Abstract
Pseudomonas aeruginosa ST274 is an international epidemic high-risk clone, mostly associated with hospital settings and appears to colonize cystic fibrosis (CF) patients worldwide. To understand the relevant mechanisms for its success, the biological and genomic characteristics of 11 ST274-P. aeruginosa strains from clinical and non-clinical origins were analyzed. The extensively drug-resistant (XDR/DTR), the non-susceptible to at least one agent (modR), and the lasR-truncated (by ISPsp7) strains showed a chronic infection phenotype characterized by loss of serotype-specific antigenicity and low motility. Furthermore, the XDR/DTR and modR strains presented low pigment production and biofilm formation, which were very high in the lasR-truncated strain. Their whole genome sequences were compared with other 14 ST274-P. aeruginosa genomes available in the NCBI database, and certain associations have been primarily detected: blaOXA-486 and blaPDC-24 genes, serotype O:3, exoS+/exoU− genotype, group V of type IV pili, and pyoverdine locus class II. Other general molecular markers highlight the absence of vqsM and pldA/tleS genes and the presence of the same mutational pattern in genes involving two-component sensor-regulator systems PmrAB and CreBD, exotoxin A, quorum-sensing RhlI, beta-lactamase expression regulator AmpD, PBP1A, or FusA2 elongation factor G. The proportionated ST274-P. aeruginosa results could serve as the basis for more specific studies focused on better antibiotic stewardship and new therapeutic developments.
Keywords:
ST274; whole genome sequencing; resistome; virulence; motility; pigment; biofilm; lasR; cystic fibrosis 1. Introduction
Pseudomonas aeruginosa is a ubiquitous Gram-negative microorganism that is adaptable and metabolically versatile and has been found in a wide variety of habitats. This species is a relevant opportunistic pathogen and one of the most frequent causes of acute nosocomial infections [1,2,3]. The increasing prevalence of multidrug-resistant (MDR) P. aeruginosa strains makes this bacterium difficult to treat, and it is consequently associated with a high risk of mortality.
P. aeruginosa strains have the capacity to develop resistance to antibiotics by the selection of genomic mutations and by exchange of transferable resistance determinants [4]. Additionally, their pathogenicity is associated with the expression of multiple virulence factors that enable evasion of the host response, such as lipases, proteases, rhamnolipids, pyocyanin, pyoverdine, catalases, and exopolysaccharides, as well as biofilm production. Most of them are under the control of quorum-sensing (QS) systems (including Las, Rhl, QscR, Pqs, and Iqs), with LasIR and RhlIR being the most dominant regulatory circuits [5,6].
P. aeruginosa commonly infects the lungs of cystic fibrosis (CF) patients, and the infections typically progress from the intermittent acquisition of single environmental strains to an extensive genetic and phenotypic adaptation to the lung environment. The characteristics associated with the transition from “acute” to a “chronic” pulmonary pathogen in CF include the downregulation of some virulence factors (motility and production of pigments, rhamnolipids, and proteases), mucoid morphotype, increased biofilm formation, and upregulation of exopolysaccharide expression, as well as the reduced quorum-sensing pathways [5,7]. Therefore, P. aeruginosa is practically impossible to eradicate in the chronic phase of infection [8,9].
Some specific sequence types (ST) of P. aeruginosa have been commonly found associated with certain antibiotic resistance, virulence, or infective characteristics [10]. In previous epidemiological CF Spanish studies, the genetic background of obtained P. aeruginosa isolates showed high genetic variability, and ST395 and ST274 were identified as endemic clones [2]. Moreover, whole genome sequence analyses have revealed the emergence of CF-adapted epidemic clones, which may result from a limited number of specific mutations with pleiotropic effects [11].
The international epidemic high-risk clone ST274 and its clonal complex (CC274) are mostly associated with hospital settings and appear to colonize CF patients worldwide [10,12,13,14,15,16,17,18,19]. Thus, the majority of studies focus on characterizing globally disseminated clones in clinical settings, but few on non-clinical strains [15,20,21]. The aim of this work was to analyze the biological and genomic characteristics of ST274-P. aeruginosa strains from different origins, to set the path in understanding the relevant mechanisms for its success in the CF setting.
2. Results
2.1. Resistance Phenotype
Nine of 11 ST274-P. aeruginosa strains were susceptible to all anti-pseudomonal agents tested (classified as multiS in this work). One non-clinical strain, recovered from a fecal sample of a healthy volunteer, showed intermediate resistance to ceftazidime and imipenem and was classified as modR. The remaining clinical strain showed non-susceptible phenotypes to piperacillin–tazobactam, ceftazidime, cefepime, imipenem, meropenem, doripenem, gentamicin, ciprofloxacin, levofloxacin, and aztreonam, and it was classified as extensively drug-resistant (XDR) and difficult-to-treat resistant (DTR) strain (Table 1 and Supplementary Materials Table S1). All strains were susceptible to colistin, cefiderocol, ceftazidime–avibactam, and ceftolozane–tazobactam. No isolate showed class A carbapenemase, extended-spectrum beta-lactamase (ESBL), or metallo-beta-lactamase (MBL) phenotypes. AmpC hyperproduction was observed in two strains (Table 1 and Supplementary Materials Table S1).

Table 1.
Characteristics of the 11 P. aeruginosa ST274 from different origins selected for this study.
2.2. Serotyping and Pulsed-Field Gel Electrophoresis (PFGE)
Table 1 presents the serotypes determined by agglutination, with the O:3 serotype found in all strains except four non-typeable strains.
Nine PFGE patterns were observed among the 11 ST274-P. aeruginosa strains studied (Figure 1 and Table 1). An indistinguishable pattern was found between two strains from river water samples that were closely related (more than 90% of identity) to the XDR-clinical strain. No association was detected by origin or resistance phenotype.
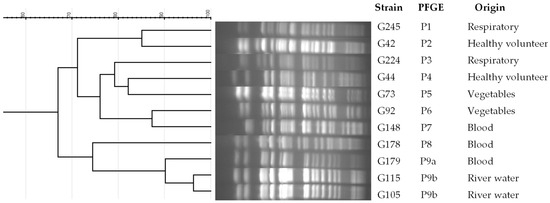
Figure 1.
Dendrogram of PFGE patterns in ST274-P. aeruginosa strains from different origins. PFGE patterns were analyzed by GelJ v2 program, using the Dice coefficient.
2.3. Detection of Virulence and Quorum-Sensing Genes
The definition of virulotypes was based on the detection of the 14 genes involved in virulence or quorum-sensing (exoS, exoU, exlA, exoY, exoT, exoA, lasA, lasB, aprA, rhlAB, rhlI, rhlR, lasI, and lasR genes). Two virulotypes were detected (Table 1). All strains harbored exoS, exoY, exoT, exoA, lasA, lasB, aprA, rhlAB, rhlI, rhlR, lasI, and lasR genes, whereas the strain G245 had the lasR gene truncated by the ISPsp7 element, an IS30-family insertion sequence. The exoU and exlA genes were absent in all of the strains.
2.4. Biofilm Formation
The production of biofilm biomass (crystal violet (CV) method) and the quantification of bacterial metabolic activity (fluorescein diacetate (FDA) method) were measured in comparison to the control strain PAO1, and the obtained results are shown in Figure 2a and Supplementary Table S1. Six strains (55%, three clinical and three non-clinical) showed less biomass production than PAO1, and eight strains (73%) had less bacterial metabolic activity than PAO1. The XDR strain showed low biofilm production (47.92% in CV and 96.41% in FDA), whereas the lasR-truncated strain had very high biofilm production (787% in CV and 753% in FDA).
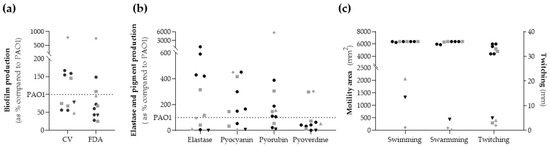
Figure 2.
Phenotypic assays of biological parameters. (a) Biofilm production: biofilm biomass (CV) and metabolic activity inside the biofilm (FDA). (b) Elastase and pigments production. (c) Motility activity: swimming and swarming in mm2 (left axis) and twitching in mm (right axis). The dotted line represents reference strain P. aeruginosa PAO1 value = 100%. Black symbol (dots and triangle) represents non-clinical strains, and grey symbols (squares, triangle and rhombus) represent clinical strains. The grey triangle corresponds to the XDR strain, the black triangle to the modR strain, and the grey rhombus to the lasR-truncated strain.
2.5. Elastase Activity and Pigment Production
Results obtained for elastase and pigment production are summarized in Figure 2b and Supplementary Table S1. The elastase production was higher than PAO1 in six strains (55%). The non-clinical P. aeruginosa strains produced more than 400%, except for both strains from healthy volunteers whose productions were lower than 7%. Additionally, the XDR strain had a low elastase production compared to PAO1 (11%).
Seven (64%) and eight (73%) strains showed higher pyocyanin and pyorubin production than PAO1, respectively. However, only two strains (G178 and G245) had a higher pyoverdine production than PAO1 (18%). The XDR strain exhibited a low production of pyocyanin pigment (8%), and the modR strain showed a very low production for all pigments. On the other hand, the lasR-truncated strain showed exceptionally high pyorubin (5714.29%), pyocyanin (450%), and pyoverdine (302%) production.
2.6. Motility
High swimming and swarming (area ranges 5892–6400 mm2) and twitching motility (diameter ranges 31–35 mm) were found in all ST274-P. aeruginosa strains except for three. These three strains were modR, XDR, and lasR-truncated ones that showed low motility values (area ≤ 2500 mm2 ranged from 34 to 2100 mm2, and twitching diameter ≤ 5 mm). G178 strain showed high swimming and swarming values (6400 mm2) but low twitching motility (3 mm) (Figure 2c and Supplementary Materials Table S1).
2.7. Genome Properties of ST274-P. aeruginosa Strains from Different Origins
The genome properties of the 11 studied ST274-P. aeruginosa strains and their assembly parameters are detailed in Table 2. Genome sizes ranged from 6,241,882 to 6,805,979 bp with 66.04–66.54% GC content. The total number of contigs ranged between 46 and 109, and the number of genes oscillated from 5734 to 6267.

Table 2.
General features of the genomes of the 11 studied ST274-P. aeruginosa strains.
2.7.1. Phylogenetic Analysis
Phylogenetic analysis was performed with a total of 27 P. aeruginosa strains, comparing our 11 studied ST274-P. aeruginosa strains with 14 reference ST274-P. aeruginosa genomes downloaded from the NCBI database, and the genomes of control strains P. aeruginosa PAO1 and PA14 (Supplementary Materials Table S2). The core genome and the SNP distance depended on the addition of PA14 and PAO1 genomes. Thus, the core genome of these 27 P. aeruginosa strains included 4,538 CDS with 90% homology and 90% coverage. By analyzing the SNPs, a range of 1 to 29,065 SNPs was detected among the 27 sequences; within the range, there were 1 to 4204 SNPs among all 25 ST274-P. aeruginosa sequences and from 1 to 3665 among the 11 ST274-P. aeruginosa sequences in this work (Supplementary Materials Table S3).
The core genome alignment was used to generate a phylogenetic tree (Figure 3), which showed no differences among strains from different origins or geographical areas. This representation can determine that ST274 is phylogenetically closer to PAO1 than to PA14 control strain. It is also remarkable to observe that the three PFGE-related strains (G105, G115, and G179) are clustered in the same clade, which is close to G178 and the reference env193 (isolated from the horse water trough, USA), AZPAE14971 (intra-abdominal tract infection, China), and AUS603 (CF, Australia) genomes.
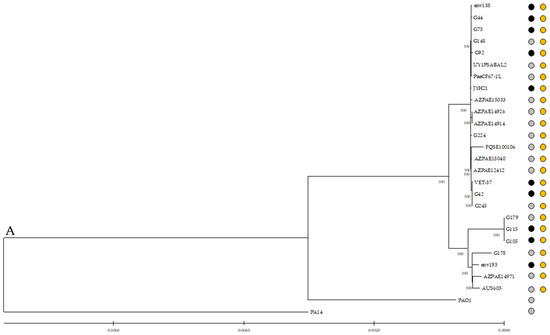
Figure 3.
Core genome phylogenetic reconstructions of P. aeruginosa. Genetic relationship between all 25 ST274-P. aeruginosa analyzed and control strains PAO1 and PA14. The phylogenetic tree was constructed using IQ-Tree 1.6.12. Orange dots represent the ST274 strains, and grey and black dots represent clinical and non-clinical strains, respectively. Bootstrap values, in percent, are shown on the internal nodes.
Pangenome was determined using Roary in all 25 ST274-P. aeruginosa strains and the control strains PAO1 and PA14. A total of 158,151 genes were analyzed, detecting between 5132 to 5296 core genes and 2594 unique genes. These unique genes represented 1.64% of the total number of genes analyzed and ranged from 0 (G105, G179, and env193) to 533 (FQSE100106) (Supplementary Materials Table S4).
There were 60 ORFs detected in all 25 ST274-P. aeruginosa strains, but not in control strains PAO1 and PA14, and were classified into 17 COG categories according to the EggNOGmapper program (Supplementary Materials Table S4).
2.7.2. Resistome
The presence of antimicrobial-acquired resistance genes and the mutational resistome was analyzed in the 11 ST274-P. aeruginosa strains from this work and the 14 reference ST274-P. aeruginosa genomes.
The blaPDC-24 and blaOXA-486 genes (β-lactam resistance) were detected in all isolates, except AZPAE15040 which harbored the blaPDC-69 variant. The aph(3′)-IIb (aminoglycoside resistance), catB7 (phenicol resistance), and fosA (fosfomycin resistance) genes were detected in all strains, excluding AZPAE14971 strain which lacked the aph(3′)-IIb gene. Two reference strains (AZPAE14926 and AZPAE14914) had additionally aph(3″)-Ib, aph(6)-Id, sul1, sul2, blaAER-1, and floR_1 genes (Supplementary Materials Table S5).
To determine the mutational resistome, the presence of non-synonymous substitutions was analyzed among a dataset of 170 PAO1 genes involved in antimicrobial resistance (Supplementary Materials Table S6). Supplementary Materials Table S7 presents the aminoacidic changes detected in each strain. The mutational resistome pattern was very similar between the ST274 strains (from this work and the reference strains) (Figure 4). The wild-type PAO1 sequence was detected in 69 genes (41%) of all studied ST274-P. aeruginosa; however, at least one missense mutation was observed in 57 genes (34%), 13 of which presented three or more non-synonymous substitutions. The highest number of aminoacidic changes was found, independently of the resistance phenotype of the strains, in PA1797, OprD, MexX, PdxB, and MexQ proteins.
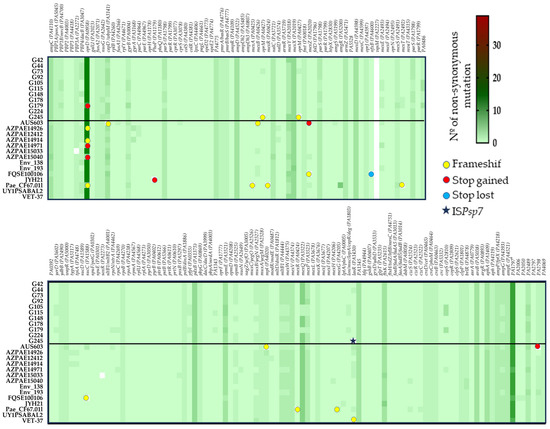
Figure 4.
Heatmap generated with GraphPad Prism 8 represents the mutational resistome (non-synonymous mutations in 170 genes involved in antimicrobial resistance) among the 11 studied ST274-P. aeruginosa strains and the 14 reference ST274-P. aeruginosa genomes. The color scale represents the number of mutational events with respect to the PAO1 genome, except mexT which was compared to PA14.
The results highlight the absence of vqsM gene and the presence of the same change/s in 26 genes (15.3%) of all ST274-P. aeruginosa strains, antimicrobial susceptible and resistant ones, that include substitutions in proteins such as the two-component sensor-regulator systems PmrAB (substitutions L71R and Y345H, respectively) and CreBD (E128G and A397V, respectively); the beta-lactamase expression regulator AmpD (R11L, G148A, D183Y); the penicillin-binding protein 1A (A615_D616insP); or the elongation factor G FusA2 (G695A) (Figure 4 and Supplementary Materials Table S7).
The XDR strain (G179) showed different missense mutations in comparison to our remaining 10 multiS and modR ST274-P. aeruginosa strains. The differences were detected in 8 of the 167 studied proteins: OprD porin (9 substitutions and a premature stop codon at position W277) implicated in carbapenem resistance; DacB/PBP4 (deletion of 12 amino acids, N454_L465del) involved in beta-lactam resistance; GyrB (S466F) related to quinolone resistance; CapD (T27S; S51G; T507A) involved in O-antigen biosynthesis and previously related with aminoglycoside resistance; MexT (T157S) and MexS (D249N; G275S) multidrug efflux pump MexEF-OprN regulators; RpoC (E386K) a DNA-directed RNA polymerase beta chain; and RpoN (insertion of 4 amino acids, I180insSLEE) the RNA polymerase sigma-54 factor that regulates many virulence genes and is linked to antibiotic resistance (Supplementary Materials Table S7). In addition, the same changes in 16 proteins were only detected in the XDR strain and its two closely related strains (G105 and G115) (Figure 4), with the exception of those substitutions in MexE, MltB1, MexW, AmpG, PA2489, and PA4069 proteins that were also detected in G178 and the reference env193, AZPAE14971, and AUS603 genomes.
The lasR-truncated strain (P. aeruginosa G245) showed unique amino acid changes in NuoG (T484A, S527N) and OprM (T173fs).
Concerning the mutome panel (Supplementary Materials Table S8), the wild type of mutS and mutL genes was detected in all 25 ST274 studied. On the other hand, the amino acidic change D61N in MutY protein was detected in 19 strains (76%), the E236D in MutT of 18 strains (72%), and D876E in PolA of all but VET-37 strain (96%).
According to the PlasmidFinder database, none of the strains harbored plasmids.
2.7.3. Detection of Virulence and Quorum-Sensing Genes
The serotype O:3 was detected in all ST274-P. aeruginosa strains using in silico serotyping with PAst 1.0.
The presence and alterations of a dataset of 247 virulence genes of PAO1, exoU gene of PA14, and exlA and exlB genes of PA7 (Supplementary Table S9) were analyzed in the 11 ST274-P. aeruginosa strains and the 14 reference ST274-P. aeruginosa genomes. Figure 5 shows the comparison of the virulence genes between PAO1 and our 11 ST274-P. aeruginosa strains. Supplementary Table S10 summarizes the complete results obtained with the 25 ST274-P. aeruginosa genomes.
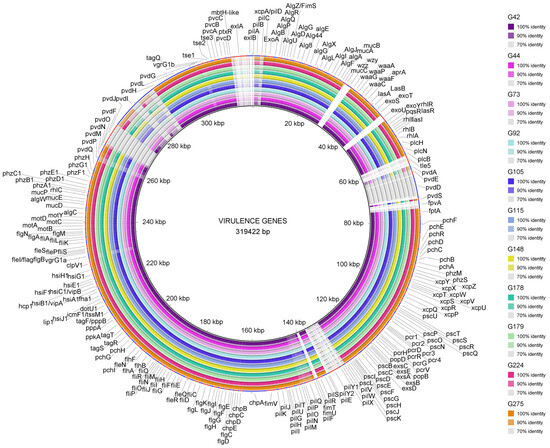
Figure 5.
Virulence genes detected among the 11 studied ST274-P. aeruginosa strains. The BRIG Tool was used to represent the genes and their identity, using the reference P. aeruginosa PAO1 genome (except for exlA and exlB genes that were compared to PA7 and exoU to PA14 genomes). Figure legend includes the name of the studied P. aeruginosa strains and the color according to the percentage of identity detected.
No changes were observed in 91 out of the 250 (36.4%) genes analyzed. On the other hand, all 25 ST274 strains shared the following characteristics in comparison with PAO1 genes: 16 genes were absent (pilA, pilE, pilV, pilW, pilX, pilY1, pilY2, fimT, fimU, pldA/tleS, fpvA, wzz, wzy, exlA, exlB, and exoU), and 13 genes, mostly involved in pyoverdine and type IV pili biosynthesis (pvdL, pvdE, pvdD, pvdJ, pvdI, pvdF, pvdM, pvdP, pvdA, pchI, pilP, pilN, and pilC), showed more than 10 equal missense changes. These genes were studied in depth as explained below.
The same mutational patterns were detected in 68 proteins (27%) of all ST274 strains, independently of their clinical/non-clinical or geographical origins. On the other hand, PscP (translocation protein in type III secretion) and LasR (quorum-sensing regulator) showed the highest number of different mutational patterns (10 and 9, respectively) among the 25 ST274-P. aeruginosa. Additionally, premature stop codons, deletions, or insertions were detected in six genes (pilC, pchF, pvdM, pcrH, pscK, and toxA) in all strains. This highlights the results observed in ToxA, the exotoxin A precursor, where two mutational patterns including deletions were found: (i) T4I, F22S, G386_A388del, I432V, D500A, detected in ST274-P. aeruginosa G105, G115, G179, and G178 and the reference env193, AZPAE14971 and AUS603 strains, and (ii) T4I, F22S, A58_T62del, and I432V observed in the remaining 18 strains.
The XDR strain (P. aeruginosa G179) showed unique amino acid changes in PilN (V30L, A31V, A33G, D40G, F43L, T44N, A45T, N49H, N51G, K75R, and a premature stop codon at position Q80) and in RhlR (Y72C) and shared the same missense mutational patterns in 23 genes only with P. aeruginosa G105, G115, G178, env193, AZPAE14971, and AUS603 strains.
The lasR-truncated strain (P. aeruginosa G245) had unique mutational patterns in FleQ, PchH, PchE (D6fs), and PvdS proteins. The ModR strain (P. aeruginosa G44) only showed the amino acid change C79I in LasR in comparison with the remaining ST274-P. aeruginosa strains.
All 40 Type III Secretion System (T3SS)-related genes and their translocated effectors were detected in all ST274 strains, with the exception of exoU gene. The genes involved in flagella, phenazines, and alginate were mostly highly conserved and homologous among all ST274 strains; however, type IV pili (T4P), pyochelin, and pyoverdine biosynthesis genes were more variable. Analyzing the 23 T4P biosynthesis genes (Supplementary Materials Table S9), there were 9 absent genes (pilA and the fimU/T/pilV/W/X/Y1/Y2/E gene cluster), and at least one missense mutation was observed in 11 genes of all ST274 strains, showing a high polymorphism in proteins, such as PilC, PilN, PilO, or PilP (from 8 to 50 modifications) (Supplementary Materials Table S10). The major subunit of T4P is a protein encoded by the pilA gene. This gene has five pili alleles that are found at a conserved chromosomal locus between the adjacent pilB and tRNA-Thr genes [23]. By analyzing the pilB/tRNA-Thr region in all ST274 strains against GenBank sequences using BLAST, we found that pilA belongs to T4P group V (pilAV), as well as the fimU/T/pilV/W/X/Y1/Y2/E gene cluster. For that reason, it was not possible to detect it when compared to PAO1-pilA which belongs to T4P group II (pilAII) (parameters used included >90% similarity).
Pyoverdine biosynthesis in P. aeruginosa is a complex process involving at least 16 different proteins (Supplementary Materials Table S9). In all ST274 strains, pvdA, pvdP, pvdD, pvdE, pvdJ, and pvdI were highly divergent genes compared with PAO1 ones, and fpvA gene (ferripyoverdine receptor) was absent. When the region between treA and pvdQ was compared to GenBank genomes using BLAST, an identity higher than 80% was detected between our ST274 strains and multiple sequences, including the reference PAO1 (class I), ATCC27853 (class II), and the ATCC BAA-2108 (class III) strains. Figure 6 shows the alignment of the pyoverdine region of an ST274 strain in comparison with class I, class II, and class III pyoverdine-producing strains. All ST274-P. aeruginosa presented a pyoverdine locus class II.
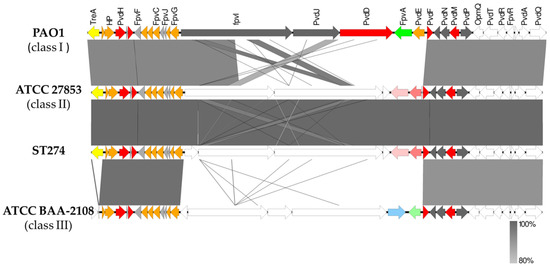
Figure 6.
Schematic representation of the genomic differences found in the pyoverdine region between P. aeruginosa PAO1 (class I), ATCC27853 (class II), ATCC BAA-2108 (class III), and ST274 P. aeruginosa strains. Colors represent the localization of genes according to the Pseudomonas webpage (www.pseudomonas.com (accessed on April 2023)): orange, cytoplasmic membrane proteins; red, cytoplasmic proteins; green, outer membrane proteins; grey, proteins of unknown function. Furthermore, the yellow arrow indicates treA gene (absent in class III); light pink represents another iron receptor different to FpvA, and dark pink represents a cyclic peptide export ABC transporter; blue, light green arrows indicate two different proteins located in that region in class III pyoverdine region and white arrows represent hypothetical proteins (unknown function). Easyfig v2.2.5 and BLAST v2.10.0 were used to build the figure.
3. Discussion
Different studies have revealed a high genetic diversity among P. aeruginosa isolates infecting CF patients. However, ST274, defined as an endemic clone in Spain and a CF epidemic clone worldwide, has been detected infecting multiple CF patients from Europe, America, Asia, and Australia [2,10,12,13,14,15,16,18,19]. Most of the reported studies characterized the ST274-P. aeruginosa isolated from CF patients or associated with hospital settings worldwide, although the majority of chronic CF lung infections are thought to be the result of colonization by P. aeruginosa from environmental sources. The investigation on the ST274-P. aeruginosa behavior, including origin, antibiotic resistance, virulence, pathogenicity, molecular typing, or environmental/infection adaptability, is necessary to know the molecular markers and biological characteristics that could explain the clonal success of the ST274 clone. In addition, the assessment of the genetic and biological markers underlying clonal success must be performed in a range of not only both resistant and susceptible strains but also both clinical and non-clinical strains.
In the current study, five clinical and six non-clinical ST274-P. aeruginosa strains have been deeply analyzed, and their whole genome sequences were compared with others included in databases. Regarding antimicrobial resistance, 9 out of 11 ST274-P. aeruginosa strains were multiS, one strain was modR, and the remaining strain was XDR and DTR. High-risk clones ST235, ST175, and ST111 are usually associated with MDR/XDR clinical strains; however, as our results show, in agreement with previous studies reported [21,24], these resistance phenotypes are not so frequent among non-clinical or ST274 P. aeruginosa strains. The unique XDR and DTR strain (G179) in our work was isolated from a blood sample, showed hyperproduction of AmpC, and possessed a different mutational resistome in comparison to the remaining 10 ST274-P. aeruginosa strains. G179 resistome showed missense mutations affecting porins and membrane proteins, multidrug efflux pumps, O-antigen biosynthesis, quinolone targets, and regulators (i.e., OprD, DacB/PBP4, GyrB, CapD, MexT, MexS, RpoC, and RpoN proteins), which likely explain the observed resistance pattern. The mutational inactivation of oprD is the most frequent cause of carbapenem resistance in P. aeruginosa, and the inactivation of dacB, which correlated with the AmpC overexpression, causes the β-lactam resistance [25,26,27]. RpoN regulates nitrogen assimilation, quorum sensing, motility, biofilm formation, and other virulence factors, but its regulation was also linked to P. aeruginosa tolerance to several antibiotics [28,29]. The insertion of four amino acids in the RpoN of the G179 strain probably induced a partial or total loss of RpoN function which is a relatively common mechanism of pathoadaptation to the CF lungs [30,31]. This strain was also classified as DTR, referring to resistance to all first-line antibiotics that include β-lactams and fluoroquinolones, and associated with higher mortality [32,33]. According to international guidelines [33], ceftolozane–tazobactam, ceftazidime–avibactam, or imipenem–relebactam are recommended for treating DTR P. aeruginosa, and our DTR ST274-P. aeruginosa strain was susceptible to all of them.
During an infection, P. aeruginosa has to adapt to a new scenario that includes the host immune response, antibiotics, and a different environment and substrate composition. There are multiple references analyzing the P. aeruginosa adaptive processes, and it is commonly observed that the gradual downregulation or loss of the function of many genes occurs, usually associated with bacterial quorum-sensing and pathogenicity [5,7,31,34,35]. Persistent colonization and chronic infection are eventually associated with resistance development, loss of O-antigen, biofilm production, and loss of motility and virulence factors. Among our analyzed 11 ST274-P. aeruginosa strains, the XDR, modR, and lasR-truncated strains showed a chronic infection phenotype characterized by loss of serotype-specific antigenicity and low motility. The remaining eight ST274-P. aeruginosa strains were fully motile in agreement with a general wild-type phenotype. Furthermore, the XDR and modR strains presented low production of pigments and reduced biofilm formation and values that were very high in the lasR-truncated strain. The lasR gene is involucrate in the control of quorum-sensing systems and is generally considered at the top of the regulatory hierarchy [36,37]. LasR showed the C79Y change in the modR strain recovered from a fecal sample of a healthy volunteer; whereas in the lasR-truncated strain, isolated from a bronchial aspirate of a patient with chronic bronchitis, the lasR was truncated by the ISPsp7 element. P. aeruginosa is frequently observed to gain mutations in lasR gene during chronic infections, including chronic CF lung infections [31,38,39], but a high prevalence of lasR-defective P. aeruginosa has been detected from not only clinical samples but also non-clinical origins [40,41,42]. This fact could suggest that the loss of lasR function gives a selective pathogenic advantage and minimizes fitness cost at a bacterial population level even outside the clinical contexts. Furthermore, there are multiple evolution studies analyzing the in vitro and in vivo effect of lasR mutants, and they have been detected that even the nutritional environment is sufficient to select those mutants, and not all of them involve a LasR-defective phenotype [39,41,43,44]. Thus, lasR mutants are neither atypical nor restricted to clinical isolates, and additionally, the lasR-loss could be offset by other interactive quorum-sensing systems and/or global regulators, such as MvaT, RpoN, and RpoS.
The vqsM and pldA/tleS genes were not detected among the 25 studied ST274 sequences. VqsM is an AraC family transcriptional regulator which activates the las quorum-sensing system and T3SS gene expression [45]. pldA/tleS gene codifies a phospholipase D1 involved in bacterial pathogenesis and persistence [46]. However, it has been reported that not all strains of P. aeruginosa carry vqsM or pldA genes; therefore, their relative infective contributions appear to be strain-specific [45,46].
According to core genome analysis, the phylogenetic tree showed all 25 ST274-P. aeruginosa interspersed and grouped in two phylogenetic clades independently of their antimicrobial resistance or origin, supporting the idea of the absence of a geographical or origin barrier for lineage evolution. P. aeruginosa G105, G115, G178, G179, env193, AZPAE14971, and AUS603 strains, which were recovered from clinical and non-clinical samples, shared the wild-type sequences of mutS, mutL, and mutT mutator genes, and the same missense mutational patterns in 7 genes of the resistome (i.e., fosA, mexE, muxB, mltB1, mexW, ampG, and PA2489) and 23 genes of the virulome (e.g., toxA, rhlC, lasI, or popN).
Interclonal sequence variation is low in the P. aeruginosa core genome [47]. Indeed, the genome clusters with the highest level of sequence diversity in P. aeruginosa are the pyoverdine locus, the flagellar regulon, T4P pilA, the T3SS effector proteins, and the O-antigen biosynthesis locus. Each cluster is present in all strains, but their involved genes are highly divergent between strains. This phenomenon could be a result of “diversifying selection”, a type of selection that maintains multiple alleles in the population [47]. In this line, and according to the results obtained in our work, the genes involved in flagella were mostly highly conserved and homologous among all ST274 strains; however, T4P and siderophores (pyoverdine and pyochelin) biosynthesis genes were highly divergent compared with PAO1 ones. T4P is associated with a number of biological activities in bacteria, such as twitching motility, bacteriophage sensitivity, attachment to biotic and abiotic surfaces, biofilm development, and DNA uptake [23]. In P. aeruginosa, T4P is composed of a single-type IVa pilin (T4aP) protein encoded by the pilA gene, for which five distinct pilin alleles (Groups I–V) have been previously reported in P. aeruginosa [23,48]. PAO1 strain possesses group II pilin, PA14 harbors group III, and all ST274 strains of our study presented a group V. Group III and V pilins have an unusual architecture and diverged from a common ancestor [23,49]. The “minor” pilins are a set of low-abundance pilin-like proteins encoded by the fimU/T/pilV/W/X/Y1/Y2/E gene cluster. In all ST274 strains, they were not detected compared with the PAO1 sequence, as observed in other works [50]. However, it has been also described that both groups III and V have identical minor pilin genes [51], which have less than 75% similarity with the group II operon [52]. In addition, the tandem tRNA genes located between the major and minor pilin operons appear to provide hot spots for genetic insertion. This information together with the fact that some P. aeruginosa pilins are more closely related to pilins of distinct bacterial species (e.g., Dichelobacter nodosus, Eikenella corrodens, Ralstonia solanacearum, and Xanthomonas campestris) strongly suggest that pilin gene diversity was generated through horizontal genetic transfer [53,54]. Group I and II pilins are the most frequently detected in P. aeruginosa strains [23,52,55], and among the scarce descriptions of isolates harboring a group V pilin, no specific differences were reported [23,55]. Further studies could determine the possible relationship between these groups of pilins, motility, and strain-specific characteristics.
In general, every P. aeruginosa isolate is able to produce one of three types of pyoverdine [56,57], and all 25 analyzed ST274-P. aeruginosa presented a pyoverdine class II. The TonB-dependent ferripyoverdine receptor encoding gene, fpvAII, is chromosomally located among the most divergent genes in the region and is not a recombination result between types. Outer membrane protein genes such as the pyoverdine receptor fpvA are common targets for entry by phage or pyocins. Pyocins are bacteriocins produced by P. aeruginosa that kill strains of the same species. This is the case for pyocin S3 which uses FpvAII as a receptor, while the pore-forming pyocin S5 utilizes the FptA ferripyochelin receptor to gain entry to the cells and kill them [58,59]. Pyocins could indeed play a particularly important role during co-colonization of the CF lung by P. aeruginosa isolates that possess different pyocin/immunity genes and favor the outcompetition of a particular P. aeruginosa clone. Thus, siderophore diversity may be also a resistance mechanism.
In conclusion, we gained insights into the ST274-P. aeruginosa isolates population structure using a combination of phenotyping and genotyping techniques, and certain associations have been primarily detected, such as the presence of blaOXA-486 and blaPDC-24 genes, O-antigen serotype O:3, exoS+/exoU− T3SS genotype, Group V of T4P, and pyoverdine locus class II. Other general molecular markers or features have also been observed linked to the analyzed ST274-P. aeruginosa sequences, highlighting the absence of vqsM and pldA/tleS genes and the presence of the same mutational pattern in genes involved in, for example, two-component sensor-regulator systems PmrAB (substitutions L71R and Y345H, respectively) and CreBD (E128G and A397V, respectively); the exotoxin A (A58 or G386 deletions in ToxA); the quorum-sensing RhlI (S62G, D83E); the beta-lactamase expression regulator AmpD (R11L, G148A, D183Y); the penicillin-binding protein 1A (A615_D616insP); or the elongation factor G FusA2 (G695A).
Whole genome sequence analyses provide detailed genome patterns that might be essential to characterize ST274-P. aeruginosa strains, although it is difficult and challenging to correlate genotypic with phenotypic variations. Sequencing alone cannot always predict phenotypic differences, due to the multifactorial nature of P. aeruginosa. The reduced number of strains and the intra-clonal variability of the parameters analyzed must be considered among the limitations of this study. This variability could be related to the activation of specific regulatory pathways, horizontal gain or loss of genetic material, or undetected punctual mutations that affect the final expression levels of key genes.
Further work is needed to improve our understanding of the basis for the global success of this and other high-risk clones. A more in-depth understanding of Pseudomonas virulence regulation, as well as the use of P. aeruginosa transcriptomics, would identify hundreds of genes specifically expressed in vitro or during adaptation to different environments. However, this study could serve as the basis for more specific studies that will prove helpful in designing novel antimicrobial approaches, better antibiotic stewardship, and new therapeutic developments.
4. Materials and Methods
4.1. Bacterial Strains
Eleven P. aeruginosa strains belonging to clone ST274 were selected from the Pseudomonas collection of the Molecular Microbiology Area (Centre for Biomedical Research in La Rioja, CIBIR, Logroño, Spain). These strains were obtained from clinical samples of non-CF patients (number of strains), respiratory (2) and blood (3), and from non-clinical samples, fecal samples of healthy volunteers (2), vegetables (2), and river water (2) (Table 1 and Supplementary Materials Table S1). P. aeruginosa PAO1, PA7, and PA14 were included as control strains in different assays.
4.2. Antimicrobial Susceptibility Testing
Susceptibility testing to 15 antipseudomonal agents was performed by MicroScan WalkAway® microdilution system (MicroScan; Beckman Coulter, Inc., Brea, CA, USA) according to CLSI [22], except cefiderocol, ceftazidime–avibactam, and ceftolozane–tazobactam that were determined by disk diffusion method [22]. Colistin resistance was also screened by colistin broth disk elution [22]. The antimicrobial categories and antipseudomonal agents tested were the following ones: aminoglycosides (gentamicin, tobramycin,), carbapenems (imipenem, meropenem, doripenem), cephalosporins (ceftazidime, cefepime, cefiderocol), fluoroquinolones (ciprofloxacin, levofloxacin), penicillin–β-lactamase inhibitor combinations (piperacillin–tazobactam, ceftazidime–avibactam, ceftolozane–tazobactam), monobactams (aztreonam), and polymyxins (colistin).
Strains were categorized as multiS, modR, MDR (multidrug-resistant), and XDR according to previously published classifications [60,61]. The strains that are non-susceptible to all of the following antibiotics, piperacillin–tazobactam, ceftazidime, cefepime, aztreonam, meropenem, imipenem, ciprofloxacin, and levofloxacin, were considered difficult-to-treat resistant (DTR) [32].
ESBL, MBL, and class A carbapenemase phenotypes were determined by double-disc synergy tests [26]. AmpC hyperproduction was determined by the phenotypic method [62].
4.3. Serotyping
Serotype identification was performed by agglutination using commercially monovalent antisera specific for 16 different P. aeruginosa O-serotypes (Bio-Rad, Temse, Belgium).
4.4. Detection of Virulence and Quorum-Sensing Genes
The presence of exoS, exoU, exoY, exoT, exoA, exlA, lasA, lasB, aprA, rhlAB, rhlI, rhlR, lasI, and lasR genes was studied by PCR as described previously [20].
4.5. Clonal Relationship
PFGE was carried out using the SpeI enzyme to digest genomic DNA [63]. PFGE patterns were analyzed by the Java program GelJ v2 using the Dice coefficient [64].
4.6. Biofilm Formation
Biofilm assays were performed by crystal violet (CV) staining to quantify the total biofilm biomass and by fluorescein diacetate (FDA) assay to determine the bacterial metabolic activity inside the biofilm structure (viable cells within the biofilm). Both methods were performed in 96-well microtiter plates using an initial inoculum of 106 cfu/mL and measured after 24 h of incubation, as previously published [20]. Absorbance measures were performed using a POLARstar Omega microplate reader (BMG Labtech, Ortenberg, Germany). All experiments were performed in triplicate, including P. aeruginosa PAO1 as the control strain. GraphPad Prism (v.8.4.3) was used to build the graphics.
4.7. Elastase and Pigment Production
Bacterial strains were grown at 37 °C in Luria-Bertani (LB) broth overnight with shaking. After centrifugation, elastase activity was determined by the Elastin-Congo-Red assay [7], and the chloroform extract method was used to quantify pyocyanin, pyoverdine, and pyorubin pigments [7]. Absorbance/fluorescence measures were performed using a POLARstar Omega microplate reader (BMG Labtech, Ortenberg, Germany). All experiments were performed in triplicate, including P. aeruginosa PAO1 as the control strain. GraphPad Prism (v.8.4.3) was used to build the graphics.
4.8. Motility
Swarming and swimming motilities were determined by placing 4 µL of bacterial suspension (1 × 109 cells in LB broth) on the middle of 0.5% (swarming) and 0.3% (swimming) LB agar plates (entire plate area was 6400 mm2) [20]. Plates were photographed with Chemi Doc system (Bio-Rad, Hercules, CA, USA), after incubation at 37 °C overnight. The images were processed with MotilityJ software (v1.0) [65].
A twitching motility assay was performed starting with overnight-grown bacteria on LB agar plates. Bacteria were stab inoculated with a sterile needle loop to the bottom of the plastic agar interface of LB broth solidified with 1% agar plates. After incubation at 37 °C for 48 h, the agar medium was removed. The twitching diameter was measured after staining with 0.05% (wt/vol) Coomassie brilliant blue (40% methanol, 10% acetic acid) for 50 min. Bacteria were classified as non-motile (diameter ≤ 5 mm), motile (diameter from >5 mm to ≤30 mm), or highly motile (diameter > 30 mm) [66,67].
All assays were performed in triplicate. GraphPad Prism (v.8.4.3) was used to build the graphics.
4.9. Whole Genome Sequencing (WGS)
Genomic DNA from the 11 ST274-P. aeruginosa strains of this study was extracted using Wizard ® Genomic DNA Purification Kit (Promega, Madison, WI, USA). Quantity and quality were assessed using a Qubit fluorimeter (Thermo Fisher Scientific, Waltham, MA, USA). Libraries were prepared using the TruSeq DNA PCR Free protocol (Illumina, San Diego, CA, USA). Then, the final quality of the libraries was assessed with a Fragment Analyzer (Std. Sens. NGS Fragment Analysis kit 1–6000 bp, AATI) and was quantified by qPCR at the Genomics and Bioinformatics Core Facility (CIBIR). Subsequent sequencing was carried out in an Illumina HiSeq 1500 (Illumina, San Diego, CA, USA).
FastQC v0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to analyze the quality of raw reads, which were subsequently trimmed and filtered by using Trim Galore v0.4.5 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Genomes were reconstructed by using PLACNETw [68]. Identification of open reading frames (ORFs) and genome annotation of the assembled genetic elements was performed by using PROKKA v1.13 [69].
Additionally, the genomes of 14 ST274-P. aeruginosa strains from different origins were downloaded from the NCBI database (January 2020) to be used as reference genomes in the different comparative analyses (Supplementary Materials Table S2). The core genome was determined as the collection of genes present in the 11 ST274-P. aeruginosa strains of this study but also in the 14 reference ST274-P. aeruginosa genomes and the control strains P. aeruginosa PAO1 and PA14. The parameters used included >90% similarity and >90% coverage, as defined by Lanza et al. (2014) [70]. A phylogenetic tree was constructed with this data using IQ-Tree 1.6.12 [71] and iTol V5.6.3 [72]. Roary v3.11.2 was used to determine and analyze the pangenome, identifying common and unique genes [73], and eggNOG-mapper v2 was used to determine the cluster of orthologous groups (COGs) and functional annotations [74].
The presence of acquired antibiotic resistance genes was evaluated using the following tools: ResFinder V3.2 [75], Comprehensive Antibiotic Resistance Database (CARD) [76], and Antibiotic Resistance Gene-ANNOTation (ARG-ANNOT) [77]. The mutational resistome was determined by analyzing the presence of mutations among a dataset of 170 genes involved in antimicrobial resistance downloaded from the Pseudomonas Genome Database (www.pseudomonas.com (Supplementary Materials Table S6)). For this purpose, variant calling was performed using PAO1 as a reference genome (Snippy V4.5.0) and confirmed with BLAST and CLUSTAL. The heatmap was constructed using the GraphPad Prism (v.8.4.3) program.
Additionally, the mutator phenotype was studied from whole genome sequence data through the analysis of genes from mutome panel [2].
Virulence genes were analyzed using the Virulence Factor DataBase (VFDB) [78], studying a dataset of 247 virulence genes of PAO1, plus exoU of PA14, and exlA and exlB of PA7. These genes are associated with flagella and pili biosynthesis/regulation, protease production, type II, IV, and VI secretion systems, protease IV, enzyme, quorum sensing, alginate production/regulation, and toxins were curated from the VFDB (Supplementary Materials Table S10). BLAST searches were used to match the virulence genes with the bacterial genomes, and BLAST Ring Image Generator (BRIG) [79] was used to represent the percentage identity between these genes and those absent. Moreover, we performed BLAST, Clustal Omega, and variant calling (Snippy V4.5.0) using PAO1 as a reference genome to find the aminoacidic changes in virulence proteins.
PlasmidFinder 1.3 was used to search the plasmid replicon types, and the Pseudomonas aeruginosa serotyper (PAst 1.0) was used for in silico serotyping (https://cge.cbs.dtu.dk/services/PAst/).
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/antibiotics12111561/s1, Table S1. Characteristics of the 11 studied ST274-Pseudomonas aeruginosa recovered from different origins; Table S2. Characteristics of the genomes of 14 reference ST274-P. aeruginosa strains and control strains PAO1 and PA14 downloaded from the NCBI database; Table S3. Single-nucleotide polymorphisms (number of SNPs) in the core genome among the 11 studied ST274-P. aeruginosa, the 14 reference ST274-P. aeruginosa genomes, and the two control genomes; Table S4. Pangenome determined using Roary in all 25 ST274-P. aeruginosa strains and the control strains PAO1 and PA14; Table S5. Acquired genes in ST274-P. aeruginosa with Resfinder prediction; Table S6. Dataset and function of the 170 antimicrobial resistance genes analyzed to determine the mutational resistome of the ST274-P. aeruginosa strains; Table S7. Mutational resistome of the 25 studied ST274-P. aeruginosa strains with respect to PAO1 genome; Table S8. Detection of alterations in the mutome genes (involved in the mutator phenotype) of the 11 studied ST274-P. aeruginosa strains and the 14 reference ST274-P. aeruginosa genomes, with respect to PAO1 genome; Table S9. Dataset and functions of the 250 virulence genes analyzed to determine the virulome of the ST274-P. aeruginosa strains; Table S10. Amino acid changes in virulome of the 25 studied ST274-P. aeruginosa strains with respect to the PAO1 genome. References [80,81,82,83,84] are cited in the supplementary materials.
Author Contributions
Conceptualization, G.C., M.L. and Y.S.; data curation, G.C., M.L., M.d.T., L.R.-R. and B.R.-B.; formal analysis, G.C., M.L., M.d.T. and Y.S.; funding acquisition, Y.S.; investigation, G.C., M.L., M.d.T., L.R.-R., B.R.-B. and Y.S.; methodology, G.C., M.L., M.d.T., L.R.-R., B.R.-B. and Y.S.; resources, G.C., M.d.T. and Y.S.; software, M.L. and M.d.T.; supervision, M.L. and Y.S.; validation, M.L., M.d.T., L.R.-R., B.R.-B. and Y.S.; visualization, G.C., M.L., B.R.-B. and Y.S.; writing—original draft, G.C., M.L. and Y.S.; writing—review and editing, G.C., M.L., M.d.T., L.R.-R., B.R.-B. and Y.S.. All authors have read and agreed to the published version of the manuscript.
Funding
This research work was supported by the Instituto de Salud Carlos III of Spain (ISCIII) (FIS project number PI20/00356) (co-funded by European Regional Development Fund (FEDER) “A way to make Europe”). Gabriela Chichón (G.C.) had a predoctoral fellowship from the Consejería de Industria, Innovación y Empleo, Gobierno de La Rioja, Spain.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All datasets are available. The whole genome data for the 11 ST274-P. aeruginosa strains have been deposited at NCBI using BioProject number PRJNA678143. The raw sequencing data were deposited at NCBI’s Sequence Read Archive (SRA) (SRR13053896 to SRR13053906).
Acknowledgments
Part of this study was presented at the 31st European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) (Abstract No. 1916, online, 9–12 July 2021) and the XXIV Congreso Nacional de la Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica (SEIMC) (Abstract No. 193, online, 2020).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Freschi, L.; Vincent, A.T.; Jeukens, J.; Emond-Rheault, J.G.; Kukavica-Ibrulj, I.; Dupont, M.J.; Charette, S.J.; Boyle, B.; Levesque, R.C. The Pseudomonas aeruginosa Pan-Genome Provides New Insights on Its Population Structure, Horizontal Gene Transfer, and Pathogenicity. Genome Biol. Evol. 2018, 11, 109–120. [Google Scholar] [CrossRef] [PubMed]
- López-Causapé, C.; de Dios-Caballero, J.; Cobo, M.; Escribano, A.; Asensio, Ó.; Oliver, A.; del Campo, R.; Cantón, R.; Solé, A.; Cortell, I.; et al. Antibiotic resistance and population structure of cystic fibrosis Pseudomonas aeruginosa isolates from a Spanish multi-centre study. Int. J. Antimicrob. Agents 2017, 50, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Diggle, S.P.; Whiteley, M. Microbe Profile: Pseudomonas aeruginosa: Opportunistic pathogen and lab rat. Microbiology 2020, 166, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Subedi, D.; Vijay, A.K.; Kohli, G.S.; Rice, S.A.; Willcox, M. Comparative genomics of clinical strains of Pseudomonas aeruginosa strains isolated from different geographic sites. Sci. Rep. 2018, 8, 15668. [Google Scholar] [CrossRef]
- Malhotra, S.; Hayes, D., Jr.; Wozniak, D.J. Cystic Fibrosis and Pseudomonas aeruginosa: The Host-Microbe Interface. Clin. Microbiol. Rev. 2019, 32, e00138-18. [Google Scholar] [CrossRef]
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef]
- Lozano, C.; Azcona-Gutiérrez, J.M.; Van Bambeke, F.; Sáenz, Y. Great phenotypic and genetic variation among successive chronic Pseudomonas aeruginosa from a cystic fibrosis patient. PLoS ONE 2018, 13, e0204167. [Google Scholar] [CrossRef]
- Mauch, R.M.; Jensen, P.Ø.; Moser, C.; Levy, C.E.; Høiby, N. Mechanisms of humoral immune response against Pseudomonas aeruginosa biofilm infection in cystic fibrosis. J. Cyst. Fibros. 2018, 17, 143–152. [Google Scholar] [CrossRef]
- Gabrielaite, M.; Johansen, H.K.; Molin, S.; Nielsen, F.C.; Marvig, R.L. Gene Loss and Acquisition in Lineages of Pseudomonas aeruginosa Evolving in Cystic Fibrosis Patient Airways. mBio 2020, 11, e02359-20. [Google Scholar] [CrossRef]
- Nageeb, W.; Amin, D.H.; Mohammedsaleh, Z.M.; Makharita, R.R. Novel Molecular Markers Linked to Pseudomonas aeruginosa Epidemic High-Risk Clones. Antibiotics 2021, 10, 35. [Google Scholar] [CrossRef]
- López-Causapé, C.; Rojo-Molinero, E.; Mulet, X.; Cabot, G.; Moyà, B.; Figuerola, J.; Togores, B.; Pérez, J.L.; Oliver, A. Clonal Dissemination, Emergence of Mutator Lineages and Antibiotic Resistance Evolution in Pseudomonas aeruginosa Cystic Fibrosis Chronic Lung Infection. PLoS ONE 2013, 8, e71001. [Google Scholar] [CrossRef] [PubMed]
- Ocampo-Sosa, A.A.; Fernández-Martínez, M.; Cabot, G.; Peña, C.; Tubau, F.; Oliver, A.; Martínez-Martínez, L. Draft Genome Sequence of the Quorum-Sensing and Biofilm-Producing Pseudomonas aeruginosa Strain Pae221, Belonging to the Epidemic High-Risk Clone Sequence Type 274. Genome Announc. 2015, 3, e01343-14. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Olmos, A.; García-Castillo, M.; Alba, J.M.; Morosini, M.I.; Lamas, A.; Romero, B.; Galán, J.C.; del Campo, R.; Cantón, R. Population structure and antimicrobial susceptibility of both nonpersistent and persistent Pseudomonas aeruginosa isolates recovered from cystic fibrosis patients. J. Clin. Microbiol. 2013, 51, 2761–2765. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Lara, S.; del Barrio-Tofiño, E.; López-Causapé, C.; Oliver, A.; Martínez-Martínez, L.; Bou, G.; Zamorano, L.; Sánchez-Diener, I.; Galán, F.; Gracia, I.; et al. Predicting Pseudomonas aeruginosa susceptibility phenotypes from whole genome sequence resistome analysis. Clin. Microbiol. Infect. 2021, 27, 1631–1637. [Google Scholar] [CrossRef] [PubMed]
- Kos, V.N.; Déraspe, M.; McLaughlin, R.E.; Whiteaker, J.D.; Roy, P.H.; Alm, R.A.; Corbeil, J.; Gardner, H. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob. Agents Chemother. 2015, 59, 427–436. [Google Scholar] [CrossRef]
- Bocharova, Y.A.; Savinova, T.A.; Lyamin, A.V.; Kondratenko, O.V.; Polikarpova, S.V.; Zhilina, S.V.; Fedorova, N.I.; Semykin, S.Y.; Chaplin, A.V.; Korostin, D.O.; et al. Genome features and antibiotic resistance of Pseudomonas aeruginosa strains isolated in patients with cystic fibrosis in the Russian Federation. Russ. Clin. Lab. Diagn. 2021, 66, 629–634. [Google Scholar] [CrossRef]
- Ahmed, M.A.S.; Hadi, H.A.; Abu Jarir, S.; Khan, F.A.; Arbab, M.A.; Hamid, J.M.; Alyazidi, M.A.; Al-Maslamani, M.A.; Skariah, S.; Sultan, A.A.; et al. Prevalence and microbiological and genetic characteristics of multidrug-resistant Pseudomonas aeruginosa over three years in Qatar. Antimicrob. Steward. Healthc. Epidemiol. 2022, 2, e96. [Google Scholar] [CrossRef]
- Van Mansfeld, R.; Willems, R.; Brimicombe, R.; Heijerman, H.; Van Berkhout, F.T.; Wolfs, T.; Van der Ent, C.; Bonten, M. Pseudomonas aeruginosa genotype prevalence in Dutch cystic fibrosis patients and age dependency of colonization by various P. aeruginosa sequence types. J. Clin. Microbiol. 2009, 47, 4096–4101. [Google Scholar] [CrossRef]
- Mitchelmore, P.J.; Randall, J.; Bull, M.J.; Moore, K.A.; O’Neill, P.A.; Paszkiewicz, K.; Mahenthiralingam, E.; Scotton, C.J.; Sheldon, C.D.; Withers, N.J.; et al. Molecular epidemiology of Pseudomonas aeruginosa in an unsegregated bronchiectasis cohort sharing hospital facilities with a cystic fibrosis cohort. Thorax 2017, 73, 677–679. [Google Scholar] [CrossRef]
- Ruiz-Roldán, L.; Rojo-Bezares, B.; de Toro, M.; López, M.; Toledano, P.; Lozano, C.; Chichón, G.; Alvarez-Erviti, L.; Torres, C.; Sáenz, Y. Antimicrobial resistance and virulence of Pseudomonas spp. among healthy animals: Concern about exolysin ExlA detection. Sci. Rep. 2020, 10, 11667. [Google Scholar] [CrossRef]
- Torres, R.T.; Cunha, M.V.; Ferreira, H.; Fonseca, C.; Palmeira, J.D. A high-risk carbapenem-resistant Pseudomonas aeruginosa clone detected in red deer (Cervus elaphus) from Portugal. Sci. Total Environ. 2022, 829, 154699. [Google Scholar] [CrossRef] [PubMed]
- CLSI 2022; Performance Standards for Antimicrobial Susceptibility Testing. 32th Informational Supplement; CLSI Document M100-S32; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022.
- Kus, J.V.; Tullis, E.; Cvitkovitch, D.G.; Burrows, L.L. Significant differences in type IV pilin allele distribution among Pseudomonas aeruginosa isolates from cystic fibrosis (CF) versus non-CF patients. Microbiology 2004, 150 Pt 5, 1315–1326. [Google Scholar] [CrossRef] [PubMed]
- Caballero, J.D.; Wheatley, R.M.; Kapel, N.; López-Causapé, C.; Van der Schalk, T.; Quinn, A.; Shaw, L.P.; Ogunlana, L.; Recanatini, C.; Xavier, B.B.; et al. Mixed strain pathogen populations accelerate the evolution of antibiotic resistance in patients. Nat. Commun. 2023, 14, 4083. [Google Scholar] [CrossRef] [PubMed]
- López-Causapé, C.; Cabot, G.; Del Barrio-Tofiño, E.; Oliver, A. The Versatile Mutational Resistome of Pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 685. [Google Scholar] [CrossRef]
- Rojo-Bezares, B.; Estepa, V.; Cebollada, R.; de Toro, M.; Somalo, S.; Seral, C.; Castillo, F.J.; Torres, C.; Sáenz, Y. Carbapenem-resistant Pseudomonas aeruginosa strains from a Spanish hospital: Characterization of metallo-beta-lactamases, porin OprD and integrons. Int. J. Med. Microbiol. 2014, 304, 405–414. [Google Scholar] [CrossRef]
- Chevalier, S.; Bouffartigues, E.; Bodilis, J.; Maillot, O.; Lesouhaitier, O.; Feuilloley, M.G.J.; Orange, N.; Dufour, A.; Cornelis, P. Structure, function and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol. Rev. 2017, 41, 698–722. [Google Scholar] [CrossRef]
- Lloyd, M.G.; Vossler, J.L.; Nomura, C.T.; Moffat, J.F. Blocking RpoN reduces virulence of Pseudomonas aeruginosa isolated from cystic fibrosis patients and increases antibiotic sensitivity in a laboratory strain. Sci. Rep. 2019, 9, 6677. [Google Scholar] [CrossRef]
- Viducic, D.; Ono, T.; Murakami, K.; Katakami, M.; Susilowati, H.; Miyake, Y. rpoN gene of Pseudomonas aeruginosa alters its susceptibility to quinolones and carbapenems. Antimicrob. Agents Chemother. 2007, 51, 1455–1462. [Google Scholar] [CrossRef]
- Winstanley, C.; O’Brien, S.; Brockhurst, M.A. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol. 2016, 24, 327–337. [Google Scholar] [CrossRef]
- Smith, E.E.; Buckley, D.G.; Wu, Z.; Saenphimmachak, C.; Hoffman, L.R.; D’argenio, D.A.; Miller, S.I.; Ramsey, B.W.; Speert, D.P.; Moskowitz, S.M.; et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 2006, 103, 8487–8492. [Google Scholar] [CrossRef]
- Kadri, S.S.; Adjemian, J.; Lai, Y.L.; Spaulding, A.B.; Ricotta, E.; Prevots, D.R.; Palmore, T.N.; Rhee, C.; Klompas, M.; Dekker, J.P.; et al. Difficult-to-Treat Resistance in Gram-Negative Bacteremia at 173 US Hospitals: Retrospective Cohort Analysis of Prevalence, Predictors, and Outcome of Resistance to All First-Line Agents. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2018, 67, 1803–1814. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Aitken, S.L.; Bonomo, R.A.; Mathers, A.J.; van Duin, D.; Clancy, C.J. Guidance on the Treatment of Extended-Spectrum β-lactamase Producing Enterobacterales (ESBL-E), Carbapenem-Resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with Difficult-to-Treat Resistance (DTR-P. aeruginosa). Clin. Infect. Dis. 2022, 75, 187–212. [Google Scholar] [CrossRef]
- Camus, L.; Vandenesch, F.; Moreau, K. From genotype to phenotype: Adaptations of Pseudomonas aeruginosa to the cystic fibrosis environment. Microb. Genom. 2021, 7, 000513. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, D.A.; Wu, M.; Hoffman, L.R.; Kulasekara, H.D.; Déziel, E.; Smith, E.E.; Nguyen, H.; Ernst, R.K.; Larson Freeman, T.J.; Spencer, D.H.; et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 2007, 64, 512–533. [Google Scholar] [CrossRef] [PubMed]
- Groleau, M.C.; de Oliveira Pereira, T.; Dekimpe, V.; Déziel, E. PqsE Is Essential for RhlR-Dependent Quorum Sensing Regulation in Pseudomonas aeruginosa. mSystems 2020, 5, e00194-20. [Google Scholar] [CrossRef] [PubMed]
- Moradali, M.F.; Ghods, S.; Rehm, B.H. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef]
- Kostylev, M.; Kim, D.Y.; Smalley, N.E.; Salukhe, I.; Greenberg, E.P.; Dandekar, A.A. Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proc. Natl. Acad. Sci. USA 2019, 116, 7027–7032. [Google Scholar] [CrossRef]
- Feltner, J.B.; Wolter, D.J.; Pope, C.E.; Groleau, M.C.; Smalley, N.E.; Greenberg, E.P.; Mayer-Hamblett, N.; Burns, J.; Déziel, E.; Hoffman, L.R.; et al. LasR Variant Cystic Fibrosis Isolates Reveal an Adaptable Quorum-Sensing Hierarchy in Pseudomonas aeruginosa. mBio 2016, 7, e01513-16. [Google Scholar] [CrossRef]
- Vincent, A.T.; Freschi, L.; Jeukens, J.; Kukavica-Ibrulj, I.; Emond-Rheault, J.-G.; Leduc, A.; Boyle, B.; Jean-Pierre, F.; Groleau, M.-C.; Déziel, E.; et al. Genomic characterization of environmental Pseudomonas aeruginosa isolated from dental unit waterlines revealed the insertion sequence ISPa11 as a chaotropic element. FEMS Microbiol. Ecol. 2017, 93, fix106. [Google Scholar] [CrossRef]
- Groleau, M.C.; Taillefer, H.; Vincent, A.T.; Constant, P.; Déziel, E. Pseudomonas aeruginosa isolates defective in function of the LasR quorum sensing regulator are frequent in diverse environmental niches. Environ. Microbiol. 2021, 24, 1062–1075. [Google Scholar] [CrossRef]
- Ruiz-Roldán, L.; Rojo-Bezares, B.; Lozano, C.; López, M.; Chichón, G.; Torres, C.; Sáenz, Y. Occurrence of Pseudomonas spp. in Raw Vegetables: Molecular and Phenotypical Analysis of Their Antimicrobial Resistance and Virulence-Related Traits. Int. J. Mol. Sci. 2021, 22, 12626. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, L.; Rao, X.; Wang, J.; Yu, H.; Jiang, J.; Zhou, W.; Xiao, Y.; Li, M.; Zhang, Y.; et al. Characterization of lasR-deficient clinical isolates of Pseudomonas aeruginosa. Sci. Rep. 2018, 8, 13344. [Google Scholar] [CrossRef]
- Scribner, M.R.; Stephens, A.C.; Huong, J.L.; Richardson, A.R.; Cooper, V.S. The Nutritional Environment Is Sufficient to Select Coexisting Biofilm and Quorum Sensing Mutants of Pseudomonas aeruginosa. J. Bacteriol. 2022, 204, e0044421. [Google Scholar] [CrossRef] [PubMed]
- Williams McMackin, E.A.; Djapgne, L.; Corley, J.M.; Yahr, T.L. Fitting Pieces into the Puzzle of Pseudomonas aeruginosa Type III Secretion System Gene Expression. J. Bacteriol. 2019, 201, e00209-19. [Google Scholar] [CrossRef] [PubMed]
- Wilderman, P.J.; Vasil, A.I.; Johnson, Z.; Vasil, M.L. Genetic and biochemical analyses of a eukaryotic-like phospholipase D of Pseudomonas aeruginosa suggest horizontal acquisition and a role for persistence in a chronic pulmonary infection model. Mol. Microbiol. 2001, 39, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.E.; Sims, E.H.; Spencer, D.H.; Kaul, R.; Olson, M.V. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa. J. Bacteriol. 2005, 187, 2138–2147. [Google Scholar] [CrossRef]
- Burrows, L.L. Pseudomonas aeruginosa twitching motility: Type IV pili in action. Annu. Rev. Microbiol. 2012, 66, 493–520. [Google Scholar] [CrossRef]
- Nguyen, Y.; Jackson, S.G.; Aidoo, F.; Junop, M.; Burrows, L.L. Structural characterization of novel Pseudomonas aeruginosa type IV pilins. J. Mol. Biol. 2010, 395, 491–503. [Google Scholar] [CrossRef]
- Kiyaga, S.; Kyany’a, C.; Muraya, A.W.; Smith, H.J.; Mills, E.G.; Kibet, C.; Mboowa, G.; Musila, L. Genetic Diversity, Distribution, and Genomic Characterization of Antibiotic Resistance and Virulence of Clinical Pseudomonas aeruginosa Strains in Kenya. Front. Microbiol. 2022, 13, 835403. [Google Scholar] [CrossRef]
- Giltner, C.L.; Rana, N.; Lunardo, M.N.; Hussain, A.Q.; Burrows, L.L. Evolutionary and functional diversity of the Pseudomonas type IVa pilin island. Environ. Microbiol. 2010, 13, 250–264. [Google Scholar] [CrossRef]
- Asikyan, M.L.; Kus, J.V.; Burrows, L.L. Novel proteins that modulate type IV pilus retraction dynamics in Pseudomonas aeruginosa. J. Bacteriol. 2008, 190, 7022–7034. [Google Scholar] [CrossRef] [PubMed]
- Kus, J.V.; Kelly, J.; Tessier, L.; Harvey, H.; Cvitkovitch, D.G.; Burrows, L.L. Modification of Pseudomonas aeruginosa Pa5196 type IV Pilins at multiple sites with D-Araf by a novel GT-C family Arabinosyltransferase, TfpW. J. Bacteriol. 2008, 190, 7464–7478. [Google Scholar] [CrossRef] [PubMed]
- Kung, V.L.; Ozer, E.A.; Hauser, A.R. The accessory genome of Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2010, 74, 621–641. [Google Scholar] [CrossRef] [PubMed]
- Horna, G.; Quezada, K.; Ramos, S.; Mosqueda, N.; Rubio, M.; Guerra, H.; Ruiz, J. Specific type IV pili groups in clinical isolates of Pseudomonas aeruginosa. Int. Microbiol. 2019, 22, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.M.; Stintzi, A.; De Vos, D.; Cornelis, P.; Tappe, R.; Taraz, K.; Budzikiewicz, H. Use of siderophores to type pseudomonads: The three Pseudomonas aeruginosa pyoverdine systems. Microbiology 1997, 143, 35–43. [Google Scholar] [CrossRef]
- Cornelis, P.; Hohnadel, D.; Meyer, J.M. Evidence for different pyoverdine-mediated iron uptake systems among Pseudomonas aeruginosa strains. Infect. Immun. 1989, 57, 3491–3497. [Google Scholar] [CrossRef]
- Baysse, C.; Meyer, J.M.; Plesiat, P.; Geoffroy, V.; Michel-Briand, Y.; Cornelis, P. Uptake of pyocin S3 occurs through the outer membrane ferripyoverdine type II receptor of Pseudomonas aeruginosa. J. Bacteriol. 1999, 181, 3849–3851. [Google Scholar] [CrossRef]
- Behrens, H.M.; Lowe, E.D.; Gault, J.; Housden, N.G.; Kaminska, R.; Weber, T.M.; Thompson, C.M.A.; Mislin, G.L.A.; Schalk, I.J.; Walker, D.; et al. Pyocin S5 Import into Pseudomonas aeruginosa Reveals a Generic Mode of Bacteriocin Transport. mBio 2020, 11, e03230-19. [Google Scholar] [CrossRef]
- Mulet, X.; Cabot, G.; Ocampo-Sosa, A.A.; Domínguez, M.A.; Zamorano, L.; Juan, C.; Tubau, F.; Rodríguez, C.; Moyà, B.; Peña, C.; et al. Biological Markers of Pseudomonas aeruginosa Epidemic High-Risk Clones. Antimicrob. Agents Chemother. 2013, 57, 5527–5535. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Rojo-Bezares, B.; Cavalié, L.; Dubois, D.; Oswald, E.; Torres, C.; Sáenz, Y. Characterization of carbapenem resistance mechanisms and integrons in Pseudomonas aeruginosa strains from blood samples in a French hospital. J. Med. Microbiol. 2016, 65, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Estepa, V.; Rojo-Bezares, B.; Torres, C.; Sáenz, Y. Faecal carriage of Pseudomonas aeruginosa in healthy humans: Antimicrobial susceptibility and global genetic lineages. FEMS Microbiol. Ecol. 2014, 89, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Heras, J.; Domínguez, C.; Mata, E.; Pascual, V.; Lozano, C.; Torres, C.; Zarazaga, M. GelJ—A tool for analyzing DNA fingerprint gel images. BMC Bioinform. 2015, 16, 270. [Google Scholar] [CrossRef] [PubMed]
- Casado-García, Á.; Chichón, G.; Domínguez, C.; García-Domínguez, M.; Heras, J.; Inés, A.; López, M.; Mata, E.; Pascual, V.; Sáenz, Y. MotilityJ: An open-source tool for the classification and segmentation of bacteria on motility images. Comput. Biol. Med. 2021, 136, 104673. [Google Scholar] [CrossRef] [PubMed]
- Semmler, A.B.; Whitchurch, C.B.; Mattick, J.S. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 1999, 145, 2863–2873. [Google Scholar] [CrossRef]
- Cullen, L.; Weiser, R.; Olszak, T.; Maldonado, R.F.; Moreira, A.S.; Slachmuylders, L.; Brackman, G.; Paunova-Krasteva, T.S.; Zarnowiec, P.; Czerwonka, G.; et al. Phenotypic characterization of an international Pseudomonas aeruginosa reference panel: Strains of cystic fibrosis (CF) origin show less in vivo virulence than non-CF strains. Microbiology 2015, 161, 1961–1977. [Google Scholar] [CrossRef]
- Vielva, L.; de Toro, M.; Lanza, V.F.; de la Cruz, F. PLACNETw: A web-based tool for plasmid reconstruction from bacterial genomes. Bioinformatics 2017, 33, 3796–3798. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Lanza, V.F.; de Toro, M.; Garcillán-Barcia, M.P.; Mora, A.; Blanco, J.; Coque, T.M.; de la Cruz, F. Plasmid flux in Escherichia coli ST131 sublineages, analyzed by Plasmid Constellation Network (PLACNET), a new method for plasmid reconstruction from whole genome sequences. PLoS Genet. 2014, 10, e1004766. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Madaha, E.L.; Mienie, C.; Gonsu, H.K.; Bughe, R.N.; Fonkoua, M.C.; Mbacham, W.F.; Alayande, K.A.; Bezuidenhout, C.C.; Ateba, C.N. Whole-genome sequence of multi-drug resistant Pseudomonas aeruginosa strains UY1PSABAL and UY1PSABAL2 isolated from human broncho-alveolar lavage, Yaoundé, Cameroon. PLoS ONE 2020, 15, e0238390. [Google Scholar] [CrossRef]
- López-Causapé, C.; Sommer, L.M.; Cabot, G.; Rubio, R.; Ocampo-Sosa, A.A.; Johansen, H.K.; Figuerola, J.; Cantón, R.; Kidd, T.J.; Molin, S.; et al. Evolution of the Pseudomonas aeruginosa mutational resistome in an international Cystic Fibrosis clone. Sci. Rep. 2017, 7, 5555. [Google Scholar] [CrossRef]
- Díaz-Caballero, J.D.; Clark, S.T.; Coburn, B.; Zhang, Y.; Wang, P.W.; Donaldson, S.L.; Tullis, D.E.; Yau, Y.C.W.; Waters, V.J.; Hwang, D.M.; et al. Selective Sweeps and Parallel Pathoadaptation Drive Pseudomonas aeruginosa Evolution in the Cystic Fibrosis Lung. mBio 2015, 6, e00981-15. [Google Scholar] [CrossRef] [PubMed]
- Winsor, G.L.; Van Rossum, T.; Lo, R.; Khaira, B.; Whiteside, M.D.; Hancock, R.E.W.; Brinkman, F.S.L. Pseudomonas Genome Database: Facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 2008, 37, D483–D488. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.G.; Urbach, J.M.; Wu, G.; Liberati, N.T.; Feinbaum, R.L.; Miyata, S.; Diggins, L.T.; He, J.; Saucier, M.; Deziel, E.; et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006, 7, R90. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).