Comprehensive Analysis of Virulence Determinants and Genomic Islands of blaNDM-1-Producing Enterobacter hormaechei Clinical Isolates from Greece
Abstract
1. Introduction
2. Results
2.1. Antimicrobial Susceptibility Testing, MLST, WGS and Type (Strain) Genome Server (TYGS) Analysis
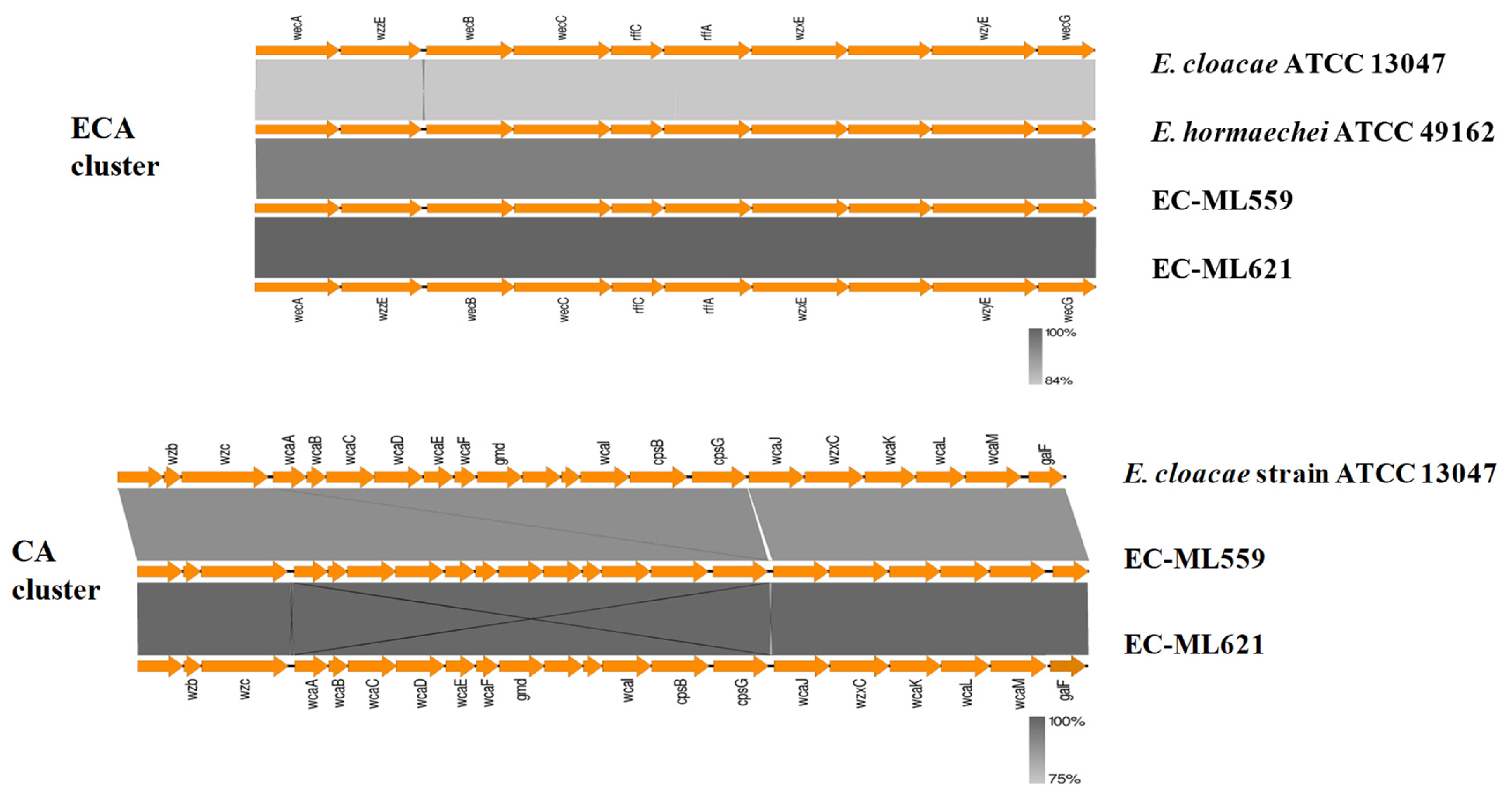
2.2. Enterobacterial Common Antigen (ECA), Colanic Acid (CA) and Lipopolysaccharide (LPS) O-Antigen
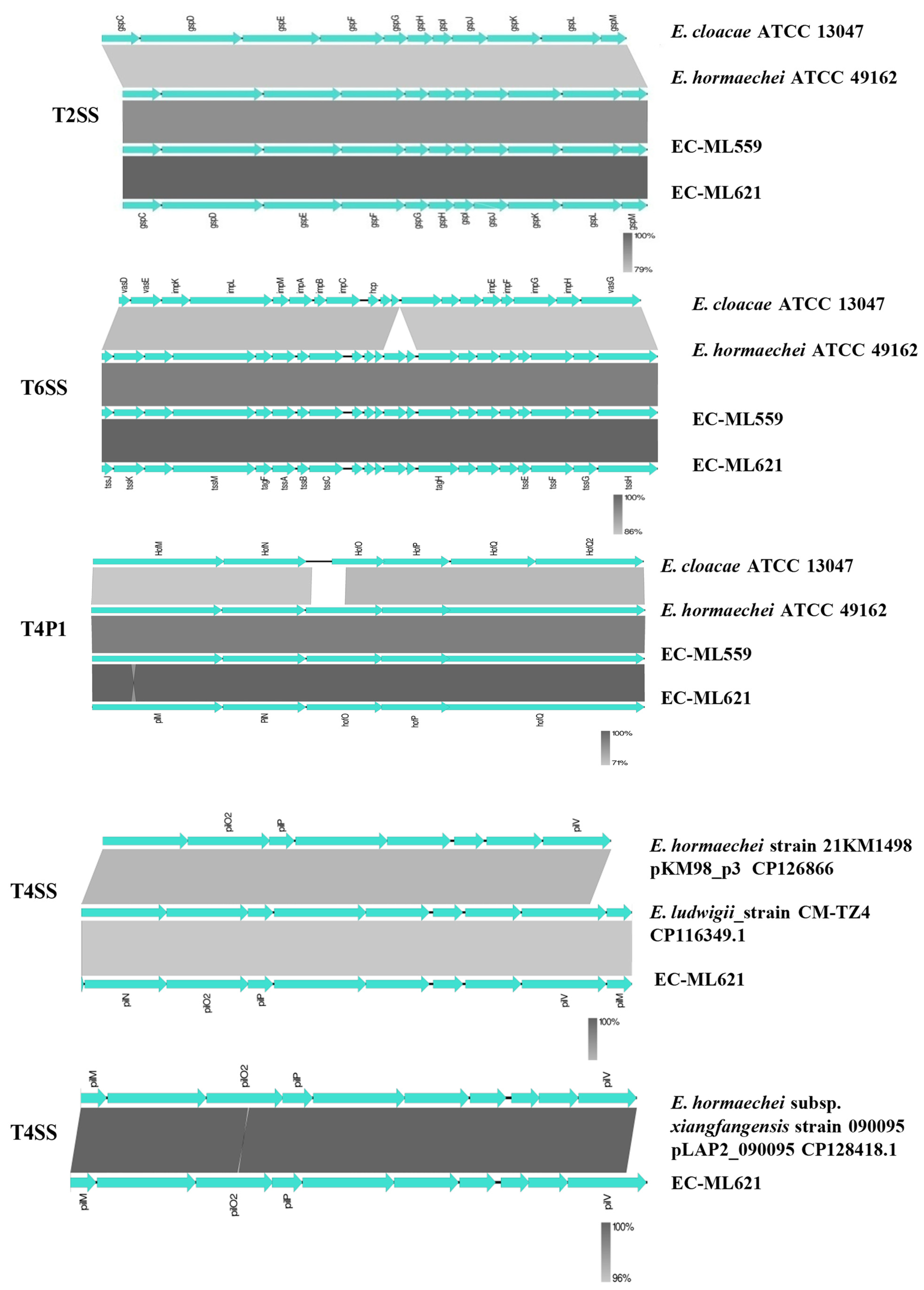
2.3. Flagella, Fimbriae and Secretion Systems
2.4. In Silico Prediction of Genomic Islands
3. Materials and Methods
3.1. Bacterial Isolates, Antimicrobial Susceptibility Testing, Whole Genome Sequencing (WGS) and MLST Typing
3.2. Whole-Genome-Based Taxonomic Analysis
3.3. In Silico Prediction of Genomic Islands, Mobile Elements, Antimicrobial Resistance and Virulence Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Davin-Regli, A.; Lavigne, J.P.; Pagès, J.M. Enterobacter spp.: Update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin. Microbiol. Rev. 2019, 32, e00002-19. [Google Scholar] [CrossRef] [PubMed]
- Paauw, A.; Caspers, M.P.M.; Schuren, F.H.J.; Hall, M.A.L.-V.; Delétoile, A.; Montijn, R.C.; Verhoef, J.; Fluit, A.C. Genomic diversity within the Enterobacter cloacae complex. PLoS ONE 2008, 3, e3018. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-Y.; Wong, C.-F.; Chung, K.M.-K.; Jiang, J.-W.; Leung, F.C.-C. Comparative genome analysis of Enterobacter cloacae. PLoS ONE 2013, 8, e74487. [Google Scholar] [CrossRef] [PubMed]
- Ciufo, S.; Kannan, S.; Sharma, S.; Badretdin, A.; Clark, K.; Turner, S.; Brover, S.; Schoch, C.L.; Kimchi, A.; DiCuccio, M. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018, 68, 2386–2392. [Google Scholar] [CrossRef] [PubMed]
- Federhen, S.; Rossello-Mora, R.; Klenk, H.-P.; Tindall, B.J.; Konstantinidis, K.T.; Whitman, W.B.; Brown, D.; Labeda, D.; Ussery, D.; Garrity, G.M.; et al. Meeting report: GenBank microbial genomic taxonomy workshop (12–13 May, 2015). Stand. Genom. Sci. 2016, 11, 15. [Google Scholar] [CrossRef]
- Mustafa, A.; Ibrahim, M.; Rasheed, M.A.; Kanwal, S.; Hussain, A.; Sami, A.; Ahmed, R.; Bo, Z. Genome-wide analysis of four Enterobacter cloacae complex type strains: Insights into virulence and niche adaptation. Sci. Rep. 2020, 10, 8150. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ma, W.; Wang, X. Insights into the structure of Escherichia coli outer membrane as the target for engineering microbial cell factories. Microb. Cell Fact. 2021, 20, 73. [Google Scholar] [CrossRef] [PubMed]
- Van der Woude, M.W.; Bäumler, A.J. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 2004, 17, 581–611. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Annavajhala, M.K.; Gomez-Simmonds, A.; Uhlemann, A.C. Multidrug-resistant Enterobacter cloacae complex emerging as a global, diversifying threat. Front. Microbiol. 2019, 10, 44. [Google Scholar] [CrossRef]
- Hall, B.G.; Barlow, M. Revised Ambler classification of β-lactamases. J. Antimicrob. Chemother. 2005, 55, 1050–1051. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; Matsumura, Y.; Adams, M.D.; Bradford, P.; Motyl, M.; Chen, L.; Kreiswirth, B.N.; Pitout, J.D. Genomic epidemiology of global carbapenemase-producing Enterobacter spp., 2008–2014. Emerg. Infect. Dis. 2018, 24, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Izdebski, R.; Baraniak, A.; Herda, M.; Fiett, J.; Bonten, M.J.; Carmeli, Y.; Goossens, H.; Hryniewicz, W.; Brun-Buisson, C.; Gniadkowski, M.; et al. MLST reveals potentially high-risk international clones of Enterobacter cloacae. J. Antimicrob. Chemother. 2015, 70, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. NDM Metallo-β-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 2019, 32, e00115-18. [Google Scholar] [CrossRef] [PubMed]
- Papagiannitsis, C.C.; Studentova, V.; Chudackova, E.; Bergerova, T.; Hrabak, J.; Radej, J.; Novak, I. Identification of a New Delhi metallo-β-lactamase-4 (NDM-4)-producing Enterobacter cloacae from a Czech patient previously hospitalized in Sri Lanka. Folia Microbiol. 2013, 58, 547–549. [Google Scholar] [CrossRef] [PubMed]
- Paskova, V.; Medvecky, M.; Skalova, A.; Chudejova, K.; Bitar, I.; Jakubu, V.; Bergerova, T.; Zemlickova, H.; Papagiannitsis, C.C.; Hrabak, J. Characterization of NDM-encoding plasmids from Enterobacteriaceae recovered From Czech Hospitals. Front. Microbiol. 2018, 9, 1549. [Google Scholar] [CrossRef] [PubMed]
- Gartzonika, K.; Politi, L.; Mavroidi, A.; Tsantes, A.G.; Spanakis, N.; Priavali, E.; Vrioni, G.; Tsakris, A. High prevalence of clonally related ST182 NDM-1-producing Enterobacter cloacae complex clinical isolates in Greece. Int. J. Antimicrob. Agents 2023, 62, 106837. [Google Scholar] [CrossRef] [PubMed]
- Costa, T.R.D.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion systems in Gram-negative bacteria: Structural and mechanistic insights. Nat. Rev. Microbiol. 2015, 13, 343–359. [Google Scholar] [CrossRef]
- Yang, M.; Zhou, X.; Bao, Y.; Zhang, Y.; Liu, B.; Gan, L.; Tao, W.; Tuo, J.; Gong, H. Comprehensive genomic analysis reveals extensive diversity of type I and Type IV secretion systems in Klebsiella pneumoniae. Curr. Microbiol. 2023, 80, 270. [Google Scholar] [CrossRef]
- Unterholzner, S.J.; Poppenberger, B.; Rozhon, W. Toxin-antitoxin systems: Biology, identification, and application. Mob. Genet. Elements 2013, 3, e26219. [Google Scholar] [CrossRef]
- Rai, A.K.; Mitchell, A.M. Enterobacterial Common Antigen: Synthesis and function of an enigmatic molecule. mBio 2020, 11, e01914-20. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, G.; Andrianopoulos, K.; Hobbs, M.; Reeves, P.R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 1996, 178, 4885–4893. [Google Scholar] [CrossRef] [PubMed]
- DebRoy, C.; Roberts, E.; Fratamico, P.M. Detection of O antigens in Escherichia coli. Anim. Health Res. Rev. 2011, 12, 169–185. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, J.; Wang, X.; Xu, C.; Han, T.; Guo, X. Genetic characterization of the O-Antigen and development of a molecular serotyping scheme for Enterobacter cloacae. Front. Microbiol. 2020, 11, 727. [Google Scholar] [CrossRef]
- Nedeljković, M.; Sastre, D.E.; Sundberg, E.J. Bacterial flagellar filament: A supramolecular multifunctional nanostructure. Int. J. Mol. Sci. 2021, 22, 7521. [Google Scholar] [CrossRef] [PubMed]
- De Maayer, P.; Pillay, T.; Coutinho, T.A. Flagella by numbers: Comparative genomic analysis of the supernumerary flagellar systems among the Enterobacterales. BMC Genom. 2020, 21, 670. [Google Scholar] [CrossRef] [PubMed]
- Mol, O.; Oudega, B. Molecular and structural aspects of fimbriae biosynthesis and assembly in Escherichia coli. FEMS Microbiol. Rev. 1996, 19, 25–52. [Google Scholar] [CrossRef]
- Low, A.S.; Holden, N.; Rosser, T.; Roe, A.J.; Constantinidou, C.; Hobman, J.L.; Smith, D.G.; Low, J.C.; Gally, D.L. Analysis of fimbrial gene clusters and their expression in enterohaemorrhagic Escherichia coli O157:H7. Environ. Microbiol. 2006, 8, 1033–1047. [Google Scholar] [CrossRef]
- Schwan, W.R. Regulation of fim genes in uropathogenic Escherichia coli. World J. Clin. Infect. Dis. 2011, 1, 17–25. [Google Scholar] [CrossRef]
- Mattick, J.S. Type IV pili and twitching motility. Annu. Rev. Microbiol. 2002, 56, 289–314. [Google Scholar] [CrossRef]
- Nudleman, E.; Kaiser, D. Pulling together with type IV pili. J. Mol. Microbiol. Biotechnol. 2004, 7, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.W.; Chen, T.L.; Chen, Y.T.; Lauderdale, T.L.; Liao, T.L.; Lee, Y.T.; Chen, C.P.; Liu, Y.M.; Lin, A.C.; Chang, Y.H.; et al. Copy number change of the NDM-1 sequence in a multidrug-resistant Klebsiella pneumoniae clinical isolate. PLoS ONE 2013, 8, e62774. [Google Scholar] [CrossRef] [PubMed]
- Merhi, G.; Amayri, S.; Bitar, I.; Araj, G.F.; Tokajian, S. Whole genome-based characterization of multidrug resistant Enterobacter and Klebsiella aerogenes isolates from Lebanon. Microbiol. Spectr. 2023, 1, e0291722. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Cattoir, V.; Soares, A.; Soussy, C.J.; Nordmann, P. Novel Ambler class A b-lactamase LAP-1 and its association with the plasmid-mediated quinolone resistance determinant QnrS1. Antimicrob. Agents Chemother. 2007, 51, 631–637. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, Z.; Mi, Z.; Wang, C. A novel b-lactamase gene, LAP-2, co-produced by an Enterobacter cloacae clinical isolate in China. J. Hosp. Infect. 2008, 70, 95–96. [Google Scholar] [CrossRef] [PubMed]
- Kocsis, B.; Kocsis, E.; Fontana, R.; Cornaglia, G.; Mazzariol, A. Identification of blaLAP-2 and qnrS1 genes in the internationally successful Klebsiella pneumoniae ST147 clone. J. Med. Microbiol. 2013, 62, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; SardàCarbasse, J.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon Fraser University Research Computing Group; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: Mobile Element Finder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.J.; Rutherford, K.M.; Berriman, M.; Rajandream, M.A.; Barrell, B.G.; Parkhill, J. ACT: The Artemis Comparison Tool. Bioinformatics 2005, 21, 3422–3423. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xie, Y.; Liu, M.; Tai, C.; Sun, J.; Deng, Z.; Ou, H.Y. oriTfinder: A web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 2018, 46, W229–W234. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Girlich, D.; Ouzani, S.; Emeraud, C.; Gauthier, L.; Bonnin, R.A.; Le Sache, N.; Mokhtari, M.; Langlois, I.; Begasse, C.; Arangia, N.; et al. Uncovering the novel Enterobacter cloacae complex species responsible for septic shock deaths in newborns: A cohort study. Lancet Microbe 2021, 2, e536–e544. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef]
- David, S.; Cohen, V.; Reuter, S.; Sheppard, A.E.; Giani, T.; Parkhill, J.; European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group; ESCMID Study Group for Epidemiological Markers (ESGEM); Rossolini, G.M.; Feil, E.J.; et al. Integrated chromosomal and plasmid sequence analyses reveal diverse modes of carbapenemase gene spread among Klebsiella pneumoniae. Proc. Natl. Acad. Sci. USA 2020, 117, 25043–25054. [Google Scholar] [CrossRef]

| Characteristics # | EC-ML559 | EC-ML621 |
|---|---|---|
| WGS Assembly | Genbank Accession no. JARUPS000000000.1 Contigs: 74; total length: 4,958,007 bp, %GC: 55.07 | Genbank Accession no. JASKGQ000000000 Contigs: 100, total length: 5,153,338 bp, %GC: 55.02 |
| MLST ST (allelic profile) | ST182 (49-20-19-44-90-24-32) | ST2143 (554-20-19-44-90-24-32) |
| Plasmids | IncFII(pECL_A), IncR IncFII(pKPX1), Col440I | IncFII(pECLA), IncR, IncFII(Yp), IncL/M(pMU407), IncFIB(pB171), IncN |
| ARGs | β-lactams: blaACT-16, blaNDM-1, blaTEM-1, blaOXA-1 aminoglycosides: aph(6)-Id, aac(6′)-Ib3, aph(3′)-Ia, aac(6′)-Ib-cr, aph(3″)Ib chloramphenicol: catB3 fosfomycin: fosA quaternary ammonium compounds: qacE quinolones: OqxA, OqxB, qnrB19 trimethoprim: dfrA14 sulfonamides: sul1, sul2 macrolides: mrx(A), mphR(A), mph(A) rifampicin: ARR-3 tetracyclines: tet(D) | β-lactams: blaACT-16, blaNDM-1, blaTEM-1, blaOXA-1, blaCTX-M-14, blaCTX-M-15, blaLAP-2 aminoglycosides: aph(6)-Id, aac(3)-IId, aadA2, aac(6′)-Ib-cr, aph(3″)-Ib chloramphenicol: catB3 fosfomycin: fosA quaternary ammonium compounds: qacE quinolones: OqxA, OqxB, qnrS1 trimethoprim: dfrA12 sulfonamides: sul1 macrolides: mrx(A), mphR(A), mph(A) |
| HMRGs | Arsenic: arsA, arsB, arsD, arsH, arsR, Copper: pcoA, pcoB, pcoC, pcoD, pcoE. pcoR, pcoS Copper/Silver: silA, silB, silC, silE, silP, silR, silS, Tellurium: terB, terC, terD, terW, terZ | Arsenic arsA, arsB, arsD, arsH, arsR, Copper: pcoA, pcoB, pcoD, pcoC, pcoE, pcoR, pcoS, Copper/Silver: silA, silB, silC, silE, silP, silR, silS, Tellurium: terC, terD, terW Mercury: merA, merC, merD, merE, merP, merR, merT |
| TA-systems | type II: CcdA, CcdB, HicA, HicB, ParD, RatA, RelB, RelE/ParE, VapB/VapC, Yaf; type IV: YeeU | type II: HicA, HicB, ParD, RatA, RelB, RelE/ParE, VapB/Vap; type IV: YeeU, CbtA |
| T1SS | HlyD, TolC, MacA, MacB | HlyD, TolC, MacA, MacB |
| T2SS | gspC, gspD, gspE, gspF, gspG, gspH, gspI, gspJ, gspK, gspL, gspM secA, secB, secD, secE, secF, secG, secM, secY tatA, tatB, tatC, tatD, ftsY, yajC, yidC | gspC, gspD, gspE, gspF, gspG, gspH, gspI, gspJ, gspK, gspL, gspM secA, secB, secD, secE, secF, secG, secM, secY, tatA, tatB, tatC, tatD, ftsY, yajC, yidC |
| T6SS | tssH, tssG, tssF, tssE, tagJ, tagH, tssC, tssB, tssA, tagF, tssM, tssK, tssJ | tssH, tssG, tssF, tssE, tagJ, tagH, tssC, tssB, tssA, tagF, tssM, tssK, tssJ |
| T4P1/T4SS | Locus 1: hofC, hofB, HofQ, HofP, HofO, HofN/PilN, HofM/PilM | Locus 1: hofC, hofB, HofQ, HofP, HofO, HofN/PilN, HofM/PilM Locus 2: pilN, pilO2, pilP, pilV, pilM Locus 3: pilM, pilN, pilO2, pilP |
| Flagella | fliZACDST | fliZACDST |
| Fimbriae | P7E32_11190, P7E32_11195, P7E32_11200, P7E32_11205, P7E32_11210, P7E32_11215, P7E32_11220, P7E32_11225, P7E32_11230 | P7F73_07975, P7F73_07980, P7F73_07985, P7F73_07990, P7F73_07995, P7F73_08000, P7F73_08005, P7F73_08010, P7F73_08015 |
| Type | Contig GenBank Accession No. | Region/Gene (s) |
|---|---|---|
| Strain EC-ML559 | ||
| oriT region | JARUPS010000033 | 2799–2897 |
| Relaxase | JARUPS010000032 | traI |
| T4CP | JARUPS010000032 | traD |
| T4SS—region 1 | JARUPS010000031 | traN, trbC, traU, traW, traC, traV, traB, traK, traL, traE, traA |
| T4SS—region 2 | JARUPS010000023 | traM, traK, traJ, traH, traF, traB, traH, traG |
| Strain EC-ML621 | ||
| oriT region | JASKGQ010000026 | 18926–18990 |
| Relaxase | JASKGQ010000026 | traI |
| T4CP | JASKGQ010000026 | traD |
| T4SS—region 1 | JASKGQ010000016 | tfc2, DotB, traH, traI, traJ, traK, traL, traM, traN, traO, traP, traQ, traR, traT, traU, traV, traW, traX, traY |
| T4SS—region 2 | JASKGQ010000022 | traH, traI, traJ, traK, traL, traM, traN, traO, traP, traQ, traR, traU, traW, traX, traY, trbC, trbB, trbA, trbN |
| T4SS—region 3 | JASKGQ010000024 | traM, traY, traA, traL, traE, traK, traB, traV, traC, traW, traU, trbC, traN, traF, traQ, traB, traH, traG, traD, traI, traX |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mavroidi, A.; Gartzonika, K.; Spanakis, N.; Froukala, E.; Kittas, C.; Vrioni, G.; Tsakris, A. Comprehensive Analysis of Virulence Determinants and Genomic Islands of blaNDM-1-Producing Enterobacter hormaechei Clinical Isolates from Greece. Antibiotics 2023, 12, 1549. https://doi.org/10.3390/antibiotics12101549
Mavroidi A, Gartzonika K, Spanakis N, Froukala E, Kittas C, Vrioni G, Tsakris A. Comprehensive Analysis of Virulence Determinants and Genomic Islands of blaNDM-1-Producing Enterobacter hormaechei Clinical Isolates from Greece. Antibiotics. 2023; 12(10):1549. https://doi.org/10.3390/antibiotics12101549
Chicago/Turabian StyleMavroidi, Angeliki, Konstantina Gartzonika, Nick Spanakis, Elisavet Froukala, Christos Kittas, Georgia Vrioni, and Athanasios Tsakris. 2023. "Comprehensive Analysis of Virulence Determinants and Genomic Islands of blaNDM-1-Producing Enterobacter hormaechei Clinical Isolates from Greece" Antibiotics 12, no. 10: 1549. https://doi.org/10.3390/antibiotics12101549
APA StyleMavroidi, A., Gartzonika, K., Spanakis, N., Froukala, E., Kittas, C., Vrioni, G., & Tsakris, A. (2023). Comprehensive Analysis of Virulence Determinants and Genomic Islands of blaNDM-1-Producing Enterobacter hormaechei Clinical Isolates from Greece. Antibiotics, 12(10), 1549. https://doi.org/10.3390/antibiotics12101549








