Identification of IncA Plasmid, Harboring blaVIM-1 Gene, in S. enterica Goldcoast ST358 and C. freundii ST62 Isolated in a Hospitalized Patient
Abstract
:1. Introduction
2. Results
2.1. Clinical Aspects and Strains Selection
2.2. Antimicrobial Susceptibility of S. enterica and C. freundii
2.3. Genomic Features of S. enterica TS1 and TS2
2.4. Genomic Features of C. freundii TS3
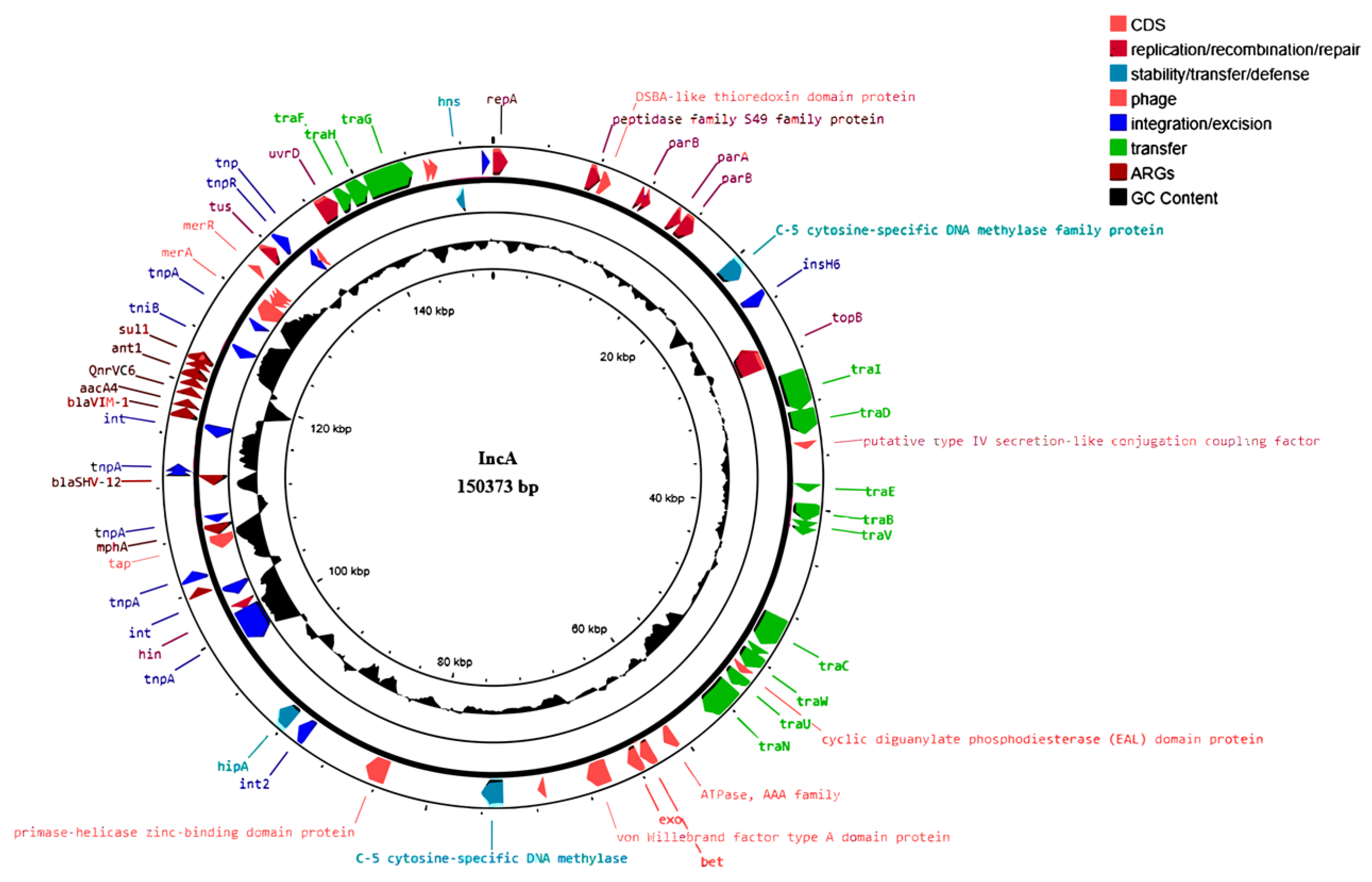
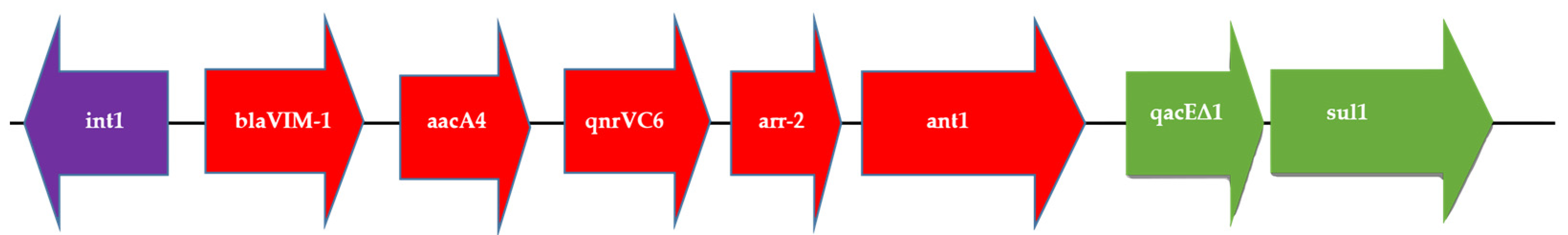
2.5. IncA Plasmid Mapping
2.6. IncA Transfer by Conjugation Assays
3. Discussion
4. Materials and Methods
4.1. Strains Selection
4.2. Antimicrobial Susceptibility Testing
4.3. Genomic and Plasmid Extraction
4.4. Whole-Genome Sequencing (WGS) and Bioinformatic Analysis
4.5. WGS-Based AMR Identification and Serotype Prediction of S. enterica TS2
4.6. WGS-Based AMR Identification of C. freundii TS3
4.7. IncA Plasmid Sequencing
4.8. Conjugation Experiments
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crump, J.A.; Sjölund-Karlsson, M.; Gordon, M.A.; Parry, C.M. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 2015, 28, 901–937. [Google Scholar] [CrossRef] [PubMed]
- EFSA—European Food Safety Authority; ECDC—European Centre for Disease Prevention and Control. The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, 6971. [Google Scholar]
- European Centre for Disease Prevention and Control. Salmonellosis. In ECDC. Annual Epidemiological Report for 2021; ECDC: Stockholm, Sweden, 2022. [Google Scholar]
- Li, P.; Liu, Q.; Luo, H.; Liang, K.; Yi, J.; Luo, Y.; Hu, Y.; Han, Y.; Kong, Q. O-Serotype conversion in Salmonella typhimurium induces protective immune responses against invasive Non-Typhoidal Salmonella infections. Front. Immunol. 2017, 8, 1647. [Google Scholar] [CrossRef] [PubMed]
- Carey, M.E.; Dyson, Z.A.; Ingle, D.J.; Amir, A.; Aworh, M.K.; Chattaway, M.A.; Chew, K.L.; Crump, J.A.; Feasey, N.A.; Howden, B.P.; et al. Global Typhoid Genomics Consortium Group Authorship. Global diversity and antimicrobial resistance of typhoid fever pathogens: Insights from a meta-analysis of 13,000 Salmonella Typhi genomes. Elife 2023, 12, e85867. [Google Scholar] [CrossRef]
- Chattaway, M.A.; Painset, A.; Godbole, G.; Gharbia, S.; Jenkins, C. Evaluation of genomic typing methods in the Salmonella reference laboratory in Public Health, England, 2012–2020. Pathogens 2023, 12, 223. [Google Scholar] [CrossRef] [PubMed]
- Marchello, C.S.; Birkhold, M.; Crump, J.A. Vacc-iNTS consortium collaborators. Complications and mortality of non-typhoidal Salmonella invasive disease: A global systematic review and meta-analysis. Lancet Infect. Dis. 2022, 22, 692–705. [Google Scholar] [CrossRef] [PubMed]
- Foley, S.L.; Lynne, A.M. Food animal-associated Salmonella challenges: Pathogenicity and antimicrobial resistance. J. Anim. Sci. 2008, 86, E173–E187. [Google Scholar] [CrossRef]
- Bremer, V.; Leitmeyer, K.; Jensen, E.; Metzel, U.; Meczulat, H.; Weise, E.; Werber, D.; Tschaepe, H.; Kreienbrock, L.; Glaser, S.; et al. Outbreak of Salmonella Goldcoast infections linked to consumption of fermented sausage, Germany 2001. Epidemiol. Infect. 2004, 132, 881–887. [Google Scholar] [CrossRef]
- Horváth, J.K.; Mengel, M.; Krisztalovics, K.; Nogrady, N.; Pászti, J.; Lenglet, A.; Takkinen, J. Investigation into an unusual increase of human cases of Salmonella Goldcoast infection in Hungary in 2009. Eurosurveillance 2013, 18, 204228. [Google Scholar] [CrossRef]
- Inns, T.; Beasley, G.; Lane, C.; Hopps, V.; Peters, T.; Pathak, K.; Perez-Moreno, R.; Adak, G.; Shankar, A. Outbreak of Salmonella enterica Goldcoast infection associated with whelk consumption, England, June to October 2013. Eurosurveillance 2013, 18, 20654. [Google Scholar] [CrossRef]
- Scavia, G.; Ciaravino, G.; Luzzi, I.; Lenglet, A.; Ricci, A.; Barco, L.; Pavan, A.; Zafanella, F.; Dionisi, A.M. A multistate epidemic outbreak of Salmonella Goldcoast infection in humans, June 2009 to March 2010: The investigation in Italy. Eurosurveillance 2013, 18, 20424. [Google Scholar] [CrossRef] [PubMed]
- EFSA—European Food Safety Authority; ECDC—European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021, 19, 6490. [Google Scholar]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Antimicrobial Resistance Collaborators Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Bonomo, R.A.; Burd, E.M.; Conly, J.; Limbago, B.M.; Poirel, L.; Segre, J.A.; Westblade, L.F. Carbapenemase-Producing Organisms: A Global Scourge. Clin. Infect. Dis. 2018, 66, 1290–1297. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. Epidemiology of β-lactamase-producing pathogens. Clin. Microbiol. Rev. 2020, 33, e00047-19. [Google Scholar] [CrossRef]
- Nobrega, D.; Peirano, G.; Matsumura, Y.; Pitout, J.D.D. Molecular epidemiology of global carbapenemase-producing Citrobacter spp. (2015–2017). Microbiol. Spectr. 2023, 11, e0414422. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.; Guerra, B.; Rodicio, M.R. Resistance to carbapenems in Non-Typhoidal Salmonella enterica serovars from humans, animals and food. Vet. Sci. 2018, 5, 40. [Google Scholar] [CrossRef]
- Miriagou, V.; Tzouvelekis, L.S.; Rossiter, S.; Tzelepi, E.; Angulo, F.J.; Whichard, J.M. Imipenem resistance in a Salmonella clinical strain due to plasmid-mediated class A carbapenemase KPC-2. Antimicrob. Agents Chemother. 2003, 47, 1297–1300. [Google Scholar] [CrossRef]
- Savard, P.; Gopinath, R.; Zhu, W.; Kitchel, B.; Rasheed, J.K.; Tekle, T.; Roberts, A.; Ross, T.; Razeq, J.; Landrum, B.M.; et al. First NDM-positive Salmonella spp. strain identified in the United States. Antimicrob. Agents Chemother. 2011, 55, 5957–5958. [Google Scholar] [CrossRef]
- Li, X.; Jiang, Y.; Wu, K.; Zhou, Y.; Liu, R.; Cao, Y.; Wu, A.; Qiu, Y. Whole-genome sequencing identification of a multidrug-resistant Salmonella enterica serovar Typhimurium strain carrying blaNDM-5 from Guangdong, China. Infect. Genet. Evol. 2017, 55, 195–198. [Google Scholar] [CrossRef]
- Seiffert, S.N.; Perreten, V.; Johannes, S.; Droz, S.; Bodmer, T.; Endimiani, A. OXA-48 carbapenemase-producing Salmonella enterica serovar Kentucky isolate of sequence type 198 in a patient transferred from Libya to Switzerland. Antimicrob. Agents Chemother. 2014, 58, 2446–2449. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L.; Mak, J.K.; White, P.A.; McIver, C.J.; Taylor, P. Multidrug-resistant Salmonella strains expressing emerging antibiotic resistance determinants. Clin. Infect. Dis. 2008, 46, 324–325. [Google Scholar] [CrossRef] [PubMed]
- Le Hello, S.; Harrois, D.; Bouchrif, B.; Sontag, L.; Elhani, D.; Guibert, V.; Zerouali, K.; Weill, F.X. Highly drug-resistant Salmonella enterica serotype Kentucky ST198-X1: A microbiological study. Lancet Infect. Dis. 2013, 13, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Roschanski, N.; Hadziabdic, S.; Borowiak, M.; Malorny, B.; Tenhagen, B.A.; Projahn, M.; Kaesbohrer, A.; Guenther, S.; Szabo, I.; Roesler, U.; et al. Detection of VIM-1-Producing Enterobacter cloacae and Salmonella enterica Serovars Infantis and Goldcoast at a breeding pig farm in Germany in 2017 and their molecular relationship to former VIM-1-Producing S. Infantis Isolates in German livestock production. mSphere 2019, 4, e00089-19. [Google Scholar]
- Fischer, J.; Rodríguez, I.; Schmoger, S.; Friese, A.; Roesler, U.; Helmuth, R.; Guerra, B. Salmonella enterica subsp. enterica producing VIM-1 carbapenemase isolated from livestock farms. J. Antimicrob. Chemother. 2013, 68, 478–480. [Google Scholar] [CrossRef] [PubMed]
- Falgenhauer, L.; Ghosh, H.; Guerra, B.; Yao, Y.; Fritzenwanker, M.; Fischer, J.; Helmuth, R.; Imirzalioglu, C.; Chakraborty, T. Comparative genome analysis of IncHI2 VIM-1 carbapenemase-encoding plasmids of Escherichia coli and Salmonella enterica isolated from a livestock farm in Germany. Vet. Microbiol. 2017, 200, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Arutyunov, D.; Arenson, B.; Manchak, J.; Frost, L.S. F plasmid TraF and TraH are components of an outer membrane complex involved in conjugation. J. Bacteriol. 2010, 192, 1730–1734. [Google Scholar] [CrossRef]
- Poole, T.L.; Edrington, T.S.; Brichta-Harhay, D.M.; Carattoli, A.; Anderson, R.C.; Nisbet, D.J. Conjugative transferability of the A/C plasmids from Salmonella enterica isolates that possess or lack bla(CMY) in the A/C plasmid backbone. Foodborne Pathog. Dis. 2009, 6, 1185–1194. [Google Scholar] [CrossRef]
- Arpin, C.; Thabet, L.; Yassine, H.; Messadi, A.; Boukadida, J.; Dubois, V.; Coulange-Mayonnove, L.; Andre, C.; Quentin, C. Evolution of an Incompatibility Group IncA/C Plasmid Harboring blaCMY-16 and qnrA6 Genes and its transfer through three clones of Providencia stuartii during a 2-year Outbreak in a Tunisian Burn Unit. Antimicrob. Agents Chemother. 2012, 56, 1342–1349. [Google Scholar] [CrossRef]
- Carattoli, A.; Villa, L.; Poirel, L.; Bonnin, R.A.; Nordmann, P. Evolution of IncA/C blaCMY-2 carrying plasmids by acquisition of the blaNDM-1 carbapenemase gene. Antimicrob. Agents Chemother. 2012, 56, 783–786. [Google Scholar] [CrossRef]
- Poulin-Laprade, D.; Carraro, N.; Burrus, V. The extended regulatory networks of SXT/R391 integrative and conjugative elements and IncA/C conjugative plasmids. Front. Microbiol. 2015, 6, 837. [Google Scholar] [CrossRef]
- Johnson, T.J.; Lang, K.S. IncA/C plasmids: An emerging threat to human and animal health? Mob. Genet. Elements 2012, 2, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Arcari, G.; Di Lella, F.M.; Bibbolino, G.; Mengoni, F.; Beccaccioli, M.; Antonelli, G.; Faino, L.; Carattoli, A. A Multispecies Cluster of VIM-1 carbapenemase-producing Enterobacterales linked by a novel, highly conjugative, and broad-host-range IncA plasmid forebodes the reemergence of VIM-1. Antimicrob. Agents Chemother. 2020, 64, e02435-19. [Google Scholar] [CrossRef] [PubMed]
- Falcone, M.; Perilli, M.; Mezzatesta, M.L.; Mancini, C.; Amicosante, G.; Stefani, S.; Venditti, M. Prolonged bacteraemia caused by VIM-1 metallo-β-lactamase-producing Proteus mirabilis: First report from Italy. Clin. Microbiol. Infect. Dis. 2010, 16, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Falcone, M.; Mezzatesta, M.L.; Perilli, M.; Forcella, C.; Giordano, A.; Cafiso, V.; Amicosante, G.; Stefani, S.; Venditti, M. Infections with VIM-1 metallo-β-lactamase-producing Enterobacter cloacae and their correlation with clinical outcome. J. Clin. Microbiol. 2009, 47, 3514–3519. [Google Scholar] [CrossRef]
- Kotsakis, S.D.; Flach, C.F.; Razavi, M.; Larsson, D.G.J. Characterization of the first OXA-10 natural variant with increased carbapenemase activity. Antimicrob. Agents Chemother. 2018, 63, e01817-18. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Song, J.; Ren, L.; Huang, D.; Wang, S.; Fu, L.; Zhao, Y.; Bai, Y.; Fan, X.; Ma, T.; et al. Complete genetic characterization of carbapenem-resistant Acinetobacter johnsonii, co-producing NDM-1, OXA-58, and PER-1 in a patient source. Front. Cell Infect. Microbiol. 2023, 13, 1227063. [Google Scholar] [CrossRef]
- Liu, J.; Yang, L.; Li, L.; Li, B.; Chen, D.; Xu, Z. Comparative genomic analyses of two novel qnrVC6 carrying multidrug-resistant Pseudomonas spp. strains. Microb. Pathog. 2018, 123, 269–274. [Google Scholar] [CrossRef]
- Bado, I.; Papa-Ezdra, R.; Cordeiro, N.; Outeda, M.; Caiata, L.; García-Fulgueiras, V.; Seija, V.; Vignoli, R. Detection of qnrVC6, within a new genetic context, in an NDM-1-producing Citrobacter freundii clinical isolate from Uruguay. J. Glob. Antimicrob. Resist. 2018, 14, 95–98. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, Z.; Chan, E.W.; Dong, N.; Xia, X.; Chen, S. Molecular Characterization of qnrVC Genes and Their Novel Alleles in Vibrio spp. Isolated from Food Products in China. Antimicrob. Agents Chemother. 2018, 62, e00529-18. [Google Scholar] [CrossRef]
- Souza, R.B.; Ferrari, R.G.; Magnani, M.; Kottwitz, L.B.; Alcocer, I.; Tognim, M.C.; Oliveira, T.C. Ciprofloxacin susceptibility reduction of Salmonella strains isolated from outbreaks. Braz. J. Microbiol. 2010, 41, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Wang, L.; Ogunremi, D. Virulence determinants of non-typhoidal Salmonellae. In Microorganisms; Intechopen: London, UK, 2019. [Google Scholar]
- Threlfall, E.J.; Hall, M.L.; Rowe, B. Salmonella Goldcoast from outbreaks of food-poisoning in the British Isles can be differentiated by plasmid profiles. J. Hyg. 1986, 97, 115–122. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from animals and food in 2020/2021. EFSA J. 2023, 21, 7867. [Google Scholar]
- Chang, Y.J.; Chen, C.L.; Yang, H.P.; Chiu, C.H. Prevalence, serotypes, and antimicrobial resistance patterns of non-typhoid Salmonella in Food in Northern Taiwan. Pathogens 2022, 11, 705. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.M.; Tang, H.L.; Ke, S.C.; Lin, Y.P.; Lu, M.C.; Lai, Y.C.; Chen, B.H.; Wang, Y.W.; Teng, R.H.; Chiou, C.S. A nosocomial salmonellosis outbreak caused by blaOXA-48-carrying, extensively drug-resistant Salmonella enterica serovar Goldcoast in a hospital respiratory care ward in Taiwan. J. Glob. Antimicrob. Resist. 2022, 29, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Ed-Dra, A.; Pan, H.; Dong, C.; Jia, C.; Yue, M. Genomic investigation of Salmonella isolates recovered from a pig slaughtering process in Hangzhou, China. Front. Microbiol. 2021, 12, 704636. [Google Scholar] [CrossRef] [PubMed]
- Sotillo, A.; Muñoz-Vélez, M.; Santamaría, M.L.; Ruiz-Carrascoso, G.; García-Bujalance, S.; Gómez-Gil, R.; Mingorance, J. Emergence of VIM-1-producing Salmonella enterica serovar Typhimurium in a paediatric patient. J. Med. Microbiol. 2015, 64, 1541–1543. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.; San José, M.; Roschanski, N.; Schmoger, S.; Baumann, B.; Irrgang, A.; Friese, A.; Roesler, U.; Helmuth, R.; Guerra, B. Spread and persistence of VIM-1 carbapenemase-producing Enterobacteriaceae in three German swine farms in 2011 and 2012. Vet. Microbiol 2017, 200, 118–123. [Google Scholar] [CrossRef]
- Liao, Y.S.; Chen, B.H.; Hong, Y.P.; Teng, R.H.; Wang, Y.W.; Liang, S.Y.; Liu, Y.Y.; Tu, Y.H.; Chen, Y.S.; Chang, J.H.; et al. Emergence of multidrug-resistant Salmonella enterica serovar Goldcoast strains in Taiwan and international spread of the ST358 clone. Antimicrob. Agents Chemother. 2019, 63, e01122-19. [Google Scholar] [CrossRef]
- Dabos, L.; Rodriguez, C.H.; Nastro, M.; Dortet, L.; Bonnin, R.A.; Famiglietti, A.; Iorga, B.I.; Vay, C.; Naas, T. LMB-1 producing Citrobacter freundii from Argentina, a novel player in the field of MBLs. Int. J. Antimicrob. Agents 2020, 55, 105857. [Google Scholar] [CrossRef]
- Yao, Y.; Falgenhauer, L.; Falgenhauer, J.; Hauri, A.M.; Heinmüller, P.; Domann, E.; Chakraborty, T.; Imirzalioglu, C. Carbapenem-Resistant Citrobacter spp. as an emerging concern in the hospital-setting: Results from a genome-based regional surveillance study. Front. Cell Infect. Microbiol. 2021, 11, 744431. [Google Scholar] [CrossRef] [PubMed]
- Piccirilli, A.; Cherubini, S.; Brisdelli, F.; Fazii, P.; Stanziale, A.; Di Valerio, S.; Chiavaroli, V.; Principe, L.; Perilli, M. Molecular Characterization by Whole-Genome Sequencing of Clinical and Environmental Serratia marcescens Strains Isolated during an Outbreak in a Neonatal Intensive Care Unit (NICU). Diagnostics 2022, 12, 2180. [Google Scholar] [CrossRef] [PubMed]
- Cherubini, S.; Perilli, M.; Segatore, B.; Fazii, P.; Parruti, G.; Frattari, A.; Amicosante, G.; Piccirilli, A. Whole-Genome Sequencing of ST2 A. baumannii causing bloodstream infections in COVID-19 Patients. Antibiotics 2022, 11, 955. [Google Scholar] [CrossRef] [PubMed]
- Cherubini, S.; Perilli, M.; Azzini, A.M.; Tacconelli, E.; Maccacaro, L.; Bazaj, A.; Naso, L.; Amicosante, G.; Ltcf-Veneto Working Group; Lo Cascio, G.; et al. Resistome and virulome of multi-drug resistant E. coli ST131 isolated from residents of Long-Term Care Facilities in the Northern Italian Region. Diagnostics 2022, 12, 213. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Nash, J.H.E. MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Bitar, I.; Caltagirone, M.; Villa, L.; Mattioni Marchetti, V.; Nucleo, E.; Sarti, M.; Migliavacca, R.; Carattoli, A. Interplay among IncA and blaKPC-carrying plasmids in Citrobacter freundii. Antimicrob. Agents Chemother. 2019, 63, e02609-18. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Piccirilli, A.; Perilli, M.; Piccirilli, V.; Segatore, B.; Amicosante, G.; Maccacaro, L.; Bazaj, A.; Naso, L.; Cascio, G.L.; Cornaglia, G. Molecular characterization of carbapenem-resistant Klebsiella pneumoniae ST14 and ST512 causing bloodstream infections in ICU and surgery wards of a tertiary university hospital of Verona (northern Italy): Co-production of KPC-3, OXA-48, and CTX-M-15 β-lactamases. Diagn. Microbiol. Infect. Dis. 2020, 96, 114968. [Google Scholar]
| Antibiotics | MIC (mg/L) | |||
|---|---|---|---|---|
| S. enterica TS1, TS2 | C. freundii TS3 | |||
| Cefotaxime | >4 | (R) | >4 | (R) |
| Ceftazidime | >128 | (R) | >128 | (R) |
| Cefepime | >16 | (R) | >16 | (R) |
| Imipenem | 4 | (I) | 4 | (I) |
| Meropenem | 2 | (S) | 1 | (S) |
| Ciprofloxacin | 0.5 | (R) | 0.12 | (S) |
| Amikacin | ≤4 | (S) | ≤4 | (S) |
| Gentamicin | 2 | (S) | 2 | (S) |
| Tigecycline | 0.25 | (S) | 0.25 | (S) |
| Genotyping of S. enterica Goldcoast TS2 Strain | ||||||
|---|---|---|---|---|---|---|
| MLST | Β-Lactam Resistance | Aminoglycoside Resistance | Macrolide Resistance | Quinolone Resistance | Rifampicin Resistance | Others |
| ST358 | blaVIM-1 blaOXA-10 blaSHV-12 | aadA1 ant(2″)-Ia aac(6′)-Iaa aac(6′)-Ib3 aac(6′)-Ib-cr | mph(A) | qnrVC6 parC T57S aac(6′)-Ib-cr | ARR-2 | sul1 tet(B) catB8 dfrA14 qacE |
| Mobile genetic elements | ||||||
| Transposons | Plasmids | pMLST | Insertion Sequences (ISs) | |||
| Tn1000 Tn6196 | IncA, IncHI2, IncHI2A, IncFII(29) | IncA:(ST = 12) IncHI2:(ST = 1) | IS6100 (IS6 family) ISEc52 IS903 ISAs22b (IS3 family; IS407 group) MITEEc1 (IS630 family) ISSty2 (IS3 family) IS421 (IS4 family; IS231 group) | |||
| VF Class | Virulence Factors | Related Genes |
|---|---|---|
| Fimbrial Adherence Determinants | Agf/Csg | csgG |
| Bcf | bcfB,C,D | |
| Fim | fimC,D,H,Z | |
| Saf | safB,C | |
| Stb | stbB,C,D | |
| Stc | stcC,D | |
| Std | stdB,C | |
| Ste | steB,C | |
| Stf | stfC,D | |
| Sth | sthC,E | |
| Sti | stiC,H | |
| Macrophage Inducible Genes | Mig-14 | mig-14 |
| Magnesium Uptake | Mg2+ transport | mgtB |
| Nonfimbrial Adherence Determinants | MisL | misL |
| RatB | ratB | |
| SinH | sinH | |
| Regulation | PhoPQ | phoQ |
| Secretion System | TTSS (SPI-1 encode) | hilA,C,D; invA,C,E,G; prgH; sipD; spaO,R,S; |
| TTSS (SPI-2 encode) | ssaC,D,L,N,Q,U,V; sseC; ssrA | |
| TTSS effectors translocated via both systems | slrP | |
| TTSS-1 translocated effectors | avrA; sipA,B,C; sopA,B,D; sptB | |
| TTSS-2 translocated effectors | pipB2; pipB; sifA,B; sopD2; sseF,G,J,K1,K2,L | |
| Toxin | Typhoid toxin | cdtB, pltA |
| Genotyping of C. freundii TS3 | ||||||
|---|---|---|---|---|---|---|
| MLST | Β-Lactam Resistance | Aminoglycoside Resistance | Macrolide Resistance | Quinolone Resistance | Rifampicin Resistance | Others |
| ST62 | blaVIM-1 blaCMY-150 blaSHV-12 | aadA1 aac(6′)-Ib3 aac(6′)-Ib-cr | mph(A) | qnrVC6 qnrB38 aac(6′)-Ib-cr | ARR-2 | sul1 dfrA14 qacE |
| Mobile genetic elements | ||||||
| Plasmids | pMLST | Insertion Sequences (ISs) | ||||
| IncA | IncA:(ST = 12) | IS6100 (IS6 family), IS5 (IS5 family), ISSen4 (IS3 family), ISAs22 (IS3 family) | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piccirilli, A.; Di Marcantonio, S.; Costantino, V.; Simonetti, O.; Busetti, M.; Luzzati, R.; Principe, L.; Di Domenico, M.; Rinaldi, A.; Cammà, C.; et al. Identification of IncA Plasmid, Harboring blaVIM-1 Gene, in S. enterica Goldcoast ST358 and C. freundii ST62 Isolated in a Hospitalized Patient. Antibiotics 2023, 12, 1659. https://doi.org/10.3390/antibiotics12121659
Piccirilli A, Di Marcantonio S, Costantino V, Simonetti O, Busetti M, Luzzati R, Principe L, Di Domenico M, Rinaldi A, Cammà C, et al. Identification of IncA Plasmid, Harboring blaVIM-1 Gene, in S. enterica Goldcoast ST358 and C. freundii ST62 Isolated in a Hospitalized Patient. Antibiotics. 2023; 12(12):1659. https://doi.org/10.3390/antibiotics12121659
Chicago/Turabian StylePiccirilli, Alessandra, Sascia Di Marcantonio, Venera Costantino, Omar Simonetti, Marina Busetti, Roberto Luzzati, Luigi Principe, Marco Di Domenico, Antonio Rinaldi, Cesare Cammà, and et al. 2023. "Identification of IncA Plasmid, Harboring blaVIM-1 Gene, in S. enterica Goldcoast ST358 and C. freundii ST62 Isolated in a Hospitalized Patient" Antibiotics 12, no. 12: 1659. https://doi.org/10.3390/antibiotics12121659
APA StylePiccirilli, A., Di Marcantonio, S., Costantino, V., Simonetti, O., Busetti, M., Luzzati, R., Principe, L., Di Domenico, M., Rinaldi, A., Cammà, C., & Perilli, M. (2023). Identification of IncA Plasmid, Harboring blaVIM-1 Gene, in S. enterica Goldcoast ST358 and C. freundii ST62 Isolated in a Hospitalized Patient. Antibiotics, 12(12), 1659. https://doi.org/10.3390/antibiotics12121659









