Ertapenem Supplemented Selective Media as a New Strategy to Distinguish β-Lactam-Resistant Enterobacterales: Application to Clinical and Wastewater Samples
Abstract
1. Introduction
2. Results
2.1. Implementation of the Technique
2.2. Application to Clinical Samples
2.3. Application to Environmental Samples
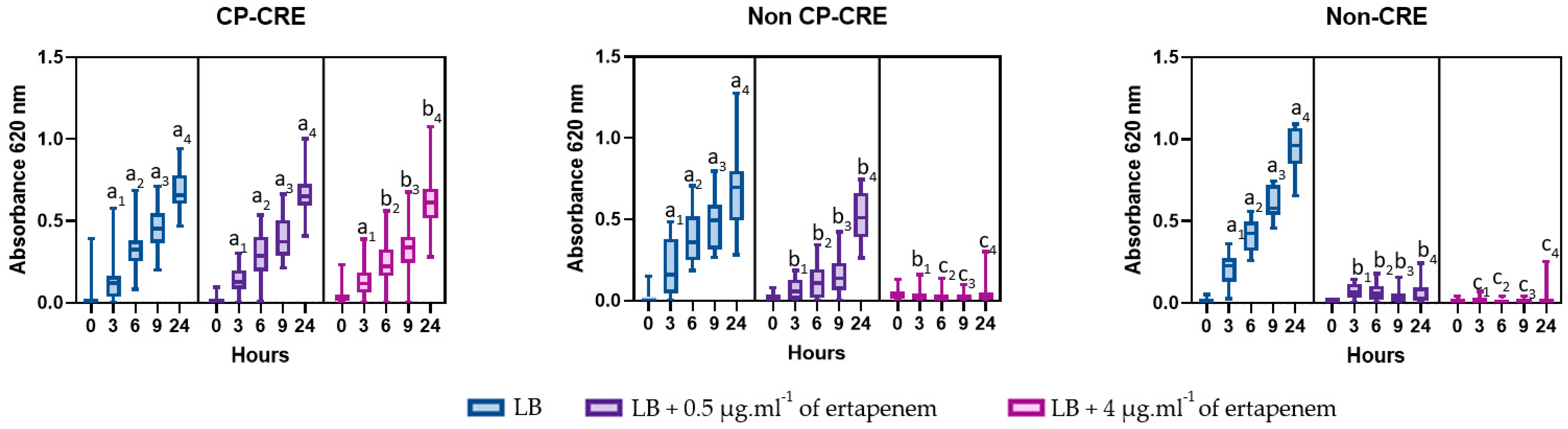
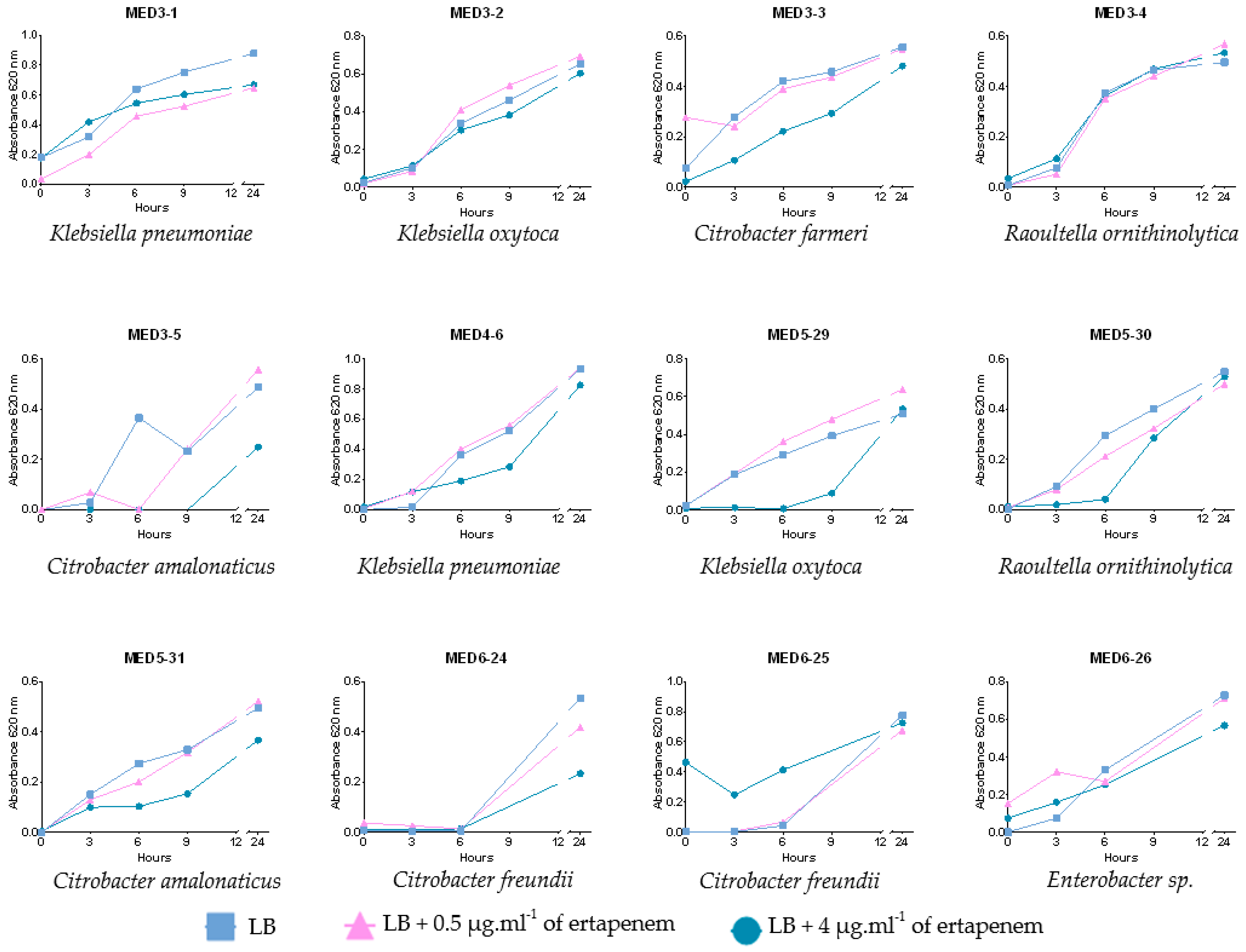
2.4. Growth over Time and Carbapenemase Class Confirmation after 9 Hours of Incubation
3. Discussion
3.1. Supplemented Ertapenem Selective Media
3.2. Application to Clinical Samples
3.3. First CP-CRE Isolated from Hospital Effluent in New Caledonia
4. Materials and Methods
4.1. Implementation of the Technique
4.1.1. Bacterial Strains Used
4.1.2. Growth Dynamics over Time in Presence of Ertapenem
4.2. Application on Clinical and Environmental Samples
4.2.1. Samples Collection
4.2.2. Growth in Presence of Ertapenem
4.2.3. Species Identification
4.2.4. Synergy Test
4.2.5. Antimicrobial Susceptibility Testing
4.2.6. Carbapenemase Class Confirmation
4.3. Growth over Time and Carbapenemase Class Confirmation after Nine Hours of Incubation
4.4. Statistical Analysis
4.5. Ethical Statement
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. The Selection and Use of Essential Medicines: Report of the WHO Expert Committee, 2017 (Including the 20th WHO Model List of Essential Medicines and the 6th Model List of Essential Medicines for Children); WHO Technical Report Series 1006; World Health Organization: Geneva, Switzerland, 2017; ISBN 978-92-4-121015-7. [Google Scholar]
- Willyard, C. The Drug-Resistant Bacteria that Pose the Greatest Health Threats. Nature 2017, 543, 15. [Google Scholar] [CrossRef] [PubMed]
- Codjoe, F.; Donkor, E. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Magiri, R.; Gaundan, S.; Choongo, K.; Zindove, T.; Bakare, A.; Okyere, E.; Okello, W.; Mutwiri, G.; Rafai, E.; Gautam, A.; et al. Antimicrobial Resistance Management in Pacific Island Countries: Current Status, Challenges, and Strategic Solutions. Int. J. One Health 2022, 8, 1–7. [Google Scholar] [CrossRef]
- Foxlee, N.; Townell, N.; McIver, L.; Lau, C. Antibiotic Resistance in Pacific Island Countries and Territories: A Systematic Scoping Review. Antibiotics 2019, 8, 29. [Google Scholar] [CrossRef]
- Loftus, M.; Stewardson, A.; Naidu, R.; Coghlan, B.; Jenney, A.; Kepas, J.; Lavu, E.; Munamua, A.; Peel, T.; Sahai, V.; et al. Antimicrobial Resistance in the Pacific Island Countries and Territories. BMJ Glob. Health 2020, 5, e002418. [Google Scholar] [CrossRef]
- Colot, J.; Fouquet, C.; Ducrocq, F.; Chevalier, S.; Le Provost, C.; Cazorla, C.; Cheval, C.; Fijalkowski, C.; Gourinat, A.-C.; Biron, A.; et al. Prevention and Control of Highly Antibiotic-Resistant Bacteria in a Pacific Territory: Feedback from New Caledonia between 2004 and 2020. Infect. Dis. Now 2022, 52, 7–12. [Google Scholar] [CrossRef]
- Kizny Gordon, A.; Phan, H.T.T.; Lipworth, S.I.; Cheong, E.; Gottlieb, T.; George, S.; Peto, T.E.A.; Mathers, A.J.; Walker, A.S.; Crook, D.W.; et al. Genomic Dynamics of Species and Mobile Genetic Elements in a Prolonged BlaIMP-4-Associated Carbapenemase Outbreak in an Australian Hospital. J. Antimicrob. Chemother. 2020, 75, 873–882. [Google Scholar] [CrossRef]
- Pongchaikul, P.; Mongkolsuk, P. Comprehensive Analysis of Imipenemase (IMP)-Type Metallo-β-Lactamase: A Global Distribution Threatening Asia. Antibiotics 2022, 11, 236. [Google Scholar] [CrossRef]
- Alizadeh, N.; Rezaee, M.A.; Kafil, H.S.; Barhaghi, M.H.S.; Memar, M.Y.; Milani, M.; Hasani, A.; Ghotaslou, R. Detection of Carbapenem-Resistant Enterobacteriaceae by Chromogenic Screening Media. J. Microbiol. Methods 2018, 153, 40–44. [Google Scholar] [CrossRef]
- Girlich, D.; Poirel, L.; Nordmann, P. Comparison of the SUPERCARBA, CHROMagar KPC, and Brilliance CRE Screening Media for Detection of Enterobacteriaceae with Reduced Susceptibility to Carbapenems. Diagn. Microbiol. Infect. Dis. 2013, 75, 214–217. [Google Scholar] [CrossRef] [PubMed]
- Goire, N.; Harnett, G.B.; O’Reilly, L.C.; Ingram, P.R.; Leung, M.J.; Speers, D.J.; Healy, P.E.; Inglis, T.J.J. The Implications of Endemic IMP-4 Carbapenemase for Clinical Laboratory Susceptibility Testing. J. Microbiol. Methods 2016, 124, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Glupczynski, Y.; Berhin, C.; Bauraing, C.; Bogaerts, P. Evaluation of a New Selective Chromogenic Agar Medium for Detection of Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae. J. Clin. Microbiol. 2007, 45, 501–505. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters; The European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2022. [Google Scholar]
- Goodman, K.; Simner, P.; Tamma, P.; Milstone, A. Infection Control Implications of Heterogeneous Resistance Mechanisms in Carbapenem-Resistant Enterobacteriaceae (CRE). Expert Rev. Anti-Infect. Ther. 2016, 14, 95–108. [Google Scholar] [CrossRef]
- Tamma, P.D.; Goodman, K.E.; Harris, A.D.; Tekle, T.; Roberts, A.; Taiwo, A.; Simner, P.J. Comparing the Outcomes of Patients with Carbapenemase-Producing and Non-Carbapenemase-Producing Carbapenem-Resistant Enterobacteriaceae Bacteremia. Clin. Infect. Dis. 2017, 64, 257–264. [Google Scholar] [CrossRef]
- Jean, S.-S.; Hsueh, P.-R. Antimicrobial Susceptibilities of the Ertapenem-Non-Susceptible Non-Carbapenemase-Producing Enterobacterales Isolates Causing Intra-Abdominal Infections in the Asia-Pacific Region during 2008–2014: Results from the Study for Monitoring the Antimicrobial Resistance Trends (SMART). J. Glob. Antimicrob. Resist. 2020, 21, 91–98. [Google Scholar] [CrossRef]
- Jean, S.-S.; Hsueh, P.-R. Distribution of ESBLs, AmpC β-Lactamases and Carbapenemases among Enterobacteriaceae Isolates Causing Intra-Abdominal and Urinary Tract Infections in the Asia-Pacific Region during 2008–2014: Results from the Study for Monitoring Antimicrobial Resistance Trends (SMART). J. Antimicrob. Chemother. 2017, 72, 166–171. [Google Scholar] [CrossRef]
- Tamma, P.D.; Huang, Y.; Opene, B.N.A.; Simner, P.J. Determining the Optimal Carbapenem MIC that Distinguishes Carbapenemase-Producing and Non-Carbapenemase-Producing Carbapenem-Resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 2016, 60, 6425–6429. [Google Scholar] [CrossRef]
- Ferguson, J.K.; Joseph, J.; Kangapu, S.; Zoleveke, H.; Townell, N.; Duke, T.; Manning, L.; Lavu, E. Quality Microbiological Diagnostics and Antimicrobial Susceptibility Testing, an Essential Component of Antimicrobial Resistance Surveillance and Control Efforts in Pacific Island Nations. WPSAR J. 2020, 11, 41–46. [Google Scholar] [CrossRef]
- Foxlee, N.D.; Townell, N.; Tosul, M.A.L.; McIver, L.; Lau, C.L. Bacteriology and Antimicrobial Resistance in Vanuatu: January 2017 to December 2019. Antibiotics 2020, 9, 151. [Google Scholar] [CrossRef]
- Garcia-Quintanilla, M.; Poirel, L.; Nordmann, P. CHROMagar MSuperCARBA and RAPIDEC® Carba NP Test for Detection of Carbapenemase-Producing Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 2018, 90, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Pfennigwerth, N.; Gatermann, S.G.; Körber-Irrgang, B.; Hönings, R. Phenotypic Detection and Differentiation of Carbapenemase Classes Including OXA-48-Like Enzymes in Enterobacterales and Pseudomonas Aeruginosa by a Highly Specialized Micronaut-S Microdilution Assay. J. Clin. Microbiol. 2020, 58, e00171-20. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Kurittu, P.; Al-Mustapha, A.I.; Heljanko, V.; Johansson, V.; Thakali, O.; Mishra, S.K.; Lehto, K.-M.; Lipponen, A.; Oikarinen, S.; et al. Wastewater Surveillance of Antibiotic-Resistant Bacterial Pathogens: A Systematic Review. Front. Microbiol. 2022, 13, 977106. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L. Defining and Combating Antibiotic Resistance from One Health and Global Health Perspectives. Nat. Microbiol. 2019, 4, 1432–1442. [Google Scholar] [CrossRef]
- Kehl, K.; Schallenberg, A.; Szekat, C.; Albert, C.; Sib, E.; Exner, M.; Zacharias, N.; Schreiber, C.; Parčina, M.; Bierbaum, G. Dissemination of Carbapenem Resistant Bacteria from Hospital Wastewater into the Environment. Sci. Total Environ. 2022, 806, 151339. [Google Scholar] [CrossRef] [PubMed]
- Urase, T.; Goto, S.; Sato, M. Monitoring Carbapenem-Resistant Enterobacterales in the Environment to Assess the Spread in the Community. Antibiotics 2022, 11, 917. [Google Scholar] [CrossRef]
- Manaia, C.M. Framework for Establishing Regulatory Guidelines to Control Antibiotic Resistance in Treated Effluents. Crit. Rev. Environ. Sci. Technol. 2022, 53, 1–26. [Google Scholar] [CrossRef]
- Hocquet, D.; Muller, A.; Bertrand, X. What Happens in Hospitals Does Not Stay in Hospitals: Antibiotic-Resistant Bacteria in Hospital Wastewater Systems. J. Hosp. Infect. 2016, 93, 395–402. [Google Scholar] [CrossRef]
- Paulshus, E.; Kühn, I.; Möllby, R.; Colque, P.; O’Sullivan, K.; Midtvedt, T.; Lingaas, E.; Holmstad, R.; Sørum, H. Diversity and Antibiotic Resistance among Escherichia Coli Populations in Hospital and Community Wastewater Compared to Wastewater at the Receiving Urban Treatment Plant. Water Res. 2019, 161, 232–241. [Google Scholar] [CrossRef]
- Cahill, N.; O’Connor, L.; Mahon, B.; Varley, Á.; McGrath, E.; Ryan, P.; Cormican, M.; Brehony, C.; Jolley, K.A.; Maiden, M.C.; et al. Hospital Effluent: A Reservoir for Carbapenemase-Producing Enterobacterales? Sci. Total Environ. 2019, 672, 618–624. [Google Scholar] [CrossRef]
- Albiger, B.; Glasner, C.; Struelens, M.J.; Grundmann, H.; Monnet, D.L.; The European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) working group. Carbapenemase-Producing Enterobacteriaceae in Europe: Assessment by National Experts from 38 Countries, May 2015. Eurosurveillance 2015, 20, 30062. [Google Scholar] [CrossRef]
- Villegas, M.V.; Pallares, C.J.; Escandón-Vargas, K.; Hernández-Gómez, C.; Correa, A.; Álvarez, C.; Rosso, F.; Matta, L.; Luna, C.; Zurita, J.; et al. Characterization and Clinical Impact of Bloodstream Infection Caused by Carbapenemase-Producing Enterobacteriaceae in Seven Latin American Countries. PLoS ONE 2016, 11, e0154092. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Ginn, A.N.; Wiklendt, A.M.; Ellem, J.; Wong, J.S.J.; Ingram, P.; Guy, S.; Garner, S.; Iredell, J.R. Emergence of BlaKPC Carbapenemase Genes in Australia. Int. J. Antimicrob. Agents 2015, 45, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Queenan, A.M.; Bush, K. Carbapenemases: The Versatile β-Lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef] [PubMed]
- Naas, T.; Dortet, L.; Iorga, B.I. Structural and Functional Aspects of Class A Carbapenemases. Curr. Drug Targets 2016, 17, 1006–1028. [Google Scholar] [CrossRef]
- Woodford, N.; Tierno, P.M.; Young, K.; Tysall, L.; Palepou, M.-F.I.; Ward, E.; Painter, R.E.; Suber, D.F.; Shungu, D.; Silver, L.L.; et al. Outbreak of Klebsiella Pneumoniae Producing a New Carbapenem- Hydrolyzing Class A β-Lactamase, KPC-3, in a New York Medical Center. Antimicrob. Agents Chemother. 2004, 48, 4793–4799. [Google Scholar] [CrossRef]
- Yigit, H.; Queenan, A.M.; Rasheed, J.K.; Biddle, J.W.; Domenech-Sanchez, A.; Alberti, S.; Bush, K.; Tenover, F.C. Carbapenem-Resistant Strain of Klebsiella Oxytoca Harboring Carbapenem-Hydrolyzing β-Lactamase KPC-2. Antimicrob. Agents Chemother. 2003, 47, 3881–3889. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Environmental Factors Influencing the Development and Spread of Antibiotic Resistance. FEMS Microbiol. Rev. 2018, 42, fux053. [Google Scholar] [CrossRef]
- Poirel, L.; Héritier, C.; Tolün, V.; Nordmann, P. Emergence of Oxacillinase-Mediated Resistance to Imipenem in Klebsiella Pneumoniae. Antimicrob. Agents Chemother. 2004, 48, 15–22. [Google Scholar] [CrossRef]
- Nayar, R. Antibiotic Impregnated Tablets for Screening ESBL and AmpC Beta Lactamases. IOSR J. Pharm. 2012, 2, 207–209. [Google Scholar] [CrossRef]
- Alatoom, A.; Elsayed, H.; Lawlor, K.; AbdelWareth, L.; El-Lababidi, R.; Cardona, L.; Mooty, M.; Bonilla, M.-F.; Nusair, A.; Mirza, I. Comparison of Antimicrobial Activity between Ceftolozane–Tazobactam and Ceftazidime–Avibactam against Multidrug-Resistant Isolates of Escherichia Coli, Klebsiella Pneumoniae, and Pseudomonas Aeruginosa. Int. J. Infect. Dis. 2017, 62, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Ling, T.K.W.; Tam, P.C.; Liu, Z.K.; Cheng, A.F.B. Evaluation of VITEK 2 Rapid Identification and Susceptibility Testing System against Gram-Negative Clinical Isolates. J. Clin. Microbiol. 2001, 39, 2964–2966. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, S.; Ledeboer, N.A.; Westblade, L.F.; Burnham, C.-A.D.; Faron, M.L.; Bergman, Y.; Yee, R.; Mesich, B.; Gerstbrein, D.; Wallace, M.A.; et al. Evaluation of NG-Test Carba 5 for Rapid Phenotypic Detection and Differentiation of Five Common Carbapenemase Families: Results of a Multicenter Clinical Evaluation. J. Clin. Microbiol. 2020, 58, e00344-20. [Google Scholar] [CrossRef]
- Han, R.; Guo, Y.; Peng, M.; Shi, Q.; Wu, S.; Yang, Y.; Zheng, Y.; Yin, D.; Hu, F. Evaluation of the Immunochromatographic NG-Test Carba 5, RESIST-5 O.O.K.N.V., and IMP K-SeT for Rapid Detection of KPC-, NDM-, IMP-, VIM-Type, and OXA-48-like Carbapenemase among Enterobacterales. Front. Microbiol. 2021, 11, 609856. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2022. [Google Scholar]

| Name | MALDI-ToF MS Identification | Phenotypic β-Lactam-Resistant Mechanisms a | Carbapenemase Class b | Ertapenem Resistance c | Presumptive Carbapenem Resistance Mechanism | |
|---|---|---|---|---|---|---|
| MIC µg·mL−1 | ||||||
| 8210610788 | Citrobacter freundii | Carbapenemase | IMP | I | 2 | CP-CRE |
| 8211020125 | Citrobacter freundii | Carbapenemase | IMP | S | ≤0.5 | CP-CRE |
| 8211020127 | Citrobacter freundii | Carbapenemase | IMP | R | >4 | CP-CRE |
| 8211320250 | Citrobacter freundii | Carbapenemase | IMP | R | 4 | CP-CRE |
| 8211510181 | Enterobacter sp. | Carbapenemase | IMP | R | >4 | CP-CRE |
| 8211760544 | Escherichia coli | Carbapenemase | IMP | S | ≤0.5 | CP-CRE |
| 8213210685 | Klebsiella pneumoniae | Carbapenemase | IMP | R | 4 | CP-CRE |
| 8211480320 | Escherichia coli | Carbapenemase | IMP | R | 4 | CP-CRE |
| CNR1 | Klebsiella pneumoniae | Carbapenemase | KPC | R | >4 | CP-CRE |
| CNR2 (ATCC 11978) | Klebsiella pneumoniae | Carbapenemase | OXA-48 | R | >4 | CP-CRE |
| AH | Enterobacter cloacae | Carbapenemase | IMP | R | >4 | CP-CRE |
| AI | Citrobacter freundii | Carbapenemase | IMP | R | 4 | CP-CRE |
| AM | Klebsiella pneumoniae | Carbapenemase | IMP | R | 2 | CP-CRE |
| BP | Klebsiella pneumoniae | Carbapenemase | IMP | R | 2 | CP-CRE |
| LHE 358.102 | Enterobacter sp. | Carbapenemase | IMP | R | 4 | CP-CRE |
| LHE 358.157 | Enterobacter sp. | Carbapenemase | IMP | R | 4 | CP-CRE |
| BF | Enterobacter sp. | AmpC | - | R | 2 | Non-CP-CRE |
| BM | Escherichia coli | AmpC | - | R | 2 | Non-CP-CRE |
| BS | Klebsiella pneumoniae | ESBL and AmpC | - | R | 4 | Non-CP-CRE |
| BR | Klebsiella aerogenes | AmpC | - | S | ≤0.5 | Non-CP-CRE |
| AY | Enterobacter sp. | ESBL and AmpC | - | R | 4 | Non-CP-CRE |
| BK | Escherichia coli | ESBL | - | S | ≤0.5 | Non-CRE |
| BQ | Enterobacter sp. | ESBL | - | S | ≤0.5 | Non-CRE |
| AD | Klebsiella pneumoniae | ESBL | - | S | ≤0.5 | Non-CRE |
| AR | Klebsiella pneumoniae | ESBL | - | S | ≤0.5 | Non-CRE |
| BO | Serratia marcescens | ESBL | - | S | ≤0.5 | Non-CRE |
| BI | Klebsiella pneumoniae | ESBL | - | S | ≤0.5 | Non-CRE |
| Sample | Ertapenem-Supplemented Selective Media | Territorial Hospital Center Laboratory | Correspondence | |
|---|---|---|---|---|
| Presumptive Resistance Mechanisms | MALDI-ToF MS Identification | Type of β-Lactamase a | ||
| CS-1 | Non-CRE (n= 3) | E. coli | ESBL | Yes |
| K. pneumoniae | - | |||
| CS-2 | Non-CP-CRE (n= 3) | Enterobacter sp. | AmpC | Yes |
| CS-3 | Non-CRE (n= 3) | E. coli | ESBL | Yes |
| K. pneumoniae | ESBL | |||
| C. freundii | ESBL | |||
| CS-4 | Non-CP-CRE (n= 3) | Enterobacter sp. | AmpC | Yes |
| CS-5 | Non-CP-CRE (n= 3) | Enterobacter sp. | AmpC | Yes |
| CS-6 | 2 Non-CRE and 1 Non-CP-CRE | K. pneumoniae | ESBL | Yes |
| E. cloacae | AmpC | |||
| CS-7 | Non-CRE (n= 5) | K. pneumoniae | ESBL | Yes |
| CS-8 | Non-CRE (n= 5) | K. pneumoniae | ESBL | Yes |
| CS-9 | 1 Non-CRE (red colony) and 4 Non-CP-CRE (green colonies) | E. coli | ESBL | Yes |
| K. aerogenes | AmpC | |||
| CS-10 | Non-CRE (n= 5) | K. pneumoniae | ESBL | Yes |
| CS-11 | Non-CRE (n= 5) | Enterobacter sp. | ESBL | Yes |
| CS-12 | Non-CRE (n= 3) | K. pneumoniae | ESBL | Yes |
| CS-13 | Non-CRE (n= 5) | Enterobacter sp. | AmpC | No |
| CS-14 | 2 Non-CRE and 1 Non-CP-CRE | Enterobacter sp. | AmpC | Supplementary phenotype |
| CS-15 | Non-CRE (n= 5) | Enterobacter sp. | AmpC | No |
| CS-16 | Non-CRE (n= 3) | K. pneumoniae | ESBL | Yes |
| CS-17 | Non-CRE (n= 3) | S. marcescens | ESBL | Yes |
| CS-18 | Non-CRE (n= 3) | K. pneumoniae | ESBL | Yes |
| CS-19 | Non-CRE (n= 3) | K. pneumoniae | ESBL | Yes |
| CS-20 | Non-CRE (n= 6) | E. coli | ESBL | Yes |
| CS-21 | Non-CRE (n= 3) | K. aerogenes | AmpC | No |
| CS-22 | Non-CP-CRE (n= 8) | Enterobacter sp. | AmpC | Yes |
| CS-23 | Non-CRE (n= 12) | K. pneumoniae | ESBL | Yes |
| CS-24 | Non-CRE (n= 8) | K. pneumoniae | ESBL | Yes |
| CS-25 | 2 Non-CRE and 2 Non-CP-CRE | Enterobacter sp. | ESBL + AmpC | Yes |
| CS-26 | Non-CRE (n= 16) | E. coli | ESBL | Yes |
| CS-27 | Non-CRE (n= 3) | C. freundii | AmpC but sensitive to ertapenem | Yes |
| CS-28 | Non-CRE (n= 16) | Enterobacter sp. | ESBL | Yes |
| CS-29 | Non-CP-CRE (n= 8) | Enterobacter sp. | AmpC | Yes |
| CS-30 | Non-CP-CRE (n= 16) | Enterobacter sp. | AmpC | Yes |
| Samples | Colony Number Tested | Presumptive CP-CRE a | Presumptive Non-CP-CRE a | Presumptive Non-CRE a |
|---|---|---|---|---|
| MED3 | 24 | 5 (21%) | 8 (33%) | 11 (46%) |
| MED4 | 23 | 1 (4.5%) | 1 (4.5%) | 21 (91%) |
| MED5 | 31 | 3 (10%) | 0 | 28 (90%) |
| MED6 | 26 | 3 (11%) | 2 (8%) | 21 (81%) |
| MED7 | 64 | 21 (33%) | 6 (9%) | 37 (58%) |
| Sample | MALDI-ToF MS Identification | Type of β-Lactamase a | Class of Carbapenemase b | Antimicrobial Susceptibility c | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ampicillin | Amoxicillin + Clavulanic Acid (Urine + Other) | Ticarcillin | Ticarcillin + Clavulanic Acid | Piperacillin | Piperacillin + Tazobactam | Cefotaxim | Ceftazidim | Cefoxitin | Imipenem | Ertapenem | Ertapenem MIC µg·mL−1 | Amikacin | Gentamicin | Nalidixic Acid | Ofloxacin | Ciprofloxacin | Levofloxacin | Nitrofurantoin | Trime Thoprim/Sulfamethoxazol | ||||
| MED3-1 | K. pneumoniae | Carbapenemase | IMP | R | R | R | R | R | R | R | R | R | R | R | 4 | I | R | R | R | R | R | I | R |
| MED3-2 | K. oxytoca | Carbapenemase | IMP | R | R | R | R | R | R | R | R | R | I | R | 2 | S | R | S | I | S | S | S | S |
| MED3-3 | C. farmeri | Carbapenemase | IMP | R | R | R | R | - | R | R | R | R | R | R | 2 | I | R | R | R | R | - | S | R |
| MED3-4 | R. ornithinolytica | Carbapenemase | IMP | R | R | R | R | - | I | R | R | R | R | R | 4 | I | R | R | R | I | - | S | S |
| MED3-5 | C. amalonaticus | ESBL | KPC | R | R | R | R | - | R | R | I | S | R | R | 2 | R | R | - | R | R | - | S | S |
| MED4-6 | K. pneumoniae | Carbapenemase | IMP | R | R | R | R | R | R | R | R | R | R | R | 4 | I | R | R | R | S | - | R | R |
| MED5-29 | K. oxytoca | Carbapenemase | IMP | R | R | R | R | R | R | R | R | R | I | R | 4 | S | R | - | R | I | - | S | R |
| MED5-30 | R. ornithinolytica | Carbapenemase | IMP | R | R | R | R | - | I | R | R | R | R | R | 4 | S | R | - | I | S | - | I | R |
| MED5-31 | C. amalonaticus | ESBL | KPC | R | R | R | R | - | R | R | I | S | R | R | 4 | R | R | - | R | R | - | S | S |
| MED6-24 | C. freundii | Carbapenemase | IMP | R | R | R | R | R | R | R | R | R | R | R | 4 | R | R | - | R | R | R | S | R |
| MED6-25 | C. freundii | Carbapenemase | IMP | R | R | R | R | R | R | R | R | R | R | R | 4 | S | R | - | R | R | R | S | R |
| MED6-26 | Enterobacter sp. | Carbapenemase | IMP | R | R | R | R | R | R | R | R | R | R | R | 4 | S | R | - | R | S | - | I | S |
| MED7-18 | K. pneumoniae | Carbapenemase | IMP | R | R | R | R | R | R | R | R | R | R | R | >4 | S | R | R | R | R | R | R | S |
| MED7-21 | K. pneumoniae | Carbapenemase | IMP | R | R | R | R | R | R | R | R | R | R | R | >4 | S | R | R | R | R | R | R | S |
| MED7-26 | K. pneumoniae | Carbapenemase | IMP | R | R | R | R | R | R | R | R | R | R | R | >4 | S | R | R | R | R | R | I | S |
| MED7-54 | K. oxytoca | Carbapenemase | IMP + KPC | R | R | R | R | R | R | R | R | R | R | R | >4 | S | R | R | R | R | R | S | R |
| MED7-57 | K. oxytoca | Carbapenemase | IMP | R | R | R | R | R | R | R | R | R | R | R | >4 | R | R | R | R | R | R | S | R |
| MED7-61 | S. marcescens | Carbapenemase | IMP | R | R | R | R | R | R | R | R | R | R | R | >4 | R | R | R | R | S | R | R | R |
| Name | Species | Presumptive CP-CRE at T9 | Immunochromatography Test (ICT) on LB + 4 µg·mL−1 Wells at T9 | ICT on LB Wells at T9 | Presumptive CP-CRE at T24 | ICT on LB + 4 µg·mL−1 Wells at T24 | Type of β-Lactamase * | ICT # |
|---|---|---|---|---|---|---|---|---|
| MED7-17 Ϯ | K. pneumoniae | Yes | - | - | Yes | IMP | - | - |
| MED7-18 Ϯ | K. pneumoniae | Yes | No band | - | Yes | IMP | Carbapenemase | - |
| MED7-19 Ϯ | K. pneumoniae | Yes | - | - | Yes | IMP | - | - |
| MED7-21 Ϯ | K. pneumoniae | Yes | IMP | - | Yes | IMP | Carbapenemase | - |
| MED7-25 Ϯ | K. pneumoniae | Yes | IMP | - | Yes | IMP | - | - |
| MED7-26 Ϯ | K. pneumoniae | Yes | No band | - | Yes | IMP | Carbapenemase | - |
| MED7-27 Ϯ | K. pneumoniae | Yes | IMP | - | Yes | IMP | - | - |
| MED7-28 Ϯ | K. pneumoniae | Yes | IMP | - | Yes | IMP | - | - |
| MED7-29 Ϯ | K. pneumoniae | Yes | IMP | - | Yes | IMP | - | - |
| MED7-30 Ϯ | K. pneumoniae | Yes | IMP | - | Yes | IMP | - | - |
| MED7-31 Ϯ | K. pneumoniae | Yes | IMP | - | Yes | IMP | - | - |
| MED7-32 Ϯ | K. pneumoniae | Yes | IMP | - | Yes | IMP | - | - |
| MED7-50 Ϯ | K. pneumoniae | Yes | No band | IMP | Yes | - | - | - |
| MED7-51 Ϯ | K. pneumoniae | Yes | No band | No band | Yes | - | Carbapenemase | IMP |
| MED7-53 Ϯ | K. pneumoniae | Yes | IMP | IMP | Yes | - | - | - |
| MED7-54 | K. oxytoca | No | - | - | Yes | - | Carbapenemase | IMP + KPC |
| MED7-57 | K. oxytoca | No | - | - | Yes | - | Carbapenemase | IMP |
| MED7-60 | K. pneumoniae | No | - | - | Yes | - | Carbapenemase | IMP |
| MED7-61 | S. marcescens | No | - | - | Yes | - | Carbapenemase | IMP |
| MED7-62 | K. oxytoca | No | - | - | Yes | - | Carbapenemase | IMP |
| MED7-64 | S. ureilytica | No | - | - | Yes | - | Carbapenemase | IMP |
| (15/21) 71% | (10/13) 77% | (21/21) 100% | (12/12) 100% | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bourles, A.; Kainiu, M.; Ukeiwe, D.; Brunet, N.; Despaux, C.; Biron, A.; Gourinat, A.-C.; Goarant, C.; Colot, J. Ertapenem Supplemented Selective Media as a New Strategy to Distinguish β-Lactam-Resistant Enterobacterales: Application to Clinical and Wastewater Samples. Antibiotics 2023, 12, 392. https://doi.org/10.3390/antibiotics12020392
Bourles A, Kainiu M, Ukeiwe D, Brunet N, Despaux C, Biron A, Gourinat A-C, Goarant C, Colot J. Ertapenem Supplemented Selective Media as a New Strategy to Distinguish β-Lactam-Resistant Enterobacterales: Application to Clinical and Wastewater Samples. Antibiotics. 2023; 12(2):392. https://doi.org/10.3390/antibiotics12020392
Chicago/Turabian StyleBourles, Alexandre, Malia Kainiu, Damaris Ukeiwe, Nina Brunet, Camille Despaux, Antoine Biron, Ann-Claire Gourinat, Cyrille Goarant, and Julien Colot. 2023. "Ertapenem Supplemented Selective Media as a New Strategy to Distinguish β-Lactam-Resistant Enterobacterales: Application to Clinical and Wastewater Samples" Antibiotics 12, no. 2: 392. https://doi.org/10.3390/antibiotics12020392
APA StyleBourles, A., Kainiu, M., Ukeiwe, D., Brunet, N., Despaux, C., Biron, A., Gourinat, A.-C., Goarant, C., & Colot, J. (2023). Ertapenem Supplemented Selective Media as a New Strategy to Distinguish β-Lactam-Resistant Enterobacterales: Application to Clinical and Wastewater Samples. Antibiotics, 12(2), 392. https://doi.org/10.3390/antibiotics12020392






