3-Substituted Coumarins Inhibit NorA and MepA Efflux Pumps of Staphylococcus aureus
Abstract
:1. Introduction
2. Results
2.1. Antibacterial Activity
2.2. Association with Antibiotics and Ethidium Bromide
2.3. Fluorescence Emission
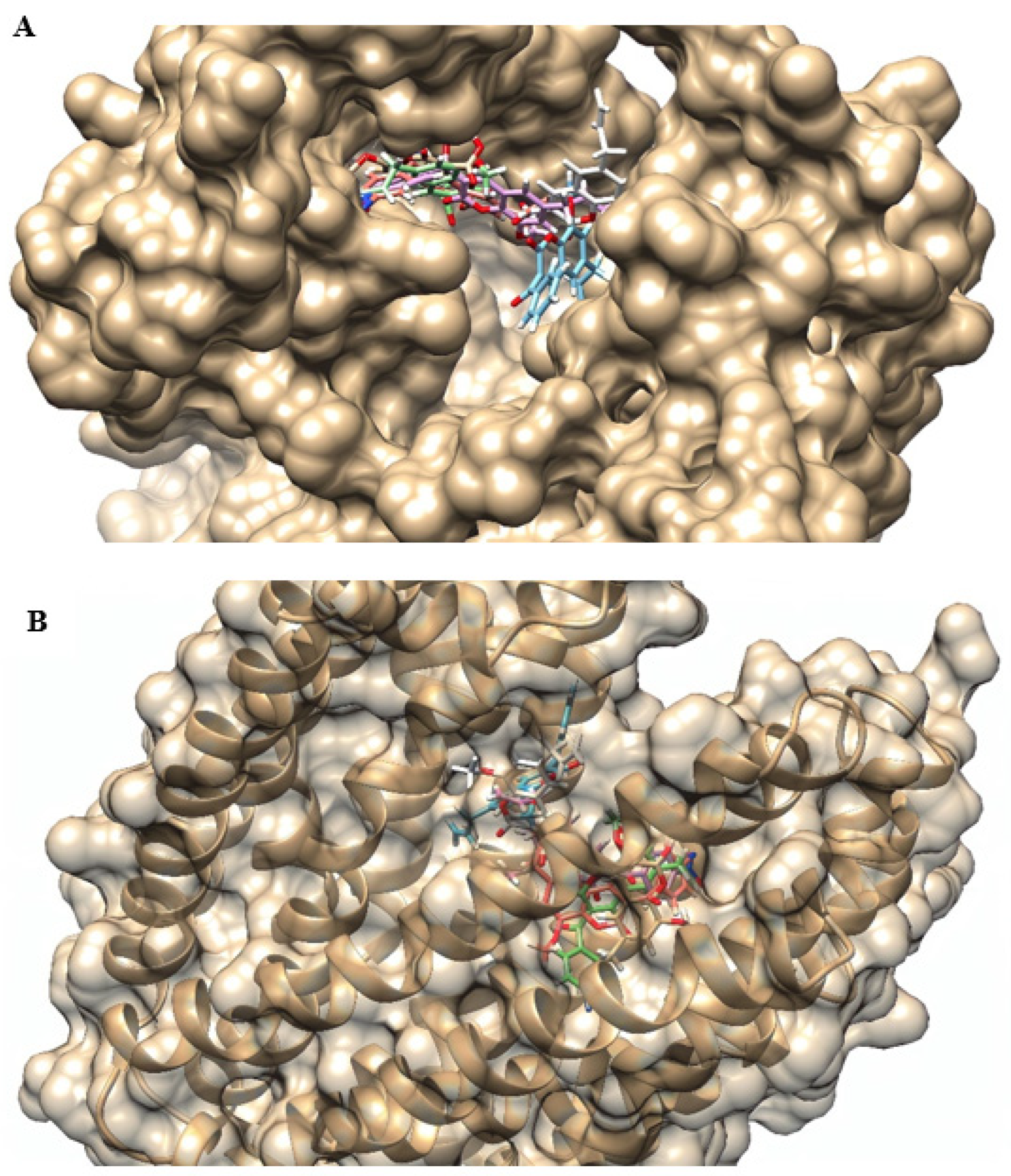
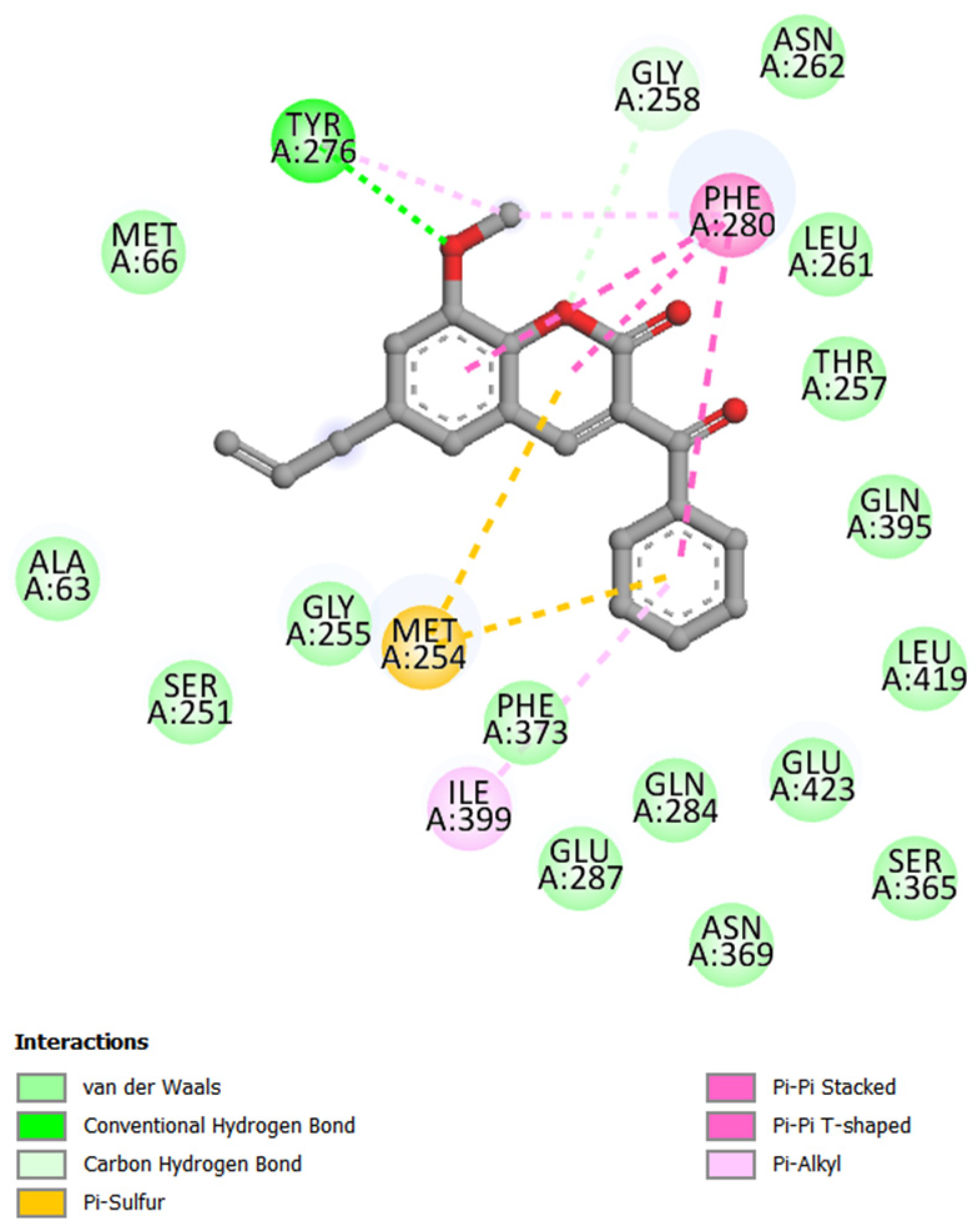
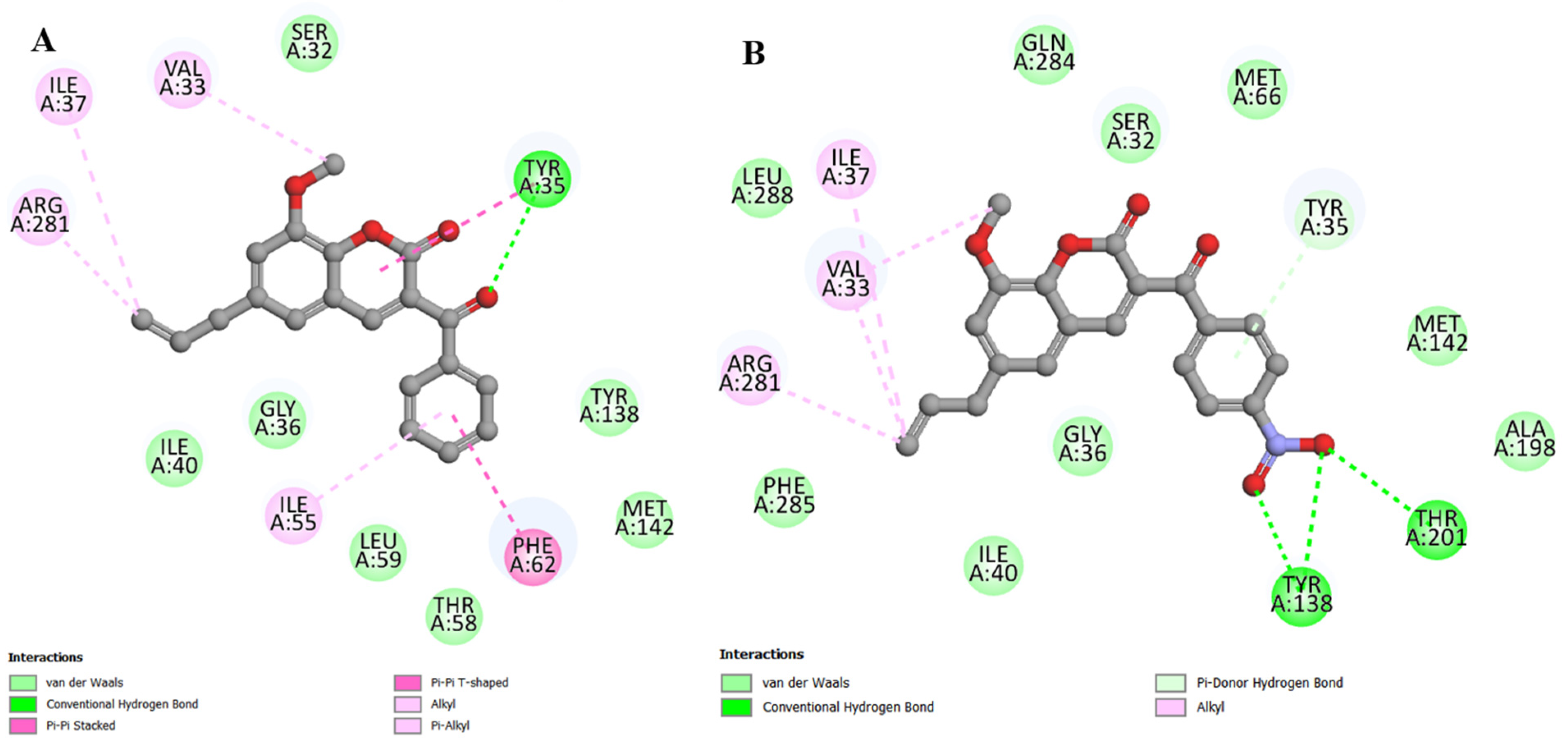
2.4. Molecular Docking
2.5. Effects on Membrane Permeability
3. Discussion
4. Materials and Methods
4.1. Obtaining and Preparing Substances
4.2. Bacterial Strains and Inoculum Preparation
4.3. Assessment of Antibacterial Activity
4.4. Association with Antibiotics and Ethidium Bromide
4.5. Fluorescence Emission Test
4.6. Molecular Docking
4.7. Effects on Membrane Permeability
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and Virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef]
- Howden, B.P.; Giulieri, S.G.; Wong Fok Lung, T.; Baines, S.L.; Sharkey, L.K.; Lee, J.Y.H.; Hachani, A.; Monk, I.R.; Stinear, T.P. Staphylococcus aureus Host Interactions and Adaptation. Nat. Rev. Microbiol. 2023, 21, 380–395. [Google Scholar] [CrossRef]
- Jadimurthy, R.; Mayegowda, S.B.; Nayak, S.C.; Mohan, C.D.; Rangappa, K.S. Escaping Mechanisms of ESKAPE Pathogens from Antibiotics and Their Targeting by Natural Compounds. Biotechnol. Rep. 2022, 34, e00728. [Google Scholar] [CrossRef]
- Mlynarczyk-Bonikowska, B.; Kowalewski, C.; Krolak-Ulinska, A.; Marusza, W. Molecular Mechanisms of Drug Resistance in Staphylococcus aureus. Int. J. Mol. Sci. 2022, 23, 8088. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wu, C.; Gao, H.; Xu, C.; Dai, M.; Huang, L.; Hao, H.; Wang, X.; Cheng, G. Bacterial Multidrug Efflux Pumps at the Frontline of Antimicrobial Resistance: An Overview. Antibiotics 2022, 11, 520. [Google Scholar] [CrossRef] [PubMed]
- Gaurav, A.; Bakht, P.; Saini, M.; Pandey, S.; Pathania, R. Role of Bacterial Efflux Pumps in Antibiotic Resistance, Virulence, and Strategies to Discover Novel Efflux Pump Inhibitors. Microbiology 2023, 169, 001333. [Google Scholar] [CrossRef]
- Stephen, J.; Salam, F.; Lekshmi, M.; Kumar, S.H.; Varela, M.F. The Major Facilitator Superfamily and Antimicrobial Resistance Efflux Pumps of the ESKAPEE Pathogen Staphylococcus aureus. Antibiotics 2023, 12, 343. [Google Scholar] [CrossRef]
- Hassanzadeh, S.; Ganjloo, S.; Pourmand, M.R.; Mashhadi, R.; Ghazvini, K. Epidemiology of Efflux Pumps Genes Mediating Resistance among Staphylococcus aureus; A Systematic Review. Microb. Pathog. 2020, 139, 103850. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Kumar, G.; Kiran Tudu, A. Tackling Multidrug-Resistant Staphylococcus aureus by Natural Products and Their Analogues Acting as NorA Efflux Pump Inhibitors. Bioorg. Med. Chem. 2023, 80, 117187. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, J.; Ahn, J. Advances in the Discovery of Efflux Pump Inhibitors as Novel Potentiators to Control Antimicrobial-Resistant Pathogens. Antibiotics 2023, 12, 1417. [Google Scholar] [CrossRef] [PubMed]
- AlMatar, M.; Albarri, O.; Makky, E.A.; Köksal, F. Efflux Pump Inhibitors: New Updates. Pharmacol. Rep. 2021, 73, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Cook, M.A.; Wright, G.D. The Past, Present, and Future of Antibiotics. Sci. Transl. Med. 2022, 14, eabo7793. [Google Scholar] [CrossRef] [PubMed]
- Tsivileva, O.M.; Koftin, O.V.; Evseeva, N.V. Coumarins as Fungal Metabolites with Potential Medicinal Properties. Antibiotics 2022, 11, 1156. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Singh, K.; Ved, A.; Hasan, S.M.; Mujeeb, S. Therapeutic Journey and Recent Advances in the Synthesis of Coumarin Derivatives. Mini Rev. Med. Chem. 2022, 22, 1314–1330. [Google Scholar] [CrossRef] [PubMed]
- Sharifi-Rad, J.; Cruz-Martins, N.; López-Jornet, P.; Lopez, E.P.-F.; Harun, N.; Yeskaliyeva, B.; Beyatli, A.; Sytar, O.; Shaheen, S.; Sharopov, F.; et al. Natural Coumarins: Exploring the Pharmacological Complexity and Underlying Molecular Mechanisms. Oxid. Med. Cell. Longev. 2021, 2021, 6492346. [Google Scholar] [CrossRef] [PubMed]
- de Araújo, R.S.A.; Barbosa-Filho, J.M.; Scotti, M.T.; Scotti, L.; da Cruz, R.M.D.; dos Falcão-Silva, V.S.; de Siqueira-Júnior, J.P.; Mendonça-Junior, F.J.B. Modulation of Drug Resistance in Staphylococcus aureus with Coumarin Derivatives. Scientifica 2016, 2016, 6894758. [Google Scholar] [CrossRef]
- Šimunović, K.; Solnier, J.; Alperth, F.; Kunert, O.; Smole Možina, S.S.; Bucar, F. Efflux Pump Inhibition and Resistance Modulation in Mycobacterium smegmatis by Peucedanum ostruthium and Its Coumarins. Antibiotics 2021, 10, 1075. [Google Scholar] [CrossRef]
- Flores-Morales, V.; Villasana-Ruíz, A.P.; Garza-Veloz, I.; González-Delgado, S.; Martinez-Fierro, M.L. Therapeutic Effects of Coumarins with Different Substitution Patterns. Molecules 2023, 28, 2413. [Google Scholar] [CrossRef]
- Ranjan Sahoo, C.; Sahoo, J.; Mahapatra, M.; Lenka, D.; Kumar Sahu, P.; Dehury, B.; Nath Padhy, R.; Kumar Paidesetty, S. Coumarin Derivatives as Promising Antibacterial Agent(s). Arab. J. Chem. 2021, 14, 102922. [Google Scholar] [CrossRef]
- Qin, H.-L.; Zhang, Z.-W.; Ravindar, L.; Rakesh, K.P. Antibacterial Activities with the Structure-Activity Relationship of Coumarin Derivatives. Eur. J. Med. Chem. 2020, 207, 112832. [Google Scholar] [CrossRef] [PubMed]
- Prabhala, P.; Sutar, S.M.; Savanur, H.M.; Joshi, S.D.; Kalkhambkar, R.G. In Vitro Antimicrobial Combat, Molecular Modelling and Structure Activity Relationship Studies of Novel Class of Aryl-Ethyne Tethered Coumarin Analogues and Some 3-Aryl Coumarin Derivatives. Eur. J. Med. Chem. Rep. 2022, 5, 100048. [Google Scholar] [CrossRef]
- Tiwari, S.; Seijas, J.; Vazquez-Tato, M.; Sarkate, A.; Karnik, K.; Nikalje, A. Facile Synthesis of Novel Coumarin Derivatives, Antimicrobial Analysis, Enzyme Assay, Docking Study, ADMET Prediction and Toxicity Study. Molecules 2017, 22, 1172. [Google Scholar] [CrossRef] [PubMed]
- Salwan, R.; Sharma, V. Bioactive Compounds of Streptomyces: Biosynthesis to Applications. In Studies in Natural Products Chemistry, 1st ed.; Rahman, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; Volume 64, pp. 467–491. [Google Scholar] [CrossRef]
- Bhatt, S.; Chatterjee, S. Fluoroquinolone Antibiotics: Occurrence, Mode of Action, Resistance, Environmental Detection, and Remediation—A Comprehensive Review. Environ. Pollut. 2022, 315, 120440. [Google Scholar] [CrossRef] [PubMed]
- Melliou, E.; Magiatis, P.; Mitaku, S.; Skaltsounis, A.-L.; Chinou, E.; Chinou, I. Natural and Synthetic 2,2-Dimethylpyranocoumarins with Antibacterial Activity. J. Nat. Prod. 2005, 68, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Baruah, P.; Basumatary, G.; Yesylevskyy, S.O.; Aguan, K.; Bez, G.; Mitra, S. Novel Coumarin Derivatives as Potent Acetylcholinesterase Inhibitors: Insight into Efficacy, Mode and Site of Inhibition. J. Biomol. Struct. Dyn. 2019, 37, 1750–1765. [Google Scholar] [CrossRef]
- Sovari, S.N.; Vojnovic, S.; Bogojevic, S.S.; Crochet, A.; Pavic, A.; Nikodinovic-Runic, J.; Zobi, F. Design, Synthesis and in Vivo Evaluation of 3-Arylcoumarin Derivatives of Rhenium(I) Tricarbonyl Complexes as Potent Antibacterial Agents against Methicillin-Resistant Staphylococcus aureus (MRSA). Eur. J. Med. Chem. 2020, 205, 112533. [Google Scholar] [CrossRef]
- Ge, Z.; Ji, Q.; Chen, C.; Liao, Q.; Wu, H.; Liu, X.; Huang, Y.; Yuan, L.; Liao, F. Synthesis and Biological Evaluation of Novel 3-Substituted Amino-4-Hydroxylcoumarin Derivatives as Chitin Synthase Inhibitors and Antifungal Agents. J. Enzym. Inhib. Med. Chem. 2016, 31, 219–228. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, V.; Pathania, R. Efflux Pump Inhibitors for Bacterial Pathogens: From Bench to Bedside. Indian J. Med. Res. 2019, 149, 129. [Google Scholar] [CrossRef]
- Martin, A.L.A.R.; De Menezes, I.R.A.; Sousa, A.K.; Farias, P.A.M.; dos Santos, F.A.V.; Freitas, T.S.; Figueredo, F.G.; Ribeiro-Filho, J.; Carvalho, D.T.; Coutinho, H.D.M.; et al. In Vitro and in Silico Antibacterial Evaluation of Coumarin Derivatives against MDR Strains of Staphylococcus aureus and Escherichia coli. Microb. Pathog. 2023, 177, 106058. [Google Scholar] [CrossRef]
- de Sousa, A.K.; Rocha, J.E.; de Freitas, T.S.; Freitas, P.R.; Pereira, R.L.S.; Júnior, F.N.P.; Brancaglion, G.A.; de Paulo, D.C.; Carvalho, D.T.; de Menezes, I.R.A.; et al. Photobiological Effect of Eugenol--derived 3--benzoylcoumarin Associated with Led Lights against MDR Microorganisms. Fundam. Clin. Pharmacol. 2023, 37, 316–323. [Google Scholar] [CrossRef]
- Madeiro, S.A.L.; Borges, N.H.P.B.; Souto, A.L.; de Figueiredo, P.T.R.; Siqueira-Junior, J.P.; Tavares, J.F. Modulation of the Antibiotic Activity against Multidrug Resistant Strains of Coumarins Isolated from Rutaceae Species. Microb. Pathog. 2017, 104, 151–154. [Google Scholar] [CrossRef]
- Martin, A.L.A.R.; Pereira, R.L.S.; Rocha, J.E.; Farias, P.A.M.; Freitas, T.S.; de Lemos Caldas, F.R.; Figueredo, F.G.; Sampaio, N.F.L.; Ribeiro-Filho, J.; Menezes, I.R. de A.; et al. In Vitro and in Silico Evidences about the Inhibition of MepA Efflux Pump by Coumarin Derivatives. Microb. Pathog. 2023, 182, 106246. [Google Scholar] [CrossRef]
- Verma, P.; Tiwari, M.; Tiwari, V. Strategies to Combat Bacterial Antimicrobial Resistance: A Focus on Mechanism of the Efflux Pumps Inhibitors. SN Compr. Clin. Med. 2021, 3, 510–527. [Google Scholar] [CrossRef]
- Roy, S.K.; Kumari, N.; Pahwa, S.; Agrahari, U.C.; Bhutani, K.K.; Jachak, S.M.; Nandanwar, H. NorA Efflux Pump Inhibitory Activity of Coumarins from Mesua ferrea. Fitoterapia 2013, 90, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Bazzaz, B.S.F.; Memariani, Z.; Khashiarmanesh, Z.; Iranshahi, M.; Naderinasab, M. Effect of Galbanic Acid, a Sesquiterpene Coumarin from Ferula Szowitsiana, as an Inhibitor of Efflux Mechanism in Resistant Clinical Isolates of Staphylococcus aureus. Braz. J. Microbiol. 2010, 41, 574–580. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.; Singh, S.; Wani, A.; Sharma, S.; Jain, S.K.; Singh, B.; Gupta, B.D.; Satti, N.K.; Koul, S.; Khan, I.A.; et al. Osthol and Curcumin as Inhibitors of Human Pgp and Multidrug Efflux Pumps of Staphylococcus aureus: Reversing the Resistance against Frontline Antibacterial Drugs. Med. Chem. Commun. 2014, 5, 1540–1547. [Google Scholar] [CrossRef]
- Rodrigues, D.F.; Borges, N.H.P.B.; Nogueira, C.E.S.; Tavares, J.F.; Arcanjo, D.D.R.; Barreto, H.M.; Siqueira-Junior, J.P. Modulation of Drug Resistance by Furanochromones in NorA Overexpressing Staphylococcus aureus. Evid.-Based Complement. Altern. Med. 2022, 2022, 9244500. [Google Scholar] [CrossRef]
- dos Santos Barbosa, C.R.; Scherf, J.R.; de Freitas, T.S.; de Menezes, I.R.A.; Pereira, R.L.S.; dos Santos, J.F.S.; de Jesus, S.S.P.; Lopes, T.P.; de Sousa Silveira, Z.; de Morais Oliveira-Tintino, C.D.; et al. Effect of Carvacrol and Thymol on NorA Efflux Pump Inhibition in Multidrug-Resistant (MDR) Staphylococcus aureus Strains. J. Bioenerg. Biomembr. 2021, 53, 489–498. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira-Tintino, C.D.M.; Tintino, S.R.; Muniz, D.F.; Rodrigues dos Santos Barbosa, C.; Pereira, R.L.S.; Begnini, I.M.; Rebelo, R.A.; da Silva, L.E.; Mireski, S.L.; Nasato, M.C.; et al. Chemical Synthesis, Molecular Docking and MepA Efflux Pump Inhibitory Effect by 1,8-Naphthyridines Sulfonamides. Eur. J. Pharm. Sci. 2021, 160, 105753. [Google Scholar] [CrossRef]
- Chen, X.; Li, X.; Li, Y. Toxicity Inhibition Strategy of Microplastics to Aquatic Organisms through Molecular Docking, Molecular Dynamics Simulation and Molecular Modification. Ecotoxicol. Environ. Saf. 2021, 226, 112870. [Google Scholar] [CrossRef]
- Catherene Tomy, P.; Mohan, C.G. Chemical Space Navigation by Machine Learning Models for Discovering Selective MAO-B Enzyme Inhibitors for Parkinson’s Disease. Artif. Intell. Chem. 2023, 1, 100012. [Google Scholar] [CrossRef]
- Coursier, D.; Coulette, D.; Leman, H.; Grenier, E.; Ichim, G. Live-Cell Imaging and Mathematical Analysis of the “Community Effect” in Apoptosis. Apoptosis 2023, 28, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Ledger, E.V.K.; Sabnis, A.; Edwards, A.M. Polymyxin and Lipopeptide Antibiotics: Membrane-Targeting Drugs of Last Resort. Microbiology 2022, 168, 001136. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Li, H.; Li, H.; Fang, S.; Liu, J.; Chen, Y.; Liu, S.; Lin, S. Development of Amphiphilic Coumarin Derivatives as Membrane-Active Antimicrobial Agents with Potent In Vivo Efficacy against Gram-Positive Pathogenic Bacteria. ACS Infect. Dis. 2021, 7, 2864–2875. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-C.; Zeng, C.-M.; Avula, S.R.; Peng, X.-M.; Geng, R.-X.; Zhou, C.-H. Novel Coumarin Aminophosphonates as Potential Multitargeting Antibacterial Agents against Staphylococcus aureus. Eur. J. Med. Chem. 2023, 245, 114891. [Google Scholar] [CrossRef] [PubMed]
- Mehta, D.; Saini, V.; Aggarwal, B.; Khan, A.; Bajaj, A. Unlocking the Bacterial Membrane as a Therapeutic Target for Next-Generation Antimicrobial Amphiphiles. Mol. Asp. Med. 2021, 81, 100999. [Google Scholar] [CrossRef] [PubMed]
- Koszelewski, D.; Kowalczyk, P.; Brodzka, A.; Hrunyk, A.; Kramkowski, K.; Ostaszewski, R. Enzymatic Synthesis of a Novel Coumarin Aminophosphonates: Antibacterial Effects and Oxidative Stress Modulation on Selected E. coli Strains. Int. J. Mol. Sci. 2023, 24, 7609. [Google Scholar] [CrossRef]
- Govêa, K.P.; Pereira, R.S.T.; de Assis, M.D.O.; Alves, P.I.; Brancaglion, G.A.; Toyota, A.E.; Machado, J.V.C.; Carvalho, D.T.; de Souza, T.C.; Beijo, L.A.; et al. Allelochemical Activity of Eugenol-Derived Coumarins on Lactuca sativa L. Plants 2020, 9, 533. [Google Scholar] [CrossRef] [PubMed]
- da Silva, M.M.C.; de Araújo-Neto, J.B.; de Araújo, A.C.J.; Freitas, P.R.; de Oliveira-Tintino, C.D.M.; Begnini, I.M.; Rebelo, R.A.; da Silva, L.E.; Mireski, S.L.; Nasato, M.C.; et al. Potentiation of Antibiotic Activity by a Meldrum’s Acid Arylamino Methylene Derivative against Multidrug-Resistant Bacterial Strains. Indian J. Microbiol. 2021, 61, 100–103. [Google Scholar] [CrossRef]
- Rocha, J.E.; de Freitas, T.S.; da Cunha Xavier, J.; Pereira, R.L.S.; Junior, F.N.P.; Nogueira, C.E.S.; Marinho, M.M.; Bandeira, P.N.; de Oliveira, M.R.; Marinho, E.S.; et al. Antibacterial and Antibiotic Modifying Activity, ADMET Study and Molecular Docking of Synthetic Chalcone (E)-1-(2-Hydroxyphenyl)-3-(2,4-Dimethoxy-3-Methylphenyl)Prop-2-En-1-One in Strains of Staphylococcus aureus Carrying NorA and MepA Efflux Pumps. Biomed. Pharmacother. 2021, 140, 111768. [Google Scholar] [CrossRef] [PubMed]
- CLSI. M100 Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; Clinical and Laboratory Standards Institute: Wayne, NJ, USA, 2020; pp. 58–66. [Google Scholar]
- Javadpour, M.M.; Juban, M.M.; Lo, W.-C.J.; Bishop, S.M.; Alberty, J.B.; Cowell, S.M.; Becker, C.L.; McLaughlin, M.L. De Novo Antimicrobial Peptides with Low Mammalian Cell Toxicity. J. Med. Chem. 1996, 39, 3107–3113. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, A.T.L.; de Araújo-Neto, J.B.; Costa da Silva, M.M.; Paulino da Silva, M.E.; Carneiro, J.N.P.; Fonseca, V.J.A.; Coutinho, H.D.M.; Bandeira, P.N.; dos Santos, H.S.; da Silva Mendes, F.R.; et al. Synthesis of Chalcones and Their Antimicrobial and Drug Potentiating Activities. Microb. Pathog. 2023, 180, 106129. [Google Scholar] [CrossRef]
- Coutinho, H.D.M.; Costa, J.G.M.; Lima, E.O.; Falcão-Silva, V.S.; Siqueira-Júnior, J.P. Enhancement of the Antibiotic Activity against a Multiresistant Escherichia coli by Mentha arvensis and Chlorpromazine. Chemotherapy 2008, 54, 328–330. [Google Scholar] [CrossRef] [PubMed]
- da Silva, L.Y.S.; Paulo, C.L.R.; Moura, T.F.; Alves, D.S.; Pessoa, R.T.; Araújo, I.M.; de Morais Oliveira-Tintino, C.D.; Tintino, S.R.; de Nonato, C.F.A.; da Costa, J.G.M.; et al. Antibacterial Activity of the Essential Oil of Piper tuberculatum Jacq. Fruits against Multidrug-Resistant Strains: Inhibition of Efflux Pumps and β-Lactamase. Plants 2023, 12, 2377. [Google Scholar] [CrossRef]
- Oliveira, M.M.; Santos, H.S.; Coutinho, H.D.M.; Bandeira, P.N.; da Silva, P.T.; Freitas, T.S.; Rocha, J.E.; Xavier, J.C.; Campina, F.F.; Barbosa, C.R.S.; et al. Spectroscopic Characterization and Efflux Pump Modulation of a Thiophene Curcumin Derivative. J. Mol. Struct. 2020, 1215, 128291. [Google Scholar] [CrossRef]
- Freitas, T.S.; Xavier, J.C.; Pereira, R.L.S.; Rocha, J.E.; Campina, F.F.; de Araújo Neto, J.B.; Silva, M.M.C.; Barbosa, C.R.S.; Marinho, E.S.; Nogueira, C.E.S.; et al. In Vitro and in Silico Studies of Chalcones Derived from Natural Acetophenone Inhibitors of NorA and MepA Multidrug Efflux Pumps in Staphylococcus aureus. Microb. Pathog. 2021, 161, 105286. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Bitencourt-Ferreira, G.; de Azevedo, W.F. Molegro Virtual Docker for Docking. In Docking Screens for Drug Discovery. Methods in Molecular Biology, 1st ed.; de Azevedo, W.F., Ed.; Humana: New York, NY, USA, 2019; Volume 2053, pp. 149–167. [Google Scholar] [CrossRef]
- de Lima Silva, M.G.; da Silva, L.Y.S.; de Freitas, T.S.; Rocha, J.E.; Pereira, R.L.S.; Tintino, S.R.; de Oliveira, M.R.C.; Bezerra Martins, A.O.B.P.; Lima, M.C.P.; Alverni da Hora, G.C.; et al. Antibacterial Effect and Evaluation of the Inhibitory Effect against Efflux Pump in Staphylococcus aureus by Abietic Acid: In Vitro and in Silico Assays. Process Biochem. 2022, 122, 363–372. [Google Scholar] [CrossRef]
- de Araújo, A.C.J.; Freitas, P.R.; dos Santos Barbosa, C.R.; Muniz, D.F.; de Almeida, R.S.; Alencar de Menezes, I.R.; Ribeiro-Filho, J.; Tintino, S.R.; Coutinho, H.D.M. In Vitro and In Silico Inhibition of Staphylococcus aureus Efflux Pump NorA by α-Pinene and Limonene. Curr. Microbiol. 2021, 78, 3388–3393. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Yuen, H.L.; Chan, S.Y.; Ding, Y.E.; Lim, S.; Tan, G.C.; Kho, C.L. Development of a Novel Antibacterial Peptide, PAM-5, via Combination of Phage Display Selection and Computer-Assisted Modification. Biomolecules 2023, 13, 466. [Google Scholar] [CrossRef] [PubMed]
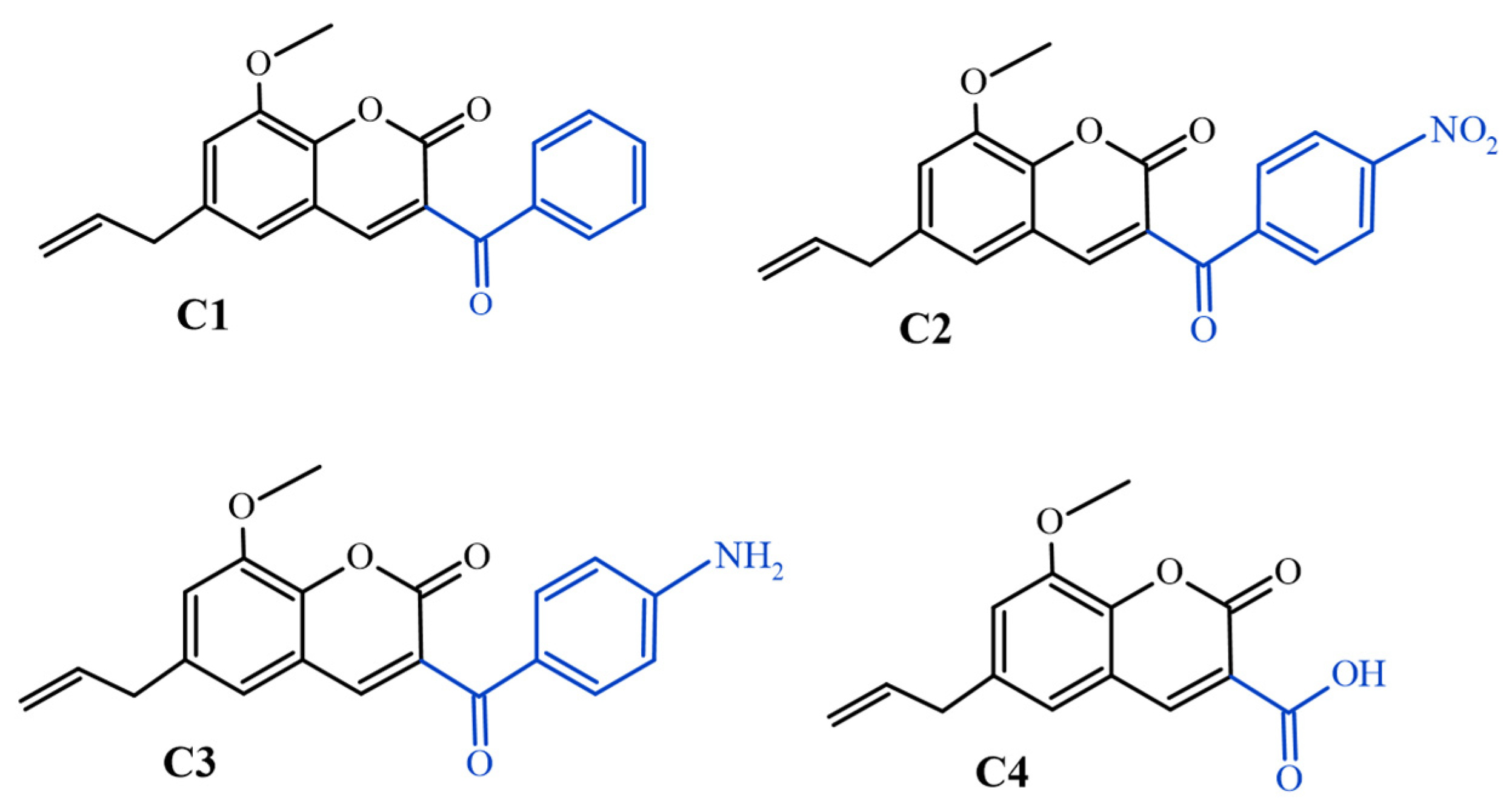

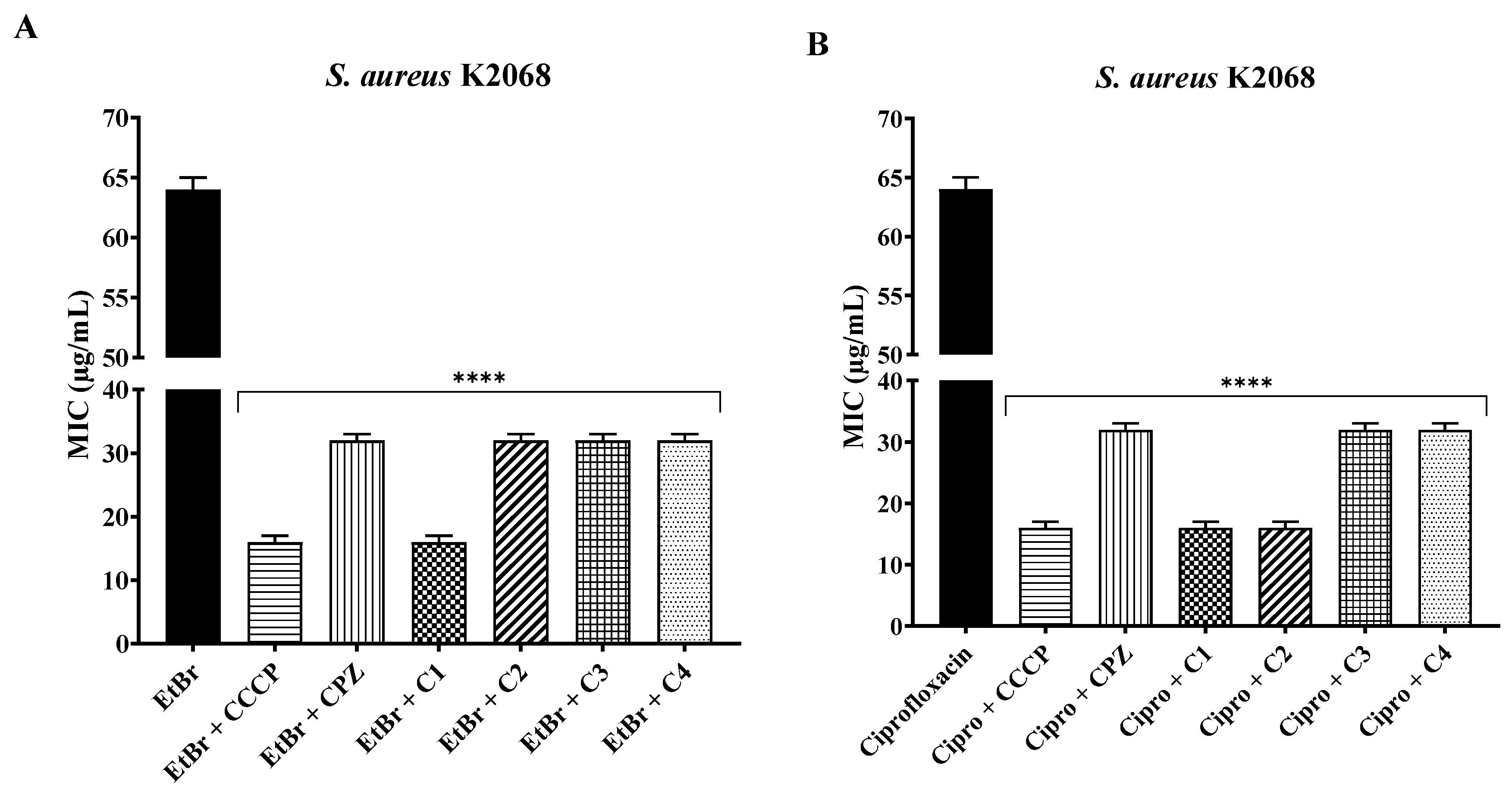



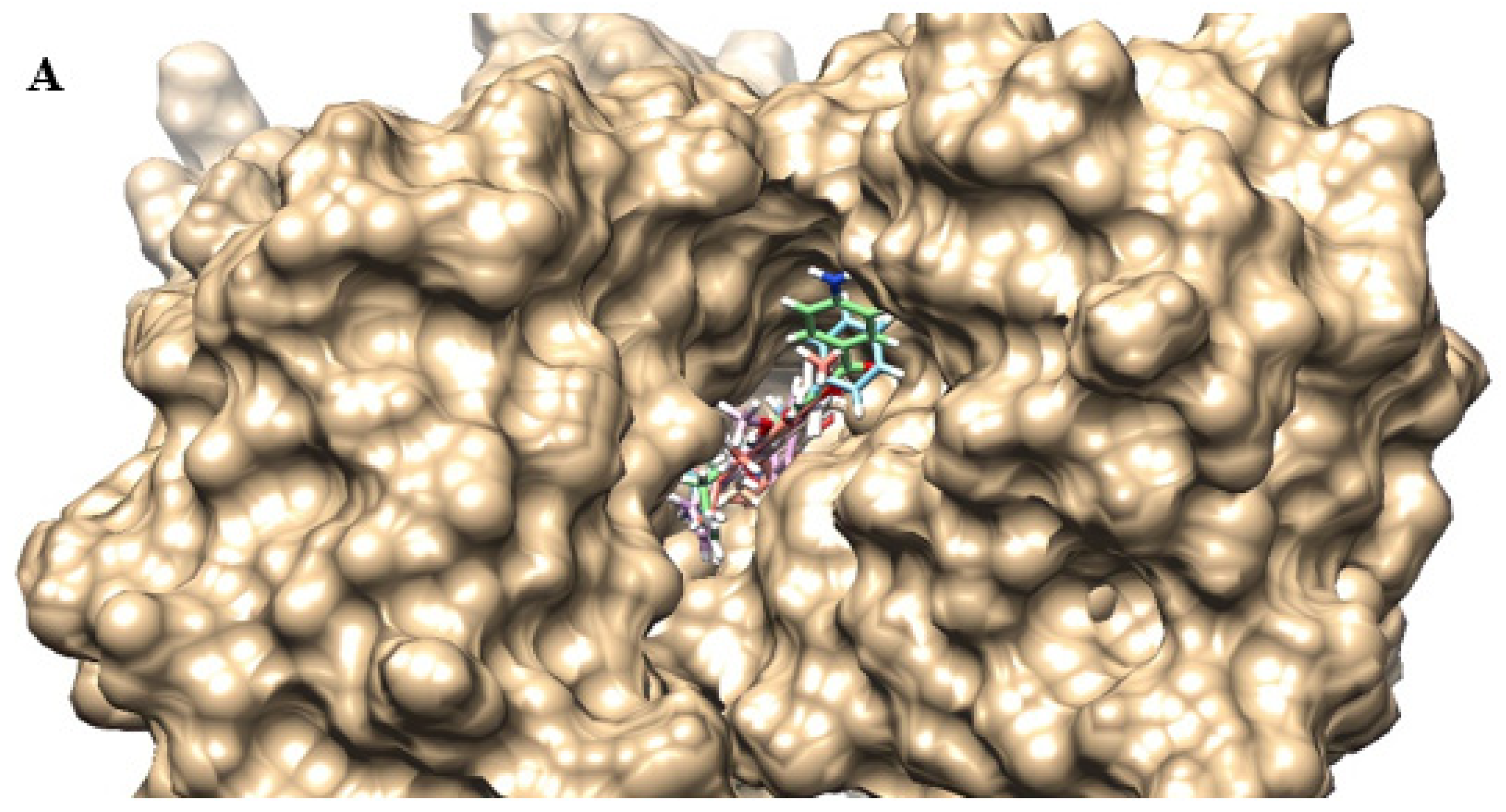
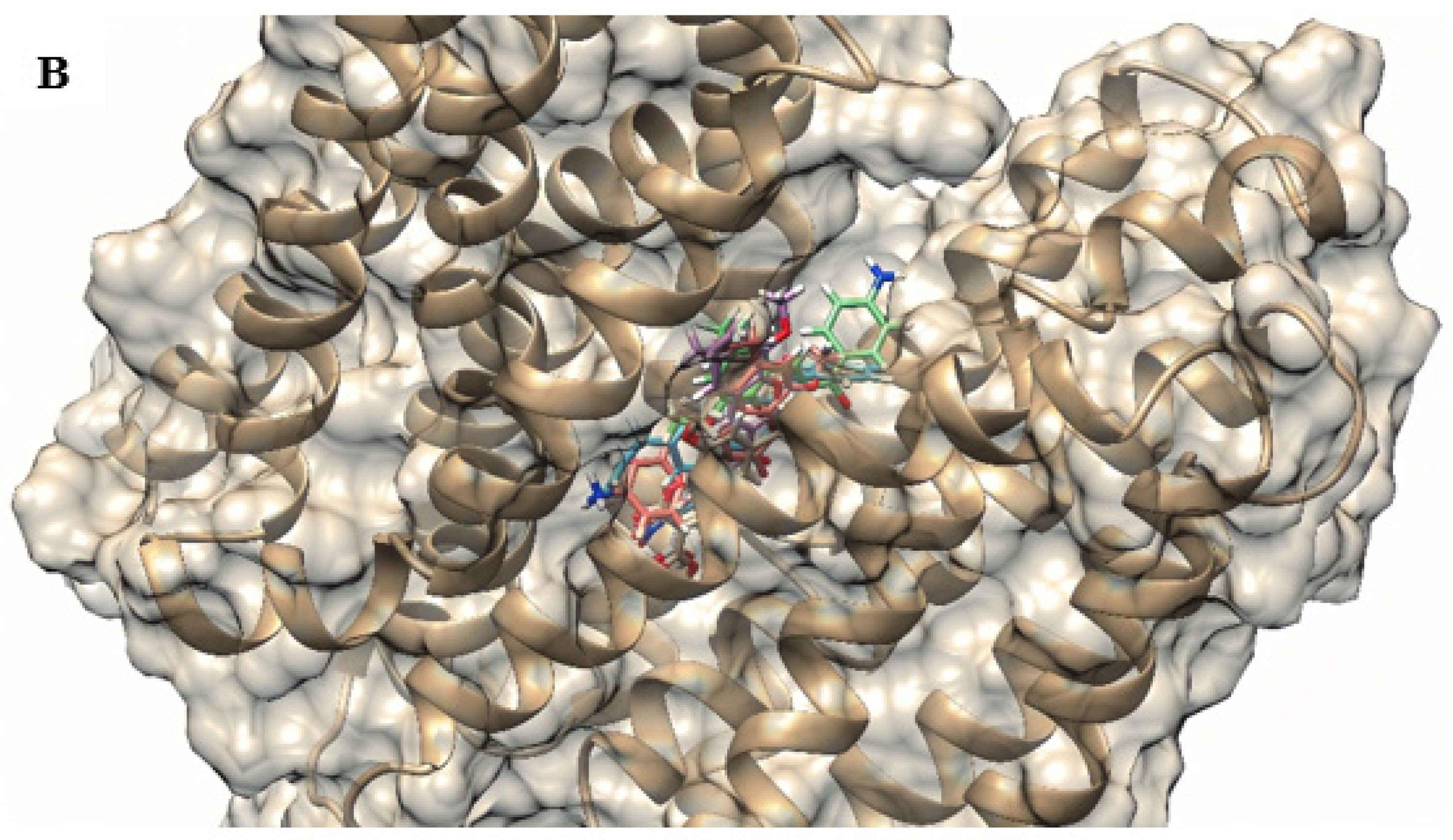
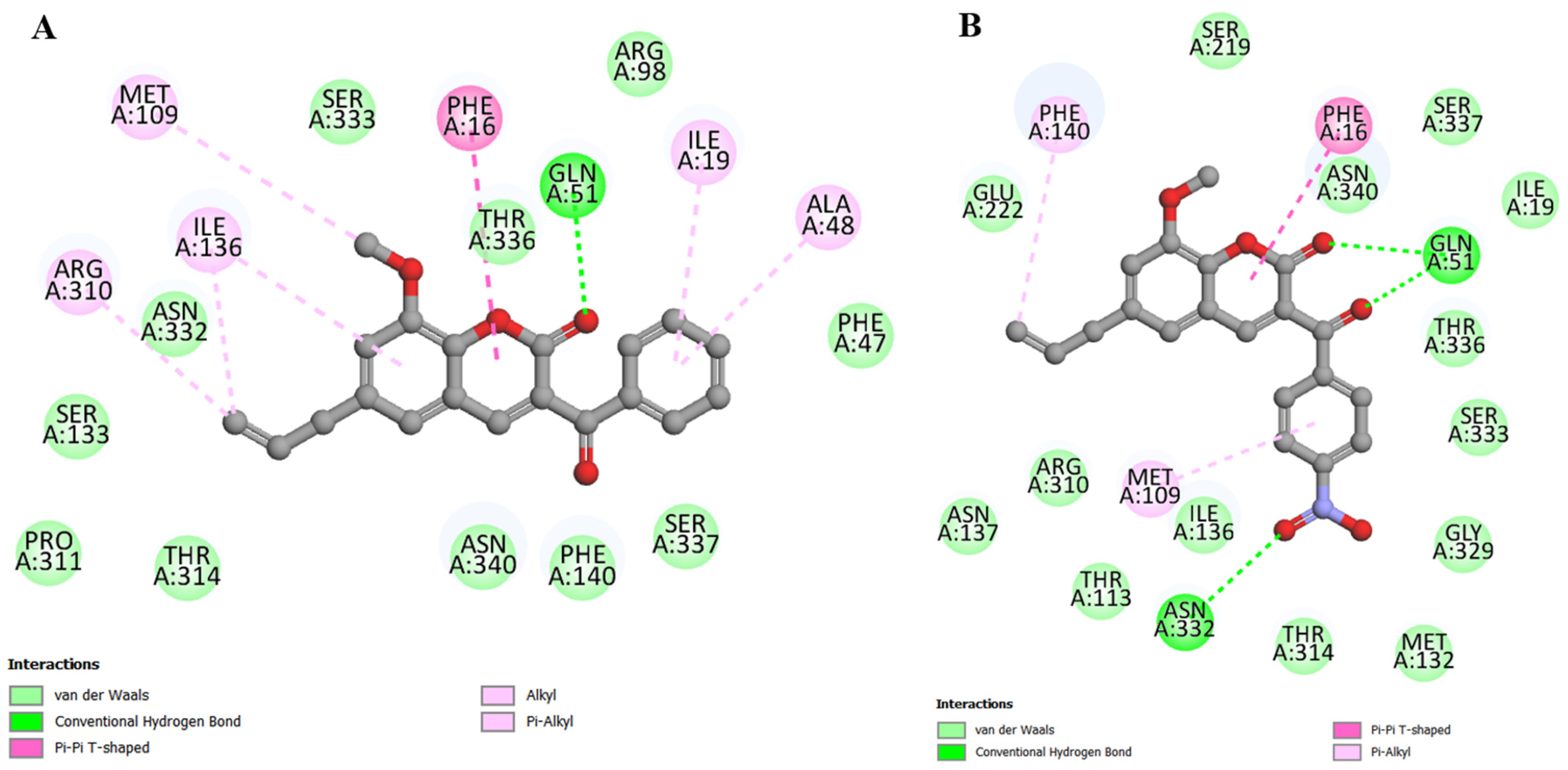
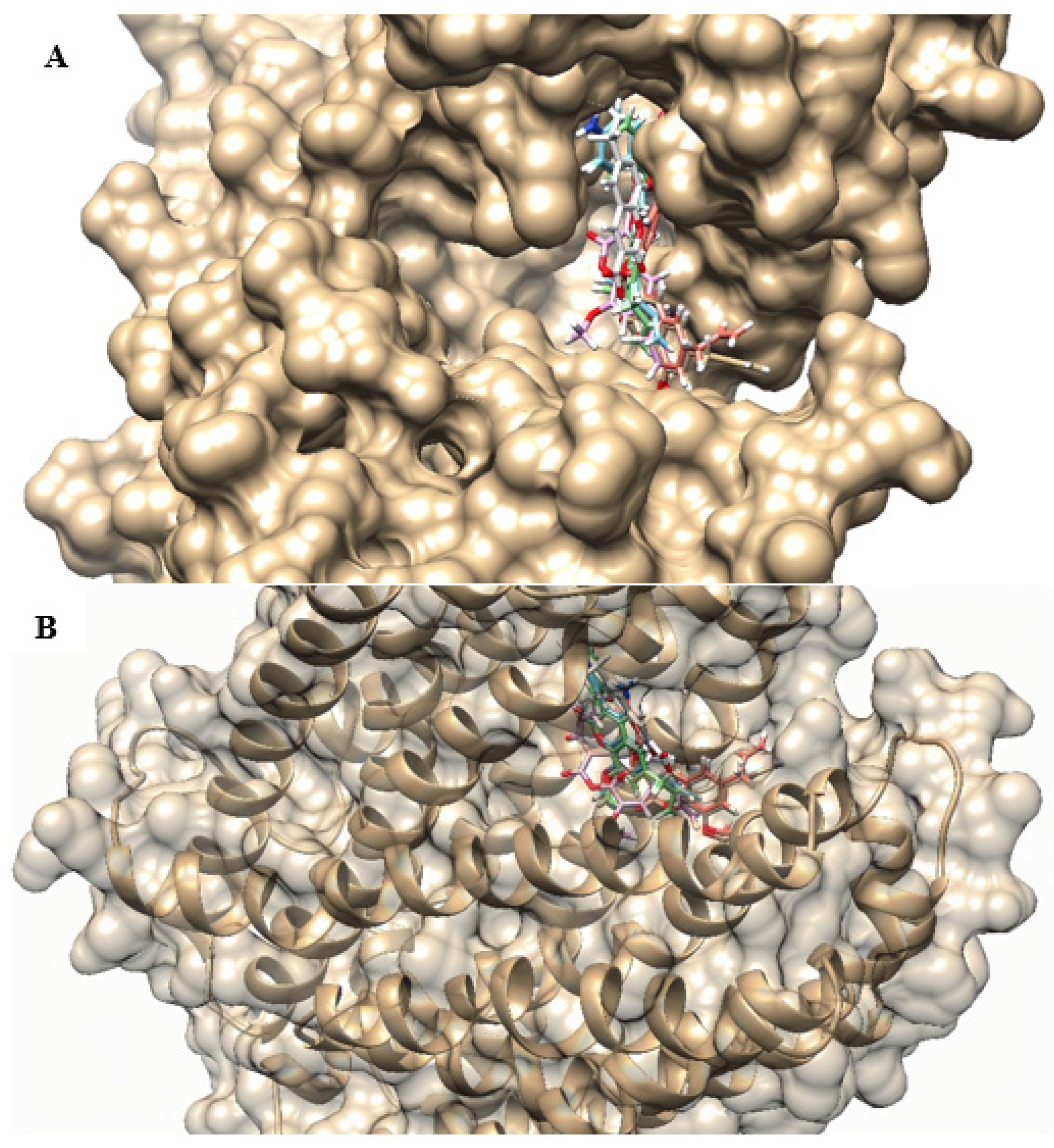

| Compounds | SA 1199B | SA K2068 |
|---|---|---|
| C1 | ≥1024 | ≥1024 |
| C2 | ≥1024 | ≥1024 |
| C3 | ≥1024 | ≥1024 |
| C4 | ≥1024 | ≥1024 |
| CCCP | 64 | 64 |
| CPZ | ≥1024 | ≥1024 |
| Coumarins | MolDock Score | Interaction | HBond | LE1 |
|---|---|---|---|---|
| C1 | −117.214 | −129.417 | −5.43272 | −4.88392 |
| C2 | −125.394 | −138.965 | −10.5218 | −4.64422 |
| C3 | −116.749 (−108.295) | −126.418 (−117.185) | −7.49231 (−0.38884) | −4.66997 (−4.33179) |
| C4 | −99.4964 (−89.4395) | −109.64 (−99.1264) | −14.7348 (−5.6347) | −5.23665 (−4.70734) |
| Coumarins | MolDock Score | Interaction | HBond | LE1 |
|---|---|---|---|---|
| C1 | −111.936 | −121.81 | −5 | −4.66402 |
| C2 | −107.692 | −119.787 | −7.4925 | −3.98861 |
| C3 | −109.268 (−108.887) | −118.995 (−119.946) | −2.5 (−4.59915) | −4.37073 (−4.35547) |
| C4 | −83.2516 (−98.6581) | −92.5696 (−107.365) | −8.88168 (−7.5) | −4.38167 (−5.19253) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araújo-Neto, J.B.d.; Oliveira-Tintino, C.D.d.M.; de Araújo, G.A.; Alves, D.S.; Ribeiro, F.R.; Brancaglion, G.A.; Carvalho, D.T.; Lima, C.M.G.; Mohammed Ali, H.S.H.; Rather, I.A.; et al. 3-Substituted Coumarins Inhibit NorA and MepA Efflux Pumps of Staphylococcus aureus. Antibiotics 2023, 12, 1739. https://doi.org/10.3390/antibiotics12121739
Araújo-Neto JBd, Oliveira-Tintino CDdM, de Araújo GA, Alves DS, Ribeiro FR, Brancaglion GA, Carvalho DT, Lima CMG, Mohammed Ali HSH, Rather IA, et al. 3-Substituted Coumarins Inhibit NorA and MepA Efflux Pumps of Staphylococcus aureus. Antibiotics. 2023; 12(12):1739. https://doi.org/10.3390/antibiotics12121739
Chicago/Turabian StyleAraújo-Neto, José B. de, Cícera D. de M. Oliveira-Tintino, Gildênia A. de Araújo, Daniel S. Alves, Fernanda R. Ribeiro, Guilherme A. Brancaglion, Diogo T. Carvalho, Clara Mariana Gonçalves Lima, Hani S. H. Mohammed Ali, Irfan A. Rather, and et al. 2023. "3-Substituted Coumarins Inhibit NorA and MepA Efflux Pumps of Staphylococcus aureus" Antibiotics 12, no. 12: 1739. https://doi.org/10.3390/antibiotics12121739
APA StyleAraújo-Neto, J. B. d., Oliveira-Tintino, C. D. d. M., de Araújo, G. A., Alves, D. S., Ribeiro, F. R., Brancaglion, G. A., Carvalho, D. T., Lima, C. M. G., Mohammed Ali, H. S. H., Rather, I. A., Wani, M. Y., Emran, T. B., Coutinho, H. D. M., Balbino, V. d. Q., & Tintino, S. R. (2023). 3-Substituted Coumarins Inhibit NorA and MepA Efflux Pumps of Staphylococcus aureus. Antibiotics, 12(12), 1739. https://doi.org/10.3390/antibiotics12121739












