Control of Escherichia coli in Fresh-Cut Mixed Vegetables Using a Combination of Bacteriophage and Carvacrol
Abstract
:1. Introduction
2. Results
2.1. Isolated Coliphages and Their Host Range
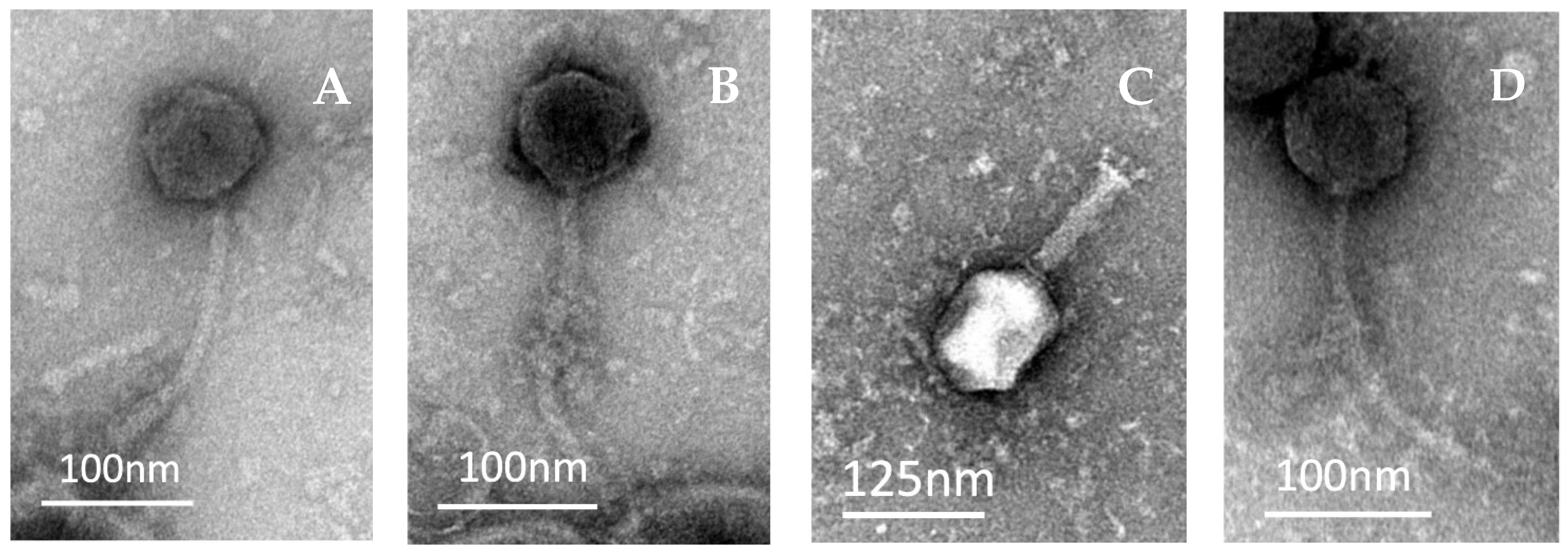
2.2. Morphological Analysis of Coliphages under TEM
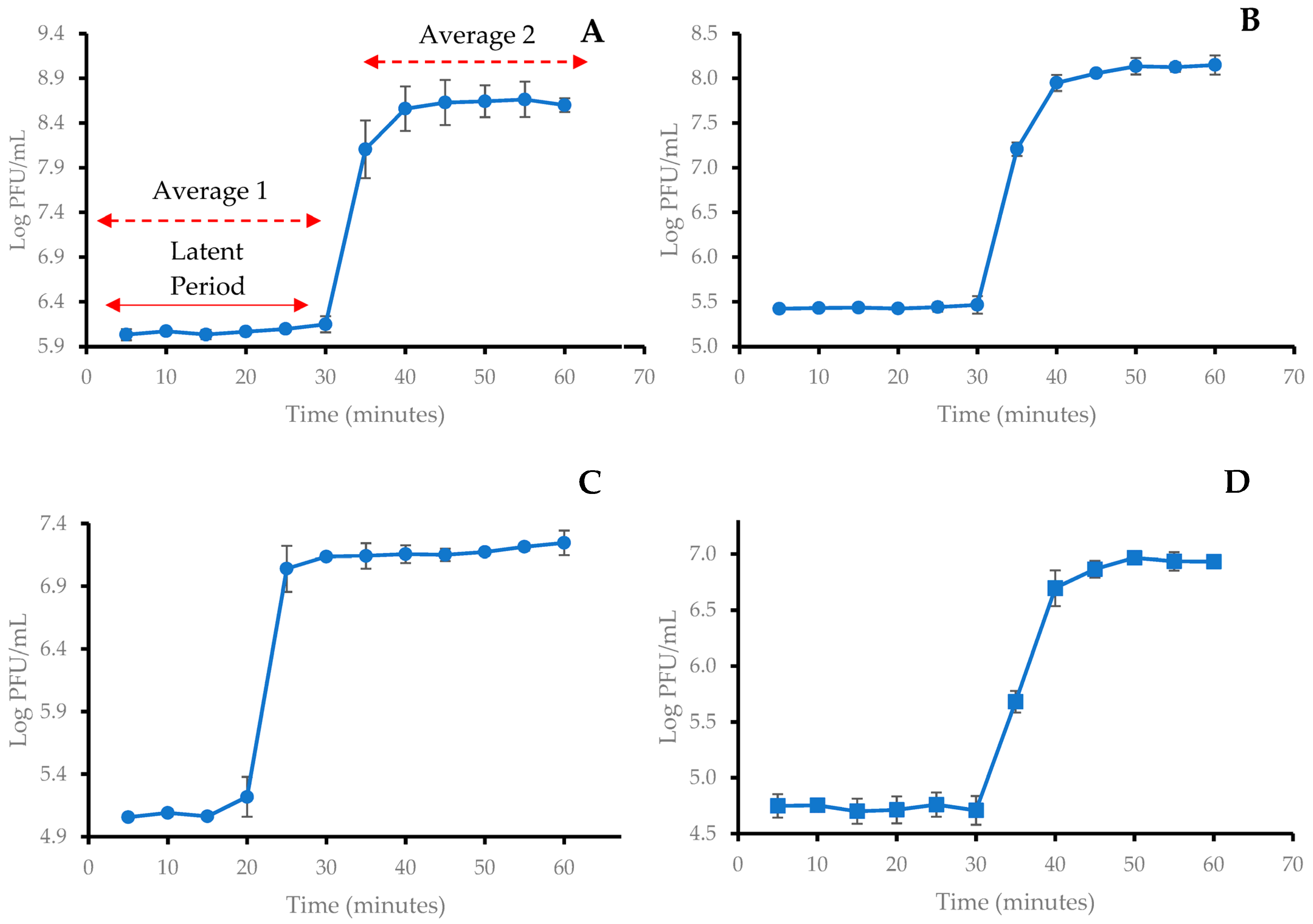
2.3. One-Step Growth Curve of Coliphages
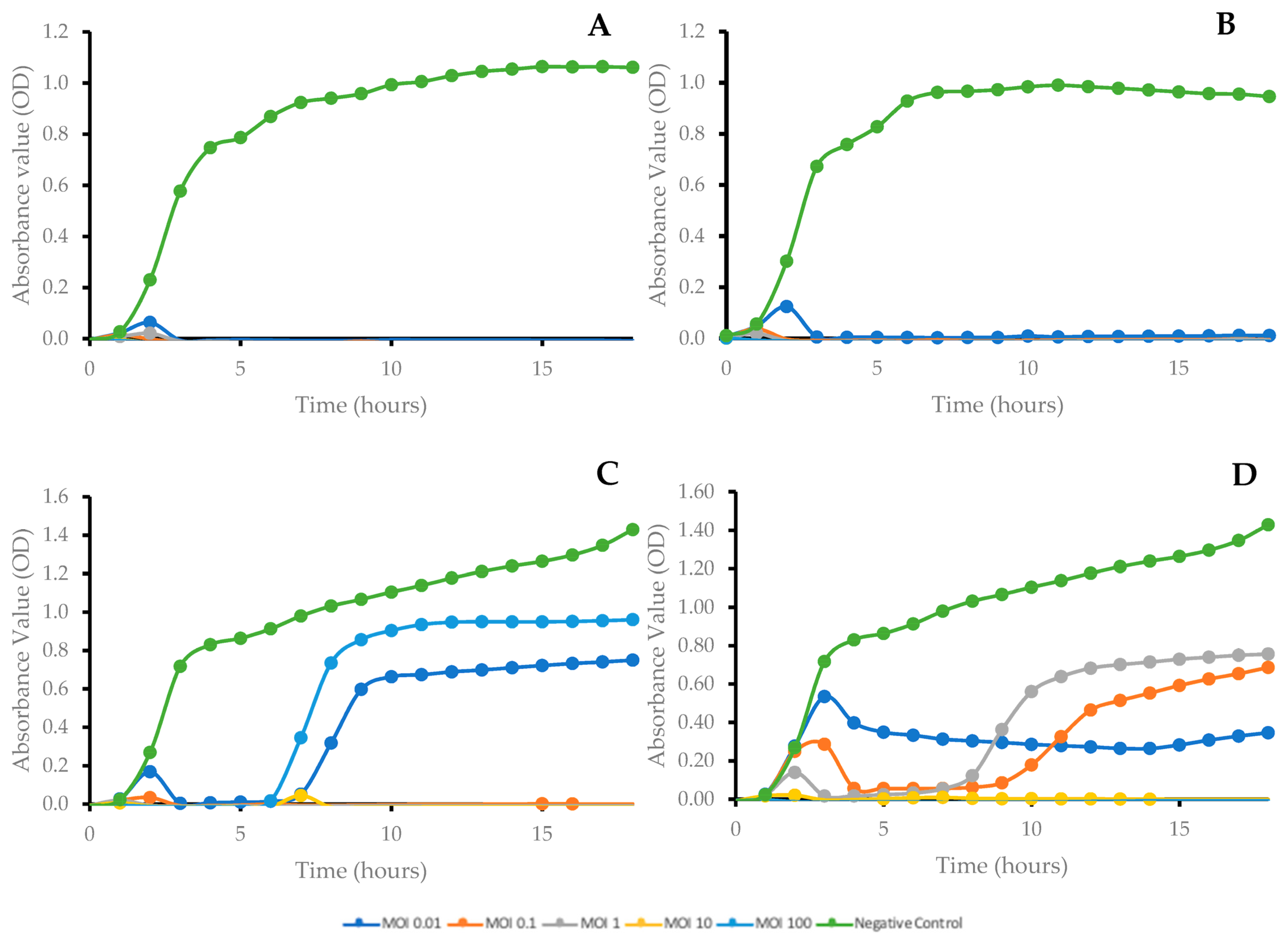
2.4. Bacterial Growth Kinetics after Phage Treatment at Different Multiplicities of Infection (MOI)
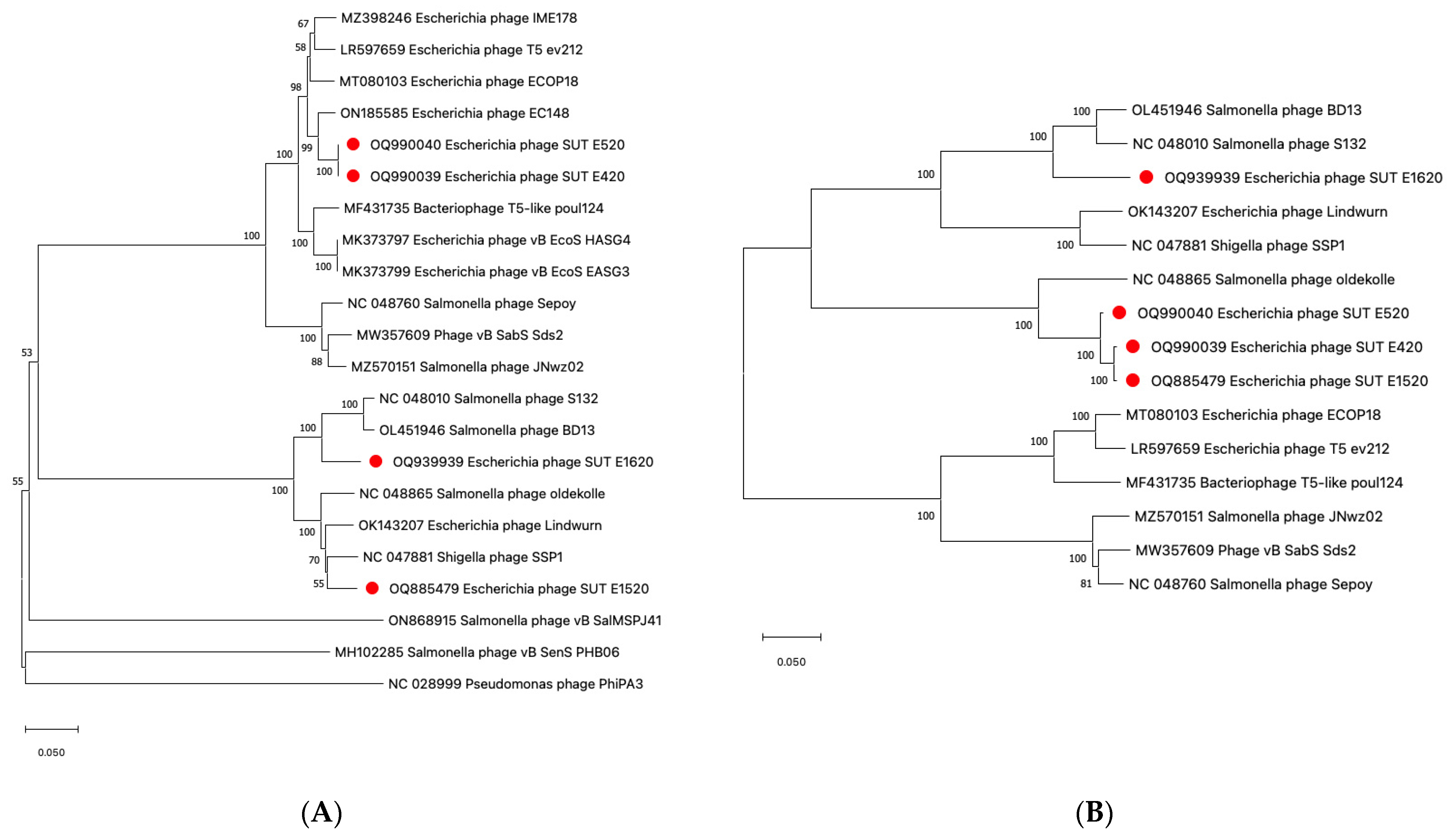
2.5. Genomic Analysis of Coliphages via Next Generation Sequencing (NGS)
2.6. Minimum Inhibitory Concentration (MIC) of Carvacrol In Vitro
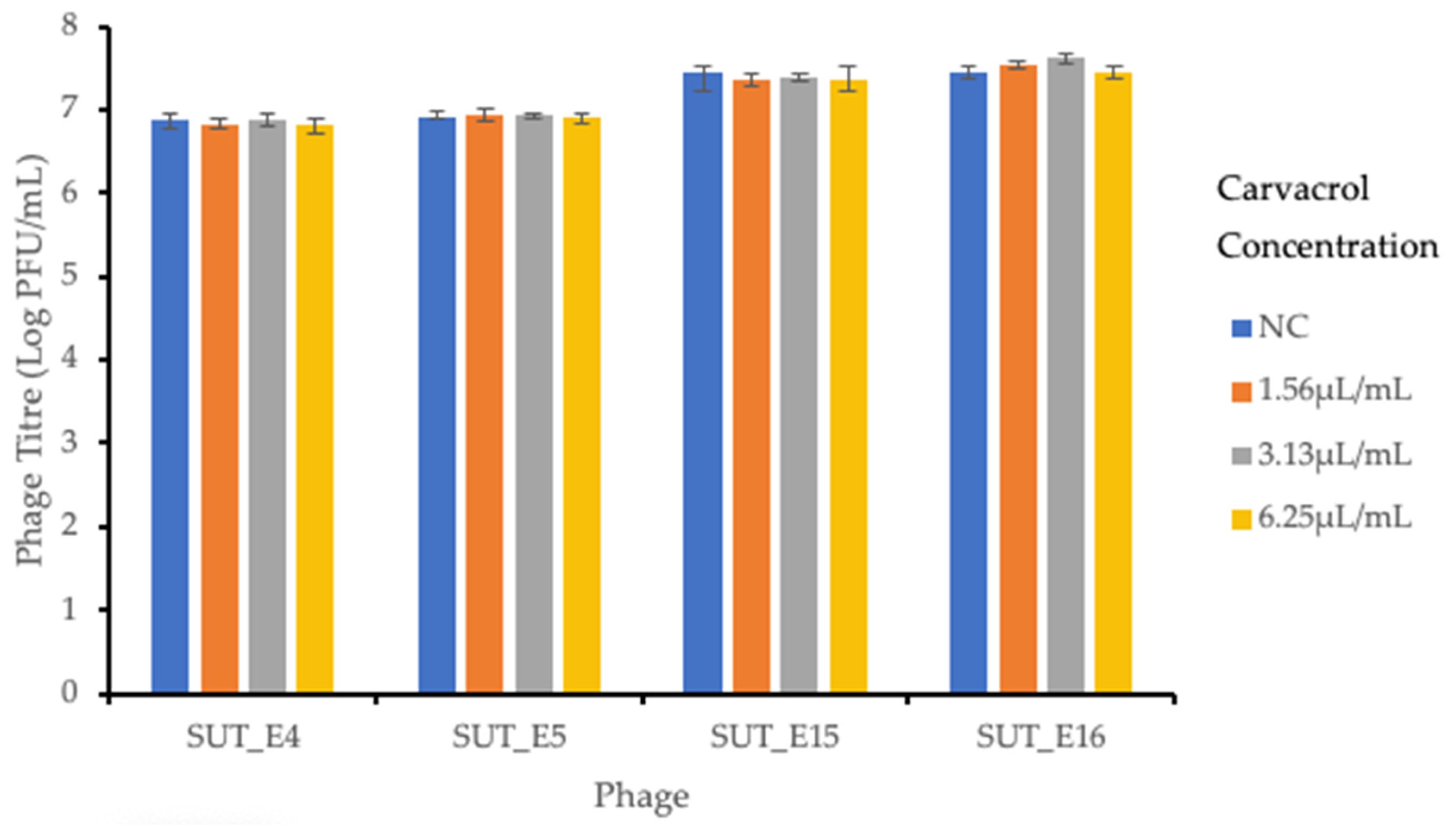
2.7. Sensitivity of Coliphages towards Carvacrol
2.8. Combined Treatment of Coliphages and Carvacrol against E. coli StrR on Fresh-Cut Mixed Vegetables
3. Discussion and Conclusions
4. Materials and Methods
4.1. Characterization of Newly Isolated Coliphages
4.1.1. Host Bacteria Strains and Culture Conditions
4.1.2. Isolation of Coliphages and Transmission Electron Microscope (TEM) Analysis
4.1.3. Host Range of Phages
4.1.4. One-Step Growth Curve of Phages
4.1.5. Growth Kinetics of Bacterial Hosts following Phage Treatment at Different MOI
4.1.6. Genomic Analysis of Phages Using Next Generation Sequencing (NGS) Technology
4.2. Analysis of Carvacrol
4.2.1. Preparation and Analysis of Carvacrol
4.2.2. Antimicrobial Activity of Carvacrol
4.2.3. Susceptibility of Phages to Carvacrol
4.3. Application of Phage Cocktail and Carvacrol to Fresh-Cut Mixed Vegetables
4.3.1. Bacterial Inoculum
4.3.2. Preparation of Phage Cocktail and Carvacrol
4.3.3. Antimicrobial Properties of Phage Cocktail and Carvacrol on Fresh-Cut Mixed Vegetables
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hall, G.; Kirk, M.D.; Becker, N.; Gregory, J.E.; Unicomb, L.; Millard, G.; Stafford, R.; Lalor, K.; Group, O.W. Estimating foodborne gastroenteritis, Australia. Emerg. Infect. Dis. 2005, 11, 1257. [Google Scholar] [CrossRef] [PubMed]
- Chang, Q.; Wang, W.; Regev-Yochay, G.; Lipsitch, M.; Hanage, W.P. Antibiotics in agriculture and the risk to human health: How worried should we be? Evol. Appl. 2015, 8, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Kuek, M.; McLean, S.K.; Palombo, E.A. Application of bacteriophages in food production and their potential as biocontrol agents in the organic farming industry. Biol. Control 2022, 165, 104817. [Google Scholar] [CrossRef]
- Smith, D.L.; Harris, A.D.; Johnson, J.A.; Silbergeld, E.K.; Morris, J.G. Animal antibiotic use has an early but important impact on the emergence of antibiotic resistance in human commensal bacteria. Proc. Natl. Acad. Sci. USA 2002, 99, 6434–6439. [Google Scholar] [CrossRef] [PubMed]
- Sulakvelidze, A.; Barrow, P. 13 Phage Therapy. In Bacteriophages: Biology and Applications; CRC Press: Boca Raton, FL, USA, 2004; p. 335. [Google Scholar]
- Abdelrahman, F.; Rezk, N.; Fayez, M.S.; Abdelmoteleb, M.; Atteya, R.; Elhadidy, M.; El-Shibiny, A. Isolation, characterization, and genomic analysis of three novel E. coli bacteriophages that effectively infect E. coli O18. Microorganisms 2022, 10, 589. [Google Scholar] [CrossRef] [PubMed]
- Garcia, P.; Martinez, B.; Obeso, J.; Rodriguez, A. Bacteriophages and their application in food safety. Lett. Appl. Microbiol. 2008, 47, 479–485. [Google Scholar] [CrossRef]
- Sarhan, W.A.; Azzazy, H.M. Phage approved in food, why not as a therapeutic? Expert Rev. Anti-Infect. Ther. 2015, 13, 91–101. [Google Scholar] [CrossRef]
- El-Shibiny, A.; El-Sahhar, S.; Adel, M. Phage applications for improving food safety and infection control in Egypt. J. Appl. Microbiol. 2017, 123, 556–567. [Google Scholar] [CrossRef]
- Kazi, M.; Annapure, U.S. Bacteriophage biocontrol of foodborne pathogens. J. Food Sci. Technol. 2016, 53, 1355–1362. [Google Scholar] [CrossRef]
- Nikolich, M.P.; Filippov, A.A. Bacteriophage therapy: Developments and directions. Antibiotics 2020, 9, 135. [Google Scholar] [CrossRef]
- Cui, H.; Zhang, C.; Li, C.; Lin, L. Antibacterial mechanism of oregano essential oil. Ind. Crops Prod. 2019, 139, 111498. [Google Scholar] [CrossRef]
- Huang, J.; Qian, C.; Xu, H.; Huang, Y. Antibacterial activity of Artemisia asiatica essential oil against some common respiratory infection causing bacterial strains and its mechanism of action in Haemophilus influenzae. Microb. Pathog. 2018, 114, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Ni, P.; Wang, L.; Deng, B.; Jiu, S.; Ma, C.; Zhang, C.; Almeida, A.; Wang, D.; Xu, W.; Wang, S. Combined application of bacteriophages and carvacrol in the control of Pseudomonas syringae pv. actinidiae planktonic and biofilm forms. Microorganisms 2020, 8, 837. [Google Scholar]
- Li, B.; Zheng, K.; Lu, J.; Zeng, D.; Xiang, Q.; Ma, Y. Antibacterial characteristics of oregano essential oil and its mechanisms against Escherichia coli O157:H7. J. Food Meas. Charact. 2022, 16, 2989–2998. [Google Scholar] [CrossRef]
- Singletary, K. Oregano: Overview of the literature on health benefits. Nutr. Today 2010, 45, 129–138. [Google Scholar] [CrossRef]
- Govaris, A.; Solomakos, N.; Pexara, A.; Chatzopoulou, P. The antimicrobial effect of oregano essential oil, nisin and their combination against Salmonella Enteritidis in minced sheep meat during refrigerated storage. Int. J. Food Microbiol. 2010, 137, 175–180. [Google Scholar] [CrossRef]
- Viazis, S.; Akhtar, M.; Feirtag, J.; Diez-Gonzalez, F. Reduction of Escherichia coli O157: H7 viability on leafy green vegetables by treatment with a bacteriophage mixture and trans-cinnamaldehyde. Food Microbiol. 2011, 28, 149–157. [Google Scholar] [CrossRef]
- Qadri, O.S.; Yousuf, B.; Srivastava, A.K. Fresh-cut fruits and vegetables: Critical factors influencing microbiology and novel approaches to prevent microbial risks—A review. Cogent Food Agric. 2015, 1, 1121606. [Google Scholar] [CrossRef]
- Abuladze, T.; Li, M.; Menetrez, M.Y.; Dean, T.; Senecal, A.; Sulakvelidze, A. Bacteriophages reduce experimental contamination of hard surfaces, tomato, spinach, broccoli, and ground beef by Escherichia coli O157:H7. Appl. Environ. Microbiol. 2008, 74, 6230–6238. [Google Scholar] [CrossRef]
- Bai, J.; Jeon, B.; Ryu, S. Effective inhibition of Salmonella Typhimurium in fresh produce by a phage cocktail targeting multiple host receptors. Food Microbiol. 2019, 77, 52–60. [Google Scholar] [CrossRef]
- Boyacioglu, O.; Sharma, M.; Sulakvelidze, A.; Goktepe, I. Biocontrol of Escherichia coli O157:H7 on fresh-cut leafy greens. Bacteriophage 2013, 3, e24620. [Google Scholar] [CrossRef] [PubMed]
- Leverentz, B.; Conway, W.S.; Alavidze, Z.; Janisiewicz, W.J.; Fuchs, Y.; Camp, M.J.; Chighladze, E.; Sulakvelidze, A. Examination of bacteriophage as a biocontrol method for Salmonella on fresh-cut fruit: A model study. J. Food Prot. 2001, 64, 1116–1121. [Google Scholar] [CrossRef] [PubMed]
- Leverentz, B.; Conway, W.S.; Camp, M.J.; Janisiewicz, W.J.; Abuladze, T.; Yang, M.; Saftner, R.; Sulakvelidze, A. Biocontrol of Listeria monocytogenes on fresh-cut produce by treatment with lytic bacteriophages and a bacteriocin. Appl. Environ. Microbiol. 2003, 69, 4519–4526. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chen, H.; Zhao, J.; Tao, Z.; Lan, W.; Zhao, Y.; Sun, X. Characterization of phage vB_SalM_SPJ41 and the reduction of risk of antibiotic-resistant Salmonella enterica contamination in two ready-to-eat foods. Antibiotics 2023, 12, 364. [Google Scholar] [CrossRef] [PubMed]
- Viazis, S.; Akhtar, M.; Feirtag, J.; Brabban, A.; Diez-Gonzalez, F. Isolation and characterization of lytic bacteriophages against enterohaemorrhagic Escherichia coli. J. Appl. Microbiol. 2011, 110, 1323–1331. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.W.; Delaquis, P.; Goodridge, L.; Lévesque, R.C.; Fong, K.; Wang, S. Inactivation of Salmonella enterica on post-harvest cantaloupe and lettuce by a lytic bacteriophage cocktail. Curr. Res. Food Sci. 2020, 2, 25–32. [Google Scholar] [CrossRef]
- Ye, J.; Kostrzynska, M.; Dunfield, K.; Warriner, K. Control of Salmonella on sprouting mung bean and alfalfa seeds by using a biocontrol preparation based on antagonistic bacteria and lytic bacteriophages. J. Food Prot. 2010, 73, 9–17. [Google Scholar] [CrossRef]
- Liu, W.; Han, L.; Song, P.; Sun, H.; Zhang, C.; Zou, L.; Cui, J.; Pan, Q.; Ren, H. Complete genome sequencing of a Tequintavirus bacteriophage with a broad host range against Salmonella Abortus equi isolates from donkeys. Front. Microbiol. 2022, 13, 2983. [Google Scholar] [CrossRef]
- Liao, Y.-T.; Salvador, A.; Harden, L.A.; Liu, F.; Lavenburg, V.M.; Li, R.W.; Wu, V.C. Characterization of a Lytic Bacteriophage as an Antimicrobial Agent for Biocontrol of Shiga Toxin-Producing Escherichia coli O145 Strains. Antibiotics 2019, 8, 74. [Google Scholar] [CrossRef]
- Wójcicki, M.; Świder, O.; Średnicka, P.; Shymialevich, D.; Ilczuk, T.; Koperski, Ł.; Cieślak, H.; Sokołowska, B.; Juszczuk-Kubiak, E. Newly Isolated Virulent Salmophages for Biocontrol of Multidrug-Resistant Salmonella in Ready-to-Eat Plant-Based Food. Int. J. Mol. Sci. 2023, 24, 10134. [Google Scholar] [CrossRef]
- Ferguson, S.; Roberts, C.; Handy, E.; Sharma, M. Lytic bacteriophages reduce Escherichia coli O157: H7 on fresh cut lettuce introduced through cross-contamination. Bacteriophage 2013, 3, e24323. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Vivas, J.; Elexpuru-Zabaleta, M.; Samano, M.L.; Barrera, A.P.; Forbes-Hernández, T.Y.; Giampieri, F.; Battino, M. Phages and Enzybiotics in Food Biopreservation. Molecules 2021, 26, 5138. [Google Scholar] [CrossRef] [PubMed]
- Synnott, A.J.; Kuang, Y.; Kurimoto, M.; Yamamichi, K.; Iwano, H.; Tanji, Y. Isolation from sewage influent and characterization of novel Staphylococcus aureus bacteriophages with wide host ranges and potent lytic capabilities. Appl. Environ. Microbiol. 2009, 75, 4483–4490. [Google Scholar] [CrossRef] [PubMed]
- Ateba, C.N.; Akindolire, M.A. Isolation and characterisation of bacteriophages with lytic activity against virulent Escherichia coli O157: H7: Potential bio-control agents. Preprints 2019, 2019, 2019010132. [Google Scholar]
- Kim, J.H.; Kim, H.J.; Jung, S.J.; Mizan, M.F.R.; Park, S.H.; Ha, S.D. Characterization of Salmonella spp.-specific bacteriophages and their biocontrol application in chicken breast meat. J. Food Sci. 2020, 85, 526–534. [Google Scholar] [CrossRef]
- Carey-Smith, G.V.; Billington, C.; Cornelius, A.J.; Hudson, J.A.; Heinemann, J.A. Isolation and characterization of bacteriophages infecting Salmonella spp. FEMS Microbiol. Lett. 2006, 258, 182–186. [Google Scholar] [CrossRef]
- Huang, C.; Virk, S.M.; Shi, J.; Zhou, Y.; Willias, S.P.; Morsy, M.K.; Abdelnabby, H.E.; Liu, J.; Wang, X.; Li, J. Isolation, characterization, and application of bacteriophage LPSE1 against Salmonella enterica in ready to eat (RTE) foods. Front. Microbiol. 2018, 9, 1046. [Google Scholar] [CrossRef]
- Park, M.; Lee, J.-H.; Shin, H.; Kim, M.; Choi, J.; Kang, D.-H.; Heu, S.; Ryu, S. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157: H7. Appl. Environ. Microbiol. 2012, 78, 58–69. [Google Scholar] [CrossRef]
- Sadekuzzaman, M.; Yang, S.; Mizan, M.F.R.; Ha, S.D. Reduction of Escherichia coli O157: H7 in biofilms using bacteriophage BPECO 19. J. Food Sci. 2017, 82, 1433–1442. [Google Scholar] [CrossRef]
- Yang, M.; Liang, Y.; Huang, S.; Zhang, J.; Wang, J.; Chen, H.; Ye, Y.; Gao, X.; Wu, Q.; Tan, Z. Isolation and Characterization of the Novel Phages vB_VpS_BA3 and vB_VpS_CA8 for Lysing Vibrio Parahaemolyticus. Front. Microbiol. 2020, 11, 259. [Google Scholar] [CrossRef]
- Hyman, P. Phages for phage therapy: Isolation, characterization, and host range breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- OFlynn, G.; Ross, R.; Fitzgerald, G.; Coffey, A. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157: H7. Appl. Environ. Microbiol. 2004, 70, 3417–3424. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M. Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef] [PubMed]
- Lundin, D.; Torrents, E.; Poole, A.M.; Sjöberg, B.-M. RNRdb, a curated database of the universal enzyme family ribonucleotide reductase, reveals a high level of misannotation in sequences deposited to Genbank. BMC Genom. 2009, 10, 1–8. [Google Scholar] [CrossRef]
- Xiang, Y.; Li, W.; Song, F.; Yang, X.; Zhou, J.; Yu, H.; Ji, X.; Wei, Y. Biological characteristics and whole-genome analysis of the Enterococcus faecalis phage PEf771. Can. J. Microbiol. 2020, 66, 505–520. [Google Scholar] [CrossRef]
- Bailly-Bechet, M.; Vergassola, M.; Rocha, E. Causes for the intriguing presence of tRNAs in phages. Genome Res. 2007, 17, 1486–1495. [Google Scholar] [CrossRef]
- Šimoliūnas, E.; Vilkaitytė, M.; Kaliniene, L.; Zajančkauskaitė, A.; Kaupinis, A.; Staniulis, J.; Valius, M.; Meškys, R.; Truncaitė, L. Incomplete LPS core-specific Felix01-like virus vB_EcoM_VpaE1. Viruses 2015, 7, 6163–6181. [Google Scholar] [CrossRef]
- Beuchat, L.R. Ecological factors influencing survival and growth of human pathogens on raw fruits and vegetables. Microbes Infect. 2002, 4, 413–423. [Google Scholar] [CrossRef]
- Snyder, A.B.; Perry, J.J.; Yousef, A.E. Developing and optimizing bacteriophage treatment to control enterohemorrhagic Escherichia coli on fresh produce. Int. J. Food Microbiol. 2016, 236, 90–97. [Google Scholar] [CrossRef]
- Center for Disease Control and Prevention (CDC). National Outbreak Reporting System. Available online: https://wwwn.cdc.gov/norsdashboard/ (accessed on 15 September 2023).
- Perera, M.N.; Abuladze, T.; Li, M.; Woolston, J.; Sulakvelidze, A. Bacteriophage cocktail significantly reduces or eliminates Listeria monocytogenes contamination on lettuce, apples, cheese, smoked salmon and frozen foods. Food Microbiol. 2015, 52, 42–48. [Google Scholar] [CrossRef]
- Burt, S. Essential oils: Their antibacterial properties and potential applications in foods—A review. Int. J. Food Microbiol. 2004, 94, 223–253. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, H.; Beuchat, L.R.; Kim, Y.; Ryu, J.-H. Synergistic antimicrobial activity of oregano and thyme thymol essential oils against Leuconostoc citreum in a laboratory medium and tomato juice. Food Microbiol. 2020, 90, 103489. [Google Scholar] [CrossRef] [PubMed]
- Hyun, J.E.; Bae, Y.M.; Song, H.; Yoon, J.H.; Lee, S.Y. Antibacterial effect of various essential oils against pathogens and spoilage microorganisms in fresh produce. J. Food Saf. 2015, 35, 206–219. [Google Scholar] [CrossRef]
- Di Pasqua, R.; Hoskins, N.; Betts, G.; Mauriello, G. Changes in membrane fatty acids composition of microbial cells induced by addiction of thymol, carvacrol, limonene, cinnamaldehyde, and eugenol in the growing media. J. Agric. Food Chem. 2006, 54, 2745–2749. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Yoon, H.; Kang, D.-H.; Chang, P.-S.; Ryu, S. Endolysin LysSA97 is synergistic with carvacrol in controlling Staphylococcus aureus in foods. Int. J. Food Microbiol. 2017, 244, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.H.; Waite-Cusic, J.; Huang, E. Control of Salmonella in chicken meat using a combination of a commercial bacteriophage and plant-based essential oils. Food Control 2020, 110, 106984. [Google Scholar] [CrossRef]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage applications for food production and processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef]
- Połaska, M.; Sokołowska, B. Bacteriophages—A new hope or a huge problem in the food industry. AIMS Microbiol. 2019, 5, 324. [Google Scholar] [CrossRef]
- Kropinski, A.M. Practical advice on the one-step growth curve. In Bacteriophages; Springer: Berlin/Heidelberg, Germany, 2018; pp. 41–47. [Google Scholar]
- Chen, L.; Liu, Q.; Fan, J.; Yan, T.; Zhang, H.; Yang, J.; Deng, D.; Liu, C.; Wei, T.; Ma, Y. Characterization and genomic analysis of ValSw3-3, a new Siphoviridae bacteriophage infecting Vibrio alginolyticus. J. Virol. 2020, 94, e00066-20. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]



| E. coli Strains | Coliphage | |||
|---|---|---|---|---|
| SUT_E420 | SUT_E520 | SUT_E1520 | SUT_E1620 | |
| ATC25922 | - | - | - | + |
| K12 | + | + | H | H |
| G106 | + | + | + | (+) |
| G131 | + | + | + | + |
| O157:H7 | H | H | - | + |
| O113:H21 | - | - | + | - |
| O111 (non-motile) | - | - | - | - |
| O130:H11 | - | - | - | - |
| O15 | - | (+) | - | + |
| O26:H11 | + | - | - | - |
| O5 (non-motile) | - | - | - | - |
| O127:H6 | - | - | - | + |
| O111 | - | - | - | - |
| O119 | - | - | - | (+) |
| O142:H6 | - | - | - | + |
| O55:H6 | - | - | - | - |
| O55:H7 | (+) | (+) | - | (+) |
| Salmonella strains | ||||
| S. Typhi | - | - | - | + |
| S. Enteritidis | - | - | - | - |
| S. Paratyphi | - | - | - | + |
| S. Hofit | - | - | - | + |
| S. Braenderup | - | - | - | - |
| S.e.s Newport | - | (+) | - | - |
| S.e.s Tallahassee | - | - | - | - |
| S.e.s Choleraesuis | - | + | - | + |
| S.e.s Anatum | - | - | - | - |
| Phage | Head Length (nm) | Head Width (nm) | Tail Length (nm) | Tail Width (nm) |
|---|---|---|---|---|
| SUT_E420 | 73 | 73 | 175 | 11 |
| SUT_E520 | 62 | 73 | 187 | 9 |
| SUT_E1520 | 109 | 90 | 102 | 24 |
| SUT_E1620 | 79 | 74 | 186 | 10 |
| Phage | Latent Period (min) | Burst Size (PFU/CFU) |
|---|---|---|
| SUT_E420 | 30 | 350 |
| SUT_E520 | 30 | 441 |
| SUT_E1520 | 20 | 116 |
| SUT_E1620 | 30 | 140 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuek, M.; McLean, S.K.; Palombo, E.A. Control of Escherichia coli in Fresh-Cut Mixed Vegetables Using a Combination of Bacteriophage and Carvacrol. Antibiotics 2023, 12, 1579. https://doi.org/10.3390/antibiotics12111579
Kuek M, McLean SK, Palombo EA. Control of Escherichia coli in Fresh-Cut Mixed Vegetables Using a Combination of Bacteriophage and Carvacrol. Antibiotics. 2023; 12(11):1579. https://doi.org/10.3390/antibiotics12111579
Chicago/Turabian StyleKuek, Maryanne, Sarah K. McLean, and Enzo A. Palombo. 2023. "Control of Escherichia coli in Fresh-Cut Mixed Vegetables Using a Combination of Bacteriophage and Carvacrol" Antibiotics 12, no. 11: 1579. https://doi.org/10.3390/antibiotics12111579
APA StyleKuek, M., McLean, S. K., & Palombo, E. A. (2023). Control of Escherichia coli in Fresh-Cut Mixed Vegetables Using a Combination of Bacteriophage and Carvacrol. Antibiotics, 12(11), 1579. https://doi.org/10.3390/antibiotics12111579







