.; Salam, M.A.; Khan, W.A.; Qadri, F.; Calderwood, S.B.; et al. Transferable quinolone resistance in Vibrio cholerae. Antimicrob. Agents Chemother. 2010, 54, 799–803. [Google Scholar] [CrossRef]
- Kotb, D.N.; Mahdy, W.K.; Mahmoud, M.S.; Khairy, R.M.M. Impact of co-existence of PMQR genes and QRDR mutations on fluoroquinolones resistance in Enterobacteriaceae strains isolated from community and hospital acquired UTIs. BMC Infect. Dis. 2019, 19, 979. [Google Scholar] [CrossRef]
- Nohejl, T.; Valcek, A.; Papousek, I.; Palkovicova, J.; Wailan, A.M.; Pratova, H.; Minoia, M.; Dolejska, M. Genomic analysis of qnr-harbouring IncX plasmids and their transferability within different hosts under induced stress. BMC Microbiol. 2022, 22, 136. [Google Scholar] [CrossRef] [PubMed]
- Van Doorslaer, X.; Dewulf, J.; Van Langenhove, H.; Demeestere, K. Fluoroquinolone antibiotics: An emerging class of environmental micropollutants. Sci. Total Environ. 2014, 500–501, 250–269. [Google Scholar] [CrossRef]
- Zhuang, M.; Achmon, Y.; Cao, Y.; Liang, X.; Chen, L.; Wang, H.; Siame, B.A.; Leung, K.Y. Distribution of antibiotic resistance genes in the environment. Environ. Pollut. 2021, 285, 117402. [Google Scholar] [CrossRef]
- Brunton, L.A.; Desbois, A.P.; Garza, M.; Wieland, B.; Mohan, C.V.; Hasler, B.; Tam, C.C.; Le, P.N.T.; Phuong, N.T.; Van, P.T.; et al. Identifying hotspots for antibiotic resistance emergence and selection, and elucidating pathways to human exposure: Application of a systems-thinking approach to aquaculture systems. Sci. Total Environ. 2019, 687, 1344–1356. [Google Scholar] [CrossRef] [PubMed]
- Belotti, P.T.; Thabet, L.; Laffargue, A.; Andre, C.; Coulange-Mayonnove, L.; Arpin, C.; Messadi, A.; M’Zali, F.; Quentin, C.; Dubois, V. Description of an original integron encompassing blaVIM-2, qnrVC1 and genes encoding bacterial group II intron proteins in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2015, 70, 2237–2240. [Google Scholar] [CrossRef] [PubMed]
- Hegde, S.S.; Vetting, M.W.; Mitchenall, L.A.; Maxwell, A.; Blanchard, J.S. Structural and biochemical analysis of the pentapeptide repeat protein EfsQnr, a potent DNA gyrase inhibitor. Antimicrob. Agents Chemother. 2011, 55, 110–117. [Google Scholar] [CrossRef]
- Jacoby, G.A.; Corcoran, M.A.; Mills, D.M.; Griffin, C.M.; Hooper, D.C. Mutational analysis of quinolone resistance protein QnrB1. Antimicrob. Agents Chemother. 2013, 57, 5733–5736. [Google Scholar] [CrossRef]
- Po, K.H.; Chan, E.W.; Chen, S. Mutational Analysis of Quinolone Resistance Protein QnrVC7 Provides Novel Insights into the Structure-Activity Relationship of Qnr Proteins. Antimicrob. Agents Chemother. 2016, 60, 1939–1942. [Google Scholar] [CrossRef]
- Yamane, K.; Wachino, J.; Suzuki, S.; Kimura, K.; Shibata, N.; Kato, H.; Shibayama, K.; Konda, T.; Arakawa, Y. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 2007, 51, 3354–3360. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).