Gut-Antimicrobial Peptides: Synergistic Co-Evolution with Antibiotics to Combat Multi-Antibiotic Resistance
Abstract
1. Introduction
2. Influence of Antibiotics on Gut Microbiota, Susceptibility to Infections, and Resistance Development
3. Interplay of Gut Microbiota with Gut AMPs
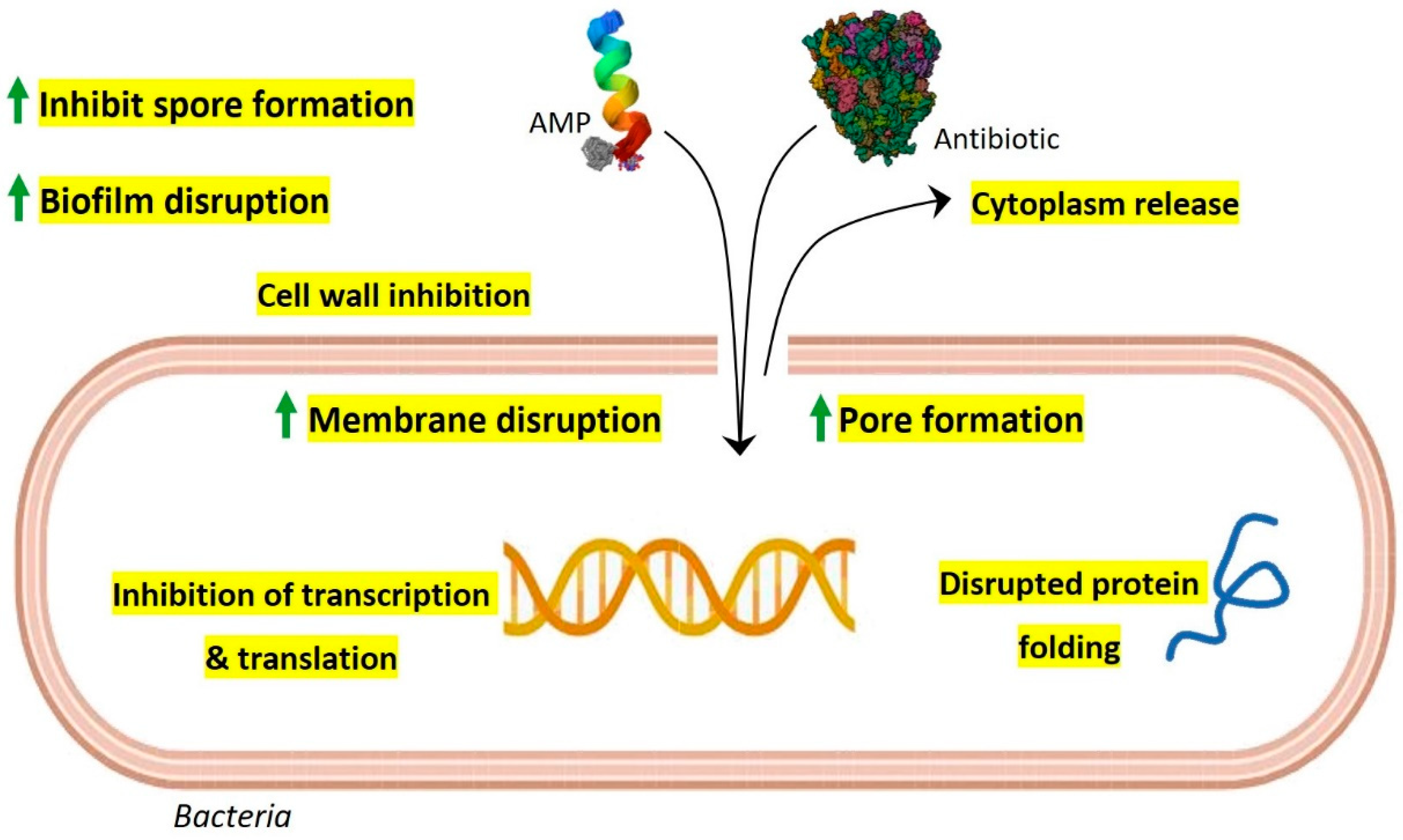
4. Synergistic Action of Gut AMPs with Conventional Antibiotics
5. Gut AMPs, Conventional Antibiotics, and Evolution of Resistance Development
6. Antimicrobial Stewardship and Modulation of Gut AMPs as a Tool to Fight against Resistance Development
7. Conclusions and Future Perspective
8. Unanswered Questions about Gut Microbiota and Gut AMPs
- What makes the gut microbiome healthy and what are the deciding bio-markers?
- What is the genetic machinery that regulates the production of gut AMPs?
- How do gut AMPs play a role in resistance development?
- Could diet help in the fight against resistance by manipulating gut microbiota? How?
- How do gut AMPs regulate the immune response to fight against resistance?
- How to reconstruct the gut microbiome and gut AMPome to counter antibiotic resistance?
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Mwangi, J.; Hao, X.; Lai, R.; Zhang, Z.Y. Antimicrobial peptides: New hope in the war against multidrug resistance. Zool. Res. 2019, 40, 488–505. [Google Scholar] [CrossRef]
- Baindara, P.; Chaudhry, V.; Mittal, G.; Liao, L.M.; Matos, C.O.; Khatri, N.; Franco, O.L.; Patil, P.B.; Korpole, S. Characterization of the antimicrobial peptide penisin, a class Ia novel lantibiotic from Paenibacillus sp. strain A3. Antimicrob. Agents Chemother. 2016, 60, 580–591. [Google Scholar] [CrossRef] [PubMed]
- Josef, J. Drug repurposing to overcome microbial resistance. Drug Discov. Today 2022, 27, 2028–2041. [Google Scholar]
- Deslouches, B.; Montelaro, R.C.; Urish, K.L.; Di, Y.P. Engineered cationic antimicrobial peptides (eCAPs) to combat multidrug-resistant bacteria. Pharmaceutics 2020, 12, 501. [Google Scholar] [CrossRef]
- Baindara, P.; Mandal, S.M. Antimicrobial Peptides and Vaccine Development to Control Multi-drug Resistant Bacteria. Protein Pept. Lett. 2019, 26, 324–331. [Google Scholar] [CrossRef]
- Mathur, H.; Field, D.; Rea, M.C.; Cotter, P.D.; Hill, C.; Ross, R.P. Fighting biofilms with lantibiotics and other groups of bacteriocins. NPJ Biofilms Microbiomes 2018, 4, 9. [Google Scholar] [CrossRef]
- Wang, S.; Thacker, P.; Watford, M.; Qiao, S. Functions of Antimicrobial Peptides in Gut Homeostasis. Curr. Protein Pept. Sci. 2015, 16, 582–591. [Google Scholar] [CrossRef]
- Lewies, A.; Du Plessis, L.H.; Wentzel, J.F. Antimicrobial Peptides: The Achilles’ Heel of Antibiotic Resistance? Probiotics Antimicrob. Proteins 2019, 11, 370–381. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.; Teixeira, C.; Gomes, P.; Martins, M.C.L. Clinical application of AMPs. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2019; Volume 1117, pp. 281–298. [Google Scholar]
- Baindara, P.; Ghosh, A.K.; Mandal, S.M. Coevolution of Resistance Against Antimicrobial Peptides. Microb. Drug Resist. 2020, 26, 880–899. [Google Scholar] [CrossRef] [PubMed]
- El Shazely, B.; Yu, G.; Johnston, P.R.; Rolff, J. Resistance Evolution Against Antimicrobial Peptides in Staphylococcus aureus Alters Pharmacodynamics Beyond the MIC. Front. Microbiol. 2020, 11, 103. [Google Scholar] [CrossRef] [PubMed]
- Dinata, R.; Baindara, P. Laterosporulin25: A probiotically produced, novel defensin-like bacteriocin and its immunogenic properties. Int. Immunopharmacol. 2023, 121, 110500. [Google Scholar] [CrossRef] [PubMed]
- Francino, M.P. Antibiotics and the human gut microbiome: Dysbioses and accumulation of resistances. Front. Microbiol. 2016, 6, 1543. [Google Scholar] [CrossRef]
- Langdon, A.; Crook, N.; Dantas, G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016, 8, 39. [Google Scholar] [CrossRef]
- Rafii, F.; Sutherland, J.B.; Cerniglia, C.E. Effects of treatment with antimicrobial agents on the human colonic microflora. Ther. Clin. Risk Manag. 2008, 4, 1343–1357. [Google Scholar] [CrossRef]
- Rashid, M.U.; Weintraub, A.; Nord, C.E. Effect of new antimicrobial agents on the ecological balance of human microflora. Anaerobe 2012, 18, 249–253. [Google Scholar] [CrossRef]
- Ivanov, I.I.; de Frutos, R.L.; Manel, N.; Yoshinaga, K.; Rifkin, D.B.; Sartor, R.B.; Finlay, B.B.; Littman, D.R. Specific Microbiota Direct the Differentiation of IL-17-Producing T-Helper Cells in the Mucosa of the Small Intestine. Cell Host Microbe 2008, 4, 337–349. [Google Scholar] [CrossRef]
- Wlodarska, M.; Willing, B.; Keeney, K.M.; Menendez, A.; Bergstrom, K.S.; Gill, N.; Russell, S.L.; Vallance, B.A.; Finlay, B.B. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect. Immun. 2011, 79, 1536–1545. [Google Scholar] [CrossRef]
- Greenwood, C.; Morrow, A.L.; Lagomarcino, A.J.; Altaye, M.; Taft, D.H.; Yu, Z.; Newburg, D.S.; Ward, D.V.; Schibler, K.R. Early empiric antibiotic use in preterm infants is associated with lower bacterial diversity and higher relative abundance of enterobacter. J. Pediatr. 2014, 165, 23–29. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Tsolis, R.M.; Bäumler, A.J. The microbiome and gut homeostasis. Science 2022, 377, eabp9960. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins-a viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Donia, M.S.; Cimermancic, P.; Schulze, C.J.; Wieland Brown, L.C.; Martin, J.; Mitreva, M.; Clardy, J.; Linington, R.G.; Fischbach, M.A. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 2014, 158, 1402–1414. [Google Scholar] [CrossRef] [PubMed]
- Corr, S.C.; Li, Y.; Riedel, C.U.; O’Toole, P.W.; Hill, C.; Gahan, C.G.M. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. USA 2007, 104, 7617–7621. [Google Scholar] [CrossRef] [PubMed]
- Rea, M.C.; Sit, C.S.; Clayton, E.; O’Connor, P.M.; Whittal, R.M.; Zheng, J.; Vederas, J.C.; Ross, R.P.; Hill, C. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl. Acad. Sci. USA 2010, 107, 9352–9357. [Google Scholar] [CrossRef]
- Gilmore, M.S.; Lebreton, F.; van Schaik, W. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr. Opin. Microbiol. 2013, 16, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Kommineni, S.; Bretl, D.J.; Lam, V.; Chakraborty, R.; Hayward, M.; Simpson, P.; Cao, Y.; Bousounis, P.; Kristich, C.J.; Salzman, N.H. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 2015, 526, 719–722. [Google Scholar] [CrossRef]
- Sassone-Corsi, M.; Nuccio, S.P.; Liu, H.; Hernandez, D.; Vu, C.T.; Takahashi, A.A.; Edwards, R.A.; Raffatellu, M. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 2016, 540, 280–283. [Google Scholar] [CrossRef]
- Riboulet-Bisson, E.; Sturme, M.H.J.; Jeffery, I.B.; O’Donnell, M.M.; Neville, B.A.; Forde, B.M.; Claesson, M.J.; Harris, H.; Gardiner, G.E.; Casey, P.G.; et al. Effect of lactobacillus salivarius bacteriocin ABP118 on the mouse and pig intestinal microbiota. PLoS ONE 2012, 7, e31113. [Google Scholar] [CrossRef]
- Ilinskaya, O.N.; Ulyanova, V.V.; Yarullina, D.R.; Gataullin, I.G. Secretome of Intestinal bacilli: A natural guard against pathologies. Front. Microbiol. 2017, 8, 1666. [Google Scholar] [CrossRef]
- O’ Connor, P.M.; O’ Shea, E.F.; Cotter, P.D.; Hill, C.; Ross, R.P. The potency of the broad spectrum bacteriocin, bactofencin A, against staphylococci is highly dependent on primary structure, N-terminal charge and disulphide formation. Sci. Rep. 2018, 8, 11833. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Teng, K.; Liu, Y.; Cao, Y.; Wang, T.; Ma, C.; Zhang, J.; Zhong, J. Bacteriocins: Potential for Human Health. Oxid. Med. Cell. Longev. 2021, 2021, 5518825. [Google Scholar] [CrossRef] [PubMed]
- Belfiore, C.; Castellano, P.; Vignolo, G. Reduction of Escherichia coli population following treatment with bacteriocins from lactic acid bacteria and chelators. Food Microbiol. 2007, 24, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Rea, M.C.; Clayton, E.; O’Connor, P.M.; Shanahan, F.; Kiely, B.; Ross, R.P.; Hill, C. Antimicrobial activity of lacticin 3147 against clinical Clostridium difficile strains. J. Med. Microbiol. 2007, 56, 940–946. [Google Scholar] [CrossRef] [PubMed]
- Borrero, J.; Brede, D.A.; Skaugen, M.; Diep, D.B.; Herranz, C.; Nes, I.F.; Cintas, L.M.; Hernández, P.E. Characterization of garvicin ML, a novel circular bacteriocin produced by lactococcus garvieae DCC43, isolated from mallard ducks (Anas platyrhynchos). Appl. Environ. Microbiol. 2011, 77, 369–373. [Google Scholar] [CrossRef]
- Millette, M.; Cornut, G.; Dupont, C.; Shareck, F.; Archambault, D.; Lacroix, M. Capacity of human nisin- and pediocin-producing lactic acid bacteria to reduce intestinal colonization by vancomycin-resistant enterococci. Appl. Environ. Microbiol. 2008, 74, 1997–2003. [Google Scholar] [CrossRef] [PubMed]
- De Kwaadsteniet, M.; Doeschate, K.T.; Dicks, L.M.T. Nisin F in the treatment of respiratory tract infections caused by Staphylococcus aureus. Lett. Appl. Microbiol. 2009, 48, 65–70. [Google Scholar] [CrossRef]
- Goldstein, B.P.; Wei, J.; Greenberg, K.; Novick, R. Activity of nisin against Streptococcus pneumoniae, in vitro, and in a mouse infection model. J. Antimicrob. Chemother. 1998, 42, 277–278. [Google Scholar] [CrossRef][Green Version]
- Norouzi, Z.; Salimi, A.; Halabian, R.; Fahimi, H. Nisin, a potent bacteriocin and anti-bacterial peptide, attenuates expression of metastatic genes in colorectal cancer cell lines. Microb. Pathog. 2018, 123, 183–189. [Google Scholar] [CrossRef]
- Dabour, N.; Zihler, A.; Kheadr, E.; Lacroix, C.; Fliss, I. In vivo study on the effectiveness of pediocin PA-1 and Pediococcus acidilactici UL5 at inhibiting Listeria monocytogenes. Int. J. Food Microbiol. 2009, 133, 225–233. [Google Scholar] [CrossRef]
- Salvucci, E.; Saavedra, L.; Hebert, E.M.; Haro, C.; Sesma, F. Enterocin CRL35 inhibits Listeria monocytogenes in a murine model. Foodborne Pathog. Dis. 2012, 9, 68–74. [Google Scholar] [CrossRef]
- Birri, D.J.; Brede, D.A.; Forberg, T.; Holo, H.; Nes, I.F. Molecular and genetic characterization of a novel bacteriocin locus in Enterococcus avium isolates from infants. Appl. Environ. Microbiol. 2010, 76, 483–492. [Google Scholar] [CrossRef]
- De Kwaadsteniet, M.; Fraser, T.; Van Reenen, C.A.; Dicks, L.M.T. Bacteriocin T8, a novel class IIa sec-dependent bacteriocin produced by Enterococcus faecium T8, isolated from vaginal secretions of children infected with human immunodeficiency virus. Appl. Environ. Microbiol. 2006, 72, 4761–4766. [Google Scholar] [CrossRef] [PubMed]
- Martin-Visscher, L.A.; van Belkum, M.J.; Garneau-Tsodikova, S.; Whittal, R.M.; Zheng, J.; McMullen, L.M.; Vederas, J.C. Isolation and characterization of carnocyclin a, a novel circular bacteriocin produced by Carnobacterium maltaromaticum UAL307. Appl. Environ. Microbiol. 2008, 74, 4756–4763. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.; Chang, H.C. Growth Inhibition of Foodborne Pathogens by Kimchi Prepared with Bacteriocin-Producing Starter Culture. J. Food Sci. 2011, 76, M72–M78. [Google Scholar] [CrossRef]
- Dey, D.; Ema, T.I.; Biswas, P.; Aktar, S.; Islam, S.; Rinik, U.R.; Firoz, M.; Ahmed, S.Z.; Al Azad, S.; Rahman, A.; et al. Antiviral effects of bacteriocin against animal-to-human transmittable mutated SARS-CoV-2: A systematic review. Front. Agric. Sci. Eng. 2021, 8, 603–622. [Google Scholar] [CrossRef]
- Filipp, D.; Brabec, T.; Vobořil, M.; Dobeš, J. Enteric α-defensins on the verge of intestinal immune tolerance and inflammation. Semin. Cell Dev. Biol. 2019, 88, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Phadke, S.M.; Deslouches, B.; Hileman, S.E.; Montelaro, R.C.; Wiesenfeld, H.C.; Mietzner, T.A. Antimicrobial peptides in mucosal secretions: The importance of local secretions in mitigating infection. J. Nutr. 2005, 135, 1289–1293. [Google Scholar] [CrossRef]
- Dutta, P.; Das, S. Mammalian Antimicrobial Peptides: Promising Therapeutic Targets Against Infection and Chronic Inflammation. Curr. Top. Med. Chem. 2015, 16, 99–129. [Google Scholar] [CrossRef]
- Ayabe, T.; Satchell, D.P.; Wilson, C.L.; Parks, W.C.; Selsted, M.E.; Ouellette, A.J. Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 2000, 1, 113–118. [Google Scholar] [CrossRef]
- Menendez, A.; Willing, B.P.; Montero, M.; Wlodarska, M.; So, C.C.; Bhinder, G.; Vallance, B.A.; Finlay, B.B. Bacterial stimulation of the TLR-MyD88 pathway modulates the homeostatic expression of ileal paneth cell α-defensins. J. Innate Immun. 2013, 5, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Salzman, N.H.; Hung, K.; Haribhai, D.; Chu, H.; Karlsson-Sjöberg, J.; Amir, E.; Teggatz, P.; Barman, M.; Hayward, M.; Eastwood, D.; et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 2010, 11, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Fusco, A.; Savio, V.; Cammarota, M.; Alfano, A.; Schiraldi, C.; Donnarumma, G. Beta-Defensin-2 and Beta-Defensin-3 Reduce Intestinal Damage Caused by Salmonella typhimurium Modulating the Expression of Cytokines and Enhancing the Probiotic Activity of Enterococcus faecium. J. Immunol. Res. 2017, 2017, 6976935. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Hu, W.; Chen, S.; Lu, Z.; Wang, Y. Cathelicidin-WA Improves Intestinal Epithelial Barrier Function and Enhances Host Defense against Enterohemorrhagic Escherichia coli O157:H7 Infection. J. Immunol. 2017, 198, 1696–1705. [Google Scholar] [CrossRef]
- Iimura, M.; Gallo, R.L.; Hase, K.; Miyamoto, Y.; Eckmann, L.; Kagnoff, M.F. Cathelicidin Mediates Innate Intestinal Defense against Colonization with Epithelial Adherent Bacterial Pathogens. J. Immunol. 2005, 174, 4901–4907. [Google Scholar] [CrossRef]
- Chromek, M.; Arvidsson, I.; Karpman, D. The Antimicrobial Peptide Cathelicidin Protects Mice from Escherichia coli O157:H7-Mediated Disease. PLoS ONE 2012, 7, e46476. [Google Scholar] [CrossRef]
- Yoshimura, T.; McLean, M.H.; Dzutsev, A.K.; Yao, X.; Chen, K.; Huang, J.; Gong, W.; Zhou, J.; Xiang, Y.; Badger, J.H.; et al. The Antimicrobial Peptide CRAMP Is Essential for Colon Homeostasis by Maintaining Microbiota Balance. J. Immunol. 2018, 200, 2174–2185. [Google Scholar] [CrossRef]
- Vaishnava, S.; Yamamoto, M.; Severson, K.M.; Ruhn, K.A.; Yu, X.; Koren, O.; Ley, R.; Wakeland, E.K.; Hooper, L.V. The antibacterial lectin RegIII gamma promotes the spatial segregation of microbiota and host in the intestine. Science 2011, 334, 255–258. [Google Scholar] [CrossRef]
- Brandl, K.; Plitas, G.; Schnabl, B.; DeMatteo, R.P.; Pamer, E.G. MyD88-mediated signals induce the bactericidal lectin RegIIIγ and protect mice against intestinal Listeria monocytogenes infection. J. Exp. Med. 2007, 204, 1891–1900. [Google Scholar] [CrossRef]
- Ju, T.; Shoblak, Y.; Gao, Y.; Yang, K.; Fouhse, J.; Finlay, B.B.; So, Y.W.; Stothard, P.; Willing, B.P. Initial gut microbial composition as a key factor driving host response to antibiotic treatment, as exemplified by the presence or absence of commensal Escherichia coli. Appl. Environ. Microbiol. 2017, 83, e01107-17. [Google Scholar] [CrossRef]
- Baindara, P.; Singh, N.; Ranjan, M.; Nallabelli, N.; Chaudhry, V.; Pathania, G.L.; Sharma, N.; Kumar, A.; Patil, P.B.; Korpole, S. Laterosporulin10: A novel defensin like class iid bacteriocin from brevibacillus sp. strain SKDU10 with inhibitory activity against microbial pathogens. Microbiology 2016, 162, 1286–1299. [Google Scholar] [CrossRef]
- Naghmouchi, K.; Le Lay, C.; Baah, J.; Drider, D. Antibiotic and antimicrobial peptide combinations: Synergistic inhibition of Pseudomonas fluorescens and antibiotic-resistant variants. Res. Microbiol. 2012, 163, 101–108. [Google Scholar] [CrossRef]
- Tong, Z.; Zhang, Y.; Ling, J.; Ma, J.; Huang, L.; Zhang, L. An in vitro study on the effects of nisin on the antibacterial activities of 18 antibiotics against Enterococcus faecalis. PLoS ONE 2014, 9, e89209. [Google Scholar] [CrossRef]
- Lebel, G.; Piché, F.; Frenette, M.; Gottschalk, M.; Grenier, D. Antimicrobial activity of nisin against the swine pathogen Streptococcus suis and its synergistic interaction with antibiotics. Peptides 2013, 50, 19–23. [Google Scholar] [CrossRef]
- Cavera, V.L.; Volski, A.; Chikindas, M.L. The Natural Antimicrobial Subtilosin A Synergizes with Lauramide Arginine Ethyl Ester (LAE), ε-Poly-l-lysine (Polylysine), Clindamycin Phosphate and Metronidazole, Against the Vaginal Pathogen Gardnerella vaginalis. Probiotics Antimicrob. Proteins 2015, 7, 164–171. [Google Scholar] [CrossRef]
- Nuding, S.; Frasch, T.; Schaller, M.; Stange, E.F.; Zabel, L.T. Synergistic effects of antimicrobial peptides and antibiotics against clostridium difficile. Antimicrob. Agents Chemother. 2014, 58, 5719–5725. [Google Scholar] [CrossRef]
- Rishi, P.; Preet, S.; Bharrhan, S.; Verma, I. In vitro and in vivo synergistic effects of cryptdin 2 and ampicillin against Salmonella. Antimicrob. Agents Chemother. 2011, 55, 4176–4182. [Google Scholar] [CrossRef]
- Kalita, A.; Verma, I.; Khuller, G.K. Role of human neutrophil peptide-1 as a possible adjunct to antituberculosis chemotherapy. J. Infect. Dis. 2004, 190, 1476–1480. [Google Scholar] [CrossRef]
- Rajasekaran, G.; Kim, E.Y.; Shin, S.Y. LL-37-derived membrane-active FK-13 analogs possessing cell selectivity, anti-biofilm activity and synergy with chloramphenicol and anti-inflammatory activity. Biochim. Biophys. Acta Biomembr. 2017, 1859, 722–733. [Google Scholar] [CrossRef]
- Lin, L.; Nonejuie, P.; Munguia, J.; Hollands, A.; Olson, J.; Dam, Q.; Kumaraswamy, M.; Rivera, H.; Corriden, R.; Rohde, M.; et al. Azithromycin Synergizes with Cationic Antimicrobial Peptides to Exert Bactericidal and Therapeutic Activity against Highly Multidrug-Resistant Gram-Negative Bacterial Pathogens. EBioMedicine 2015, 2, 690–698. [Google Scholar] [CrossRef]
- Ruden, S.; Rieder, A.; Chis Ster, I.; Schwartz, T.; Mikut, R.; Hilpert, K. Synergy Pattern of Short Cationic Antimicrobial Peptides against Multidrug-Resistant Pseudomonas aeruginosa. Front. Microbiol. 2019, 10, 2740. [Google Scholar] [CrossRef]
- Herrmann, G.; Yang, L.; Wu, H.; Song, Z.; Wang, H.; Høiby, N.; Ulrich, M.; Molin, S.; Riethmüller, J.; Döring, G. Colistin-tobramycin combinations are superior to monotherapy concerning the killing of biofilm Pseudomonas aeruginosa. J. Infect. Dis. 2010, 202, 1585–1592. [Google Scholar] [CrossRef] [PubMed]
- Brumfitt, W.; Salton, M.R.J.; Hamilton-Miller, J.M.T. Nisin, alone and combined with peptidoglycan-modulating antibiotics: Activity against methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci. J. Antimicrob. Chemother. 2002, 50, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Giacometti, A.; Cirioni, O.; Barchiesi, F.; Fortuna, M.; Scalise, G. In-vitro activity of cationic peptides alone and in combination with clinically used antimicrobial agents against Pseudomonas aeruginosa. J. Antimicrob. Chemother. 1999, 44, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Draper, L.A.; Cotter, P.D.; Hill, C.; Ross, R.P. The two peptide lantibiotic lacticin 3147 acts synergistically with polymyxin to inhibit Gram negative bacteria. BMC Microbiol. 2013, 13, 212. [Google Scholar] [CrossRef] [PubMed]
- Mathur, H.; O’Connor, P.M.; Hill, C.; Cotter, P.D.; Ross, R.P. Analysis of anti-clostridium difficile activity of thuricin CD, vancomycin, metronidazole, ramoplanin, and actagardine, both singly and in paired combinations. Antimicrob. Agents Chemother. 2013, 57, 2882–2886. [Google Scholar] [CrossRef]
- Lobos, O.; Padilla, A.; Padilla, C. In vitro antimicrobial effect of bacteriocin PsVP-10 in combination with chlorhexidine and triclosan against Streptococcus mutans and Streptococcus sobrinus strains. Arch. Oral Biol. 2009, 54, 230–234. [Google Scholar] [CrossRef]
- Sharma, A.; Srivastava, S. Anti-Candida activity of two-peptide bacteriocins, plantaricins (Pln E/F and J/K) and their mode of action. Fungal Biol. 2014, 118, 264–275. [Google Scholar] [CrossRef]
- Larsen, J.; Raisen, C.L.; Ba, X.; Sadgrove, N.J.; Padilla-González, G.F.; Simmonds, M.S.J.; Loncaric, I.; Kerschner, H.; Apfalter, P.; Hartl, R.; et al. Emergence of methicillin resistance predates the clinical use of antibiotics. Nature 2022, 602, 135–141. [Google Scholar] [CrossRef]
- O’Toole, R.F.; Leong, K.W.C.; Cumming, V.; Van Hal, S.J. Vancomycin-resistant Enterococcus faecium and the emergence of new sequence types associated with hospital infection. Res. Microbiol. 2023, 174, 104046. [Google Scholar] [CrossRef]
- Forslund, K.; Sunagawa, S.; Kultima, J.R.; Mende, D.R.; Arumugam, M.; Typas, A.; Bork, P. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 2013, 23, 1163–1169. [Google Scholar] [CrossRef] [PubMed]
- Anthony, W.E.; Burnham, C.A.D.; Dantas, G.; Kwon, J.H. The gut microbiome as a reservoir for antimicrobial resistance. J. Infect. Dis. 2021, 223, S209–S213. [Google Scholar] [CrossRef] [PubMed]
- Baur, D.; Gladstone, B.P.; Burkert, F.; Carrara, E.; Foschi, F.; Döbele, S.; Tacconelli, E. Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and Clostridium difficile infection: A systematic review and meta-analysis. Lancet Infect. Dis. 2017, 17, 990–1001. [Google Scholar] [CrossRef] [PubMed]
- Ya, K.Z.; Win, P.T.N.; Bielicki, J.; Lambiris, M.; Fink, G. Association Between Antimicrobial Stewardship Programs and Antibiotic Use Globally: A Systematic Review and Meta-Analysis. JAMA Netw. Open 2023, 6, E2253806. [Google Scholar] [CrossRef]
- Chanderraj, R.; Baker, J.M.; Kay, S.G.; Brown, C.A.; Hinkle, K.J.; Fergle, D.J.; McDonald, R.A.; Falkowski, N.R.; Metcalf, J.D.; Kaye, K.S.; et al. In critically ill patients, anti-anaerobic antibiotics increase risk of adverse clinical outcomes. Eur. Respir. J. 2023, 61, 2200910. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, A.; Pultz, N.J.; Ray, A.J.; Hoyen, C.K.; Eckstein, E.C.; Donskey, C.J. Antianaerobic Antibiotic Therapy Promotes Overgrowth of Antibiotic-Resistant, Gram-Negative Bacilli and Vancomycin-Resistant Enterococci in the Stool of Colonized Patients. Infect. Control Hosp. Epidemiol. 2003, 24, 644–649. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, B.G.; Hall, L.; White, N.; Barnett, A.G.; Halton, K.; Paterson, D.L.; Riley, T.V.; Gardner, A.; Page, K.; Farrington, A.; et al. An environmental cleaning bundle and health-care-associated infections in hospitals (REACH): A multicentre, randomised trial. Lancet Infect. Dis. 2019, 19, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Shahi, F.; Redeker, K.; Chong, J. Rethinking antimicrobial stewardship paradigms in the context of the gut microbiome. JAC—Antimicrob. Resist. 2019, 1, dlz015. [Google Scholar] [CrossRef]
- Leo, S.; Lazarevic, V.; von Dach, E.; Kaiser, L.; Prendki, V.; Schrenzel, J.; Huttner, B.D.; Huttner, A. Effects of antibiotic duration on the intestinal microbiota and resistome: The PIRATE RESISTANCE project, a cohort study nested within a randomized trial. EBioMedicine 2021, 71, 103566. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, Y.; Zhou, Y.; Buckley, T.; Wang, H.H. Antibiotic administration routes significantly influence the levels of antibiotic resistance in gut microbiota. Antimicrob. Agents Chemother. 2013, 57, 3659–3666. [Google Scholar] [CrossRef]
- Wistrand-Yuen, E.; Knopp, M.; Hjort, K.; Koskiniemi, S.; Berg, O.G.; Andersson, D.I. Evolution of high-level resistance during low-level antibiotic exposure. Nat. Commun. 2018, 9, 1599. [Google Scholar] [CrossRef]
- Mojsoska, B.; Jenssen, H. Peptides and peptidomimetics for antimicrobial drug design. Pharmaceuticals 2015, 8, 366–415. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Xu, H.; Xia, J.; Ma, J.; Xu, J.; Li, Y.; Feng, J. D- and Unnatural Amino Acid Substituted Antimicrobial Peptides with Improved Proteolytic Resistance and Their Proteolytic Degradation Characteristics. Front. Microbiol. 2020, 11, 563030. [Google Scholar] [CrossRef] [PubMed]
- Fadaka, A.O.; Sibuyi, N.R.S.; Madiehe, A.M.; Meyer, M. Nanotechnology-based delivery systems for antimicrobial peptides. Pharmaceutics 2021, 13, 1795. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Bevins, C.L.; Salzman, N.H. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 2011, 9, 356–368. [Google Scholar] [CrossRef]
- Cullen, T.W.; Schofield, W.B.; Barry, N.A.; Putnam, E.E.; Rundell, E.A.; Trent, M.S.; Degnan, P.H.; Booth, C.J.; Yu, H.; Goodman, A.L. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 2015, 347, 170–175. [Google Scholar] [CrossRef]
- Yan, H.; Hancock, R.E.W. Synergistic interactions between mammalian antimicrobial defense peptides. Antimicrob. Agents Chemother. 2001, 45, 1558–1560. [Google Scholar] [CrossRef]

| Gut AMPs | Producing Bacteria | Targeted Pathogens or Diseases | References |
|---|---|---|---|
| Bacteriocin Abp118 | L. salivarius | Listeriosis | [30] |
| Bacteriocin OR-7 | L. salivarius NRRLB | Campylobacter jejuni | [31] |
| Bactofencin A | L. salivarius | Antilisterial, antistaphylococcal | [32] |
| Lactocin AL705 | L. curvatus | Listeriosis | [33] |
| Lactocin 160 | L. rhamnosus | Escherichia coli Bordetella pertussis | [34] |
| Lacticin3147 | Lactococcus lactis DPC3147 | C. difficile-associated diarrhea (CDAD) | [35] |
| Garvicin ML | L. garvieae | Streptococcus pneumonia | [36] |
| Nisin Z | L. lactis | Immunomodulatory effect | [37] |
| Nisin F | L. lactis | Respiratory infection | [38] |
| Nisin | L. lactis | Meningitis, sepsis, pneumonia | [39] |
| Nisin Z | L. lactis | Enteric pathogens | [37] |
| Nisin A | L. lactis | Colorectal cancer | [40] |
| Pediocin PA1 | Pediococcus acidilactici | Listeriosis | [41] |
| Pediocin AcH | P. acidilactici | Enteric pathogens | [37] |
| Enterocin CRL35 | Enterococcus mundtii RL35 | Listeriosis | [42] |
| Avicin | E. avium | Listeriosis | [43] |
| Enterocin P | E. faecium P13 | Enteric pathogens | [44] |
| Piscicolin 126, carnobacteriocin | Carnobacterium maltaromaticum | Listeriosis | [45] |
| Kimchichin | Leuconostoc citreum GJ7 | Salmonella typhi | [46] |
| Erwinaocin NA4 | Erwinia carotovora NA4 | Coliphage | [47] |
| Gut AMPs | Antibiotics | Target | References |
|---|---|---|---|
| Nisin | Ramoplanin | MRSA | [74] |
| Polymyxin E Clarithromycin | P. aeruginosa | [75] | |
| Amoxicillin Penicillin Streptomycin Ceftiofur Tetracycline | S. suis | [65] | |
| Nisin Z | Ampicillin Chloramphenicol Kanamycin Lincomycin Penicillin G Rifampicin Streptomycin Tetracycline Vancomycin | P. fluorescens LRC-R73 and its Penicillin-resistant/Streptomycin-resistant/Lincomycin-resistant/Rifampicin-resistant variant | [63] |
| Lacticin 3147 | Polymyxin B | S. aureus 5247 | [76] |
| Actagardine | Ramoplanin Metronidazole Vancomycin | C. difficile | [77] |
| Thuricin CD | Ramoplanin | C. difficile | [77] |
| Vancomycin | C. difficile | [77] | |
| Subtilosin A | Clindamycin phosphate Metronidazole | G. vaginalis | [66] |
| Lauramide arginate Ester poly-lysine | G. vaginalis | [66] | |
| PsVP-10 | Chlorhexidine | S. mutans S. sobrinus | [78] |
| Plantaricin E, F, J, K | Several antibiotics | C. albicans | [79] |
| Colistin | Tobramycin | P. aeruginosa | [73] |
| Cryptdin 2 | Ampicillin | S. typhimurium | [68] |
| Laterosporulin10 | Rifampicin | M. tuberculosis H37Rv | [62] |
| Colistin | Azithromycin | A. baumannii K. pneumoniae P. aeruginosa | [71] |
| LL-37 | Azithromycin | A. baumannii K. pneumoniae P. aeruginosa | [71] |
| Human defensin 5 (HD5) | Meropenem | C. difficile | [67] |
| Human neutrophil peptide-1 (HNP1) | Rifampicin | M. tuberculosis H37Rv | [69] |
| Human β-defensin 3 (HBD3) | Meropenem Moxifloxacin Piperacillin-Tazobactam Tigecycline | C. difficile | [67] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baindara, P.; Mandal, S.M. Gut-Antimicrobial Peptides: Synergistic Co-Evolution with Antibiotics to Combat Multi-Antibiotic Resistance. Antibiotics 2023, 12, 1732. https://doi.org/10.3390/antibiotics12121732
Baindara P, Mandal SM. Gut-Antimicrobial Peptides: Synergistic Co-Evolution with Antibiotics to Combat Multi-Antibiotic Resistance. Antibiotics. 2023; 12(12):1732. https://doi.org/10.3390/antibiotics12121732
Chicago/Turabian StyleBaindara, Piyush, and Santi M. Mandal. 2023. "Gut-Antimicrobial Peptides: Synergistic Co-Evolution with Antibiotics to Combat Multi-Antibiotic Resistance" Antibiotics 12, no. 12: 1732. https://doi.org/10.3390/antibiotics12121732
APA StyleBaindara, P., & Mandal, S. M. (2023). Gut-Antimicrobial Peptides: Synergistic Co-Evolution with Antibiotics to Combat Multi-Antibiotic Resistance. Antibiotics, 12(12), 1732. https://doi.org/10.3390/antibiotics12121732






