Molecular Epidemiologic and Geo-Spatial Characterization of Staphylococcus aureus Cultured from Skin and Soft Tissue Infections from United States-Born and Immigrant Patients Living in New York City
Abstract
1. Introduction
2. Results
2.1. Overview of Enrollment Population
2.2. Concordance between S. aureus Wound and Carriage
2.3. Comparison between US-Born and Non-US-Born
2.4. Risk Factors for S. aureus MRSA SSTI and USA300 SSTI by Birthplace
2.5. Molecular Features of S. aureus MRSA SSTI and USA300 SSTI by Birthplace and Characteristics of Current Residence
2.6. Risk for S. aureus USA300 Skin and Soft Tissue Infection Based on Immigration Status (US-Born vs. Non-US-Born)
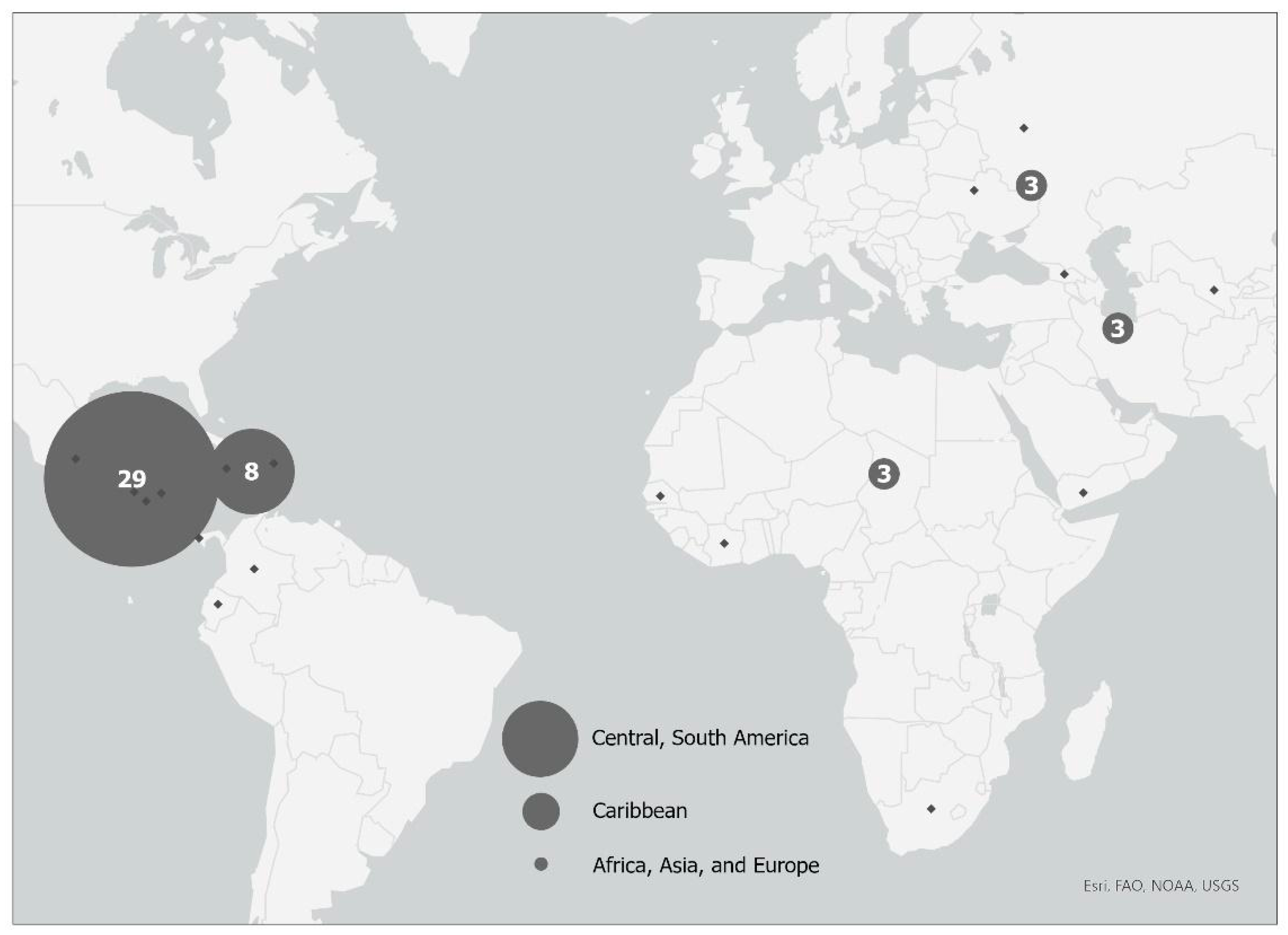
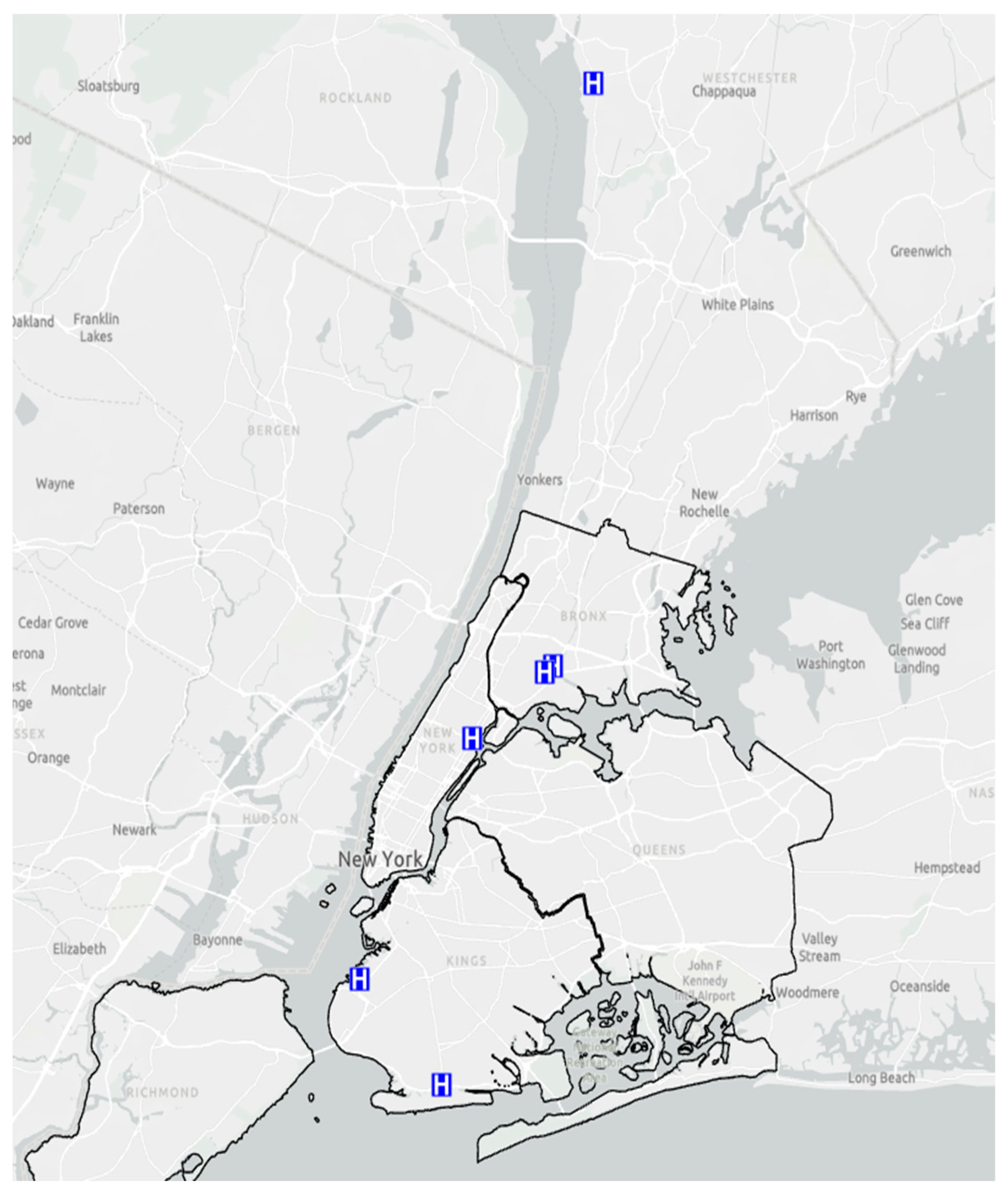
2.7. Geo-Spatial Distribution of Patients with S. aureus Skin and Soft Tissue Infections
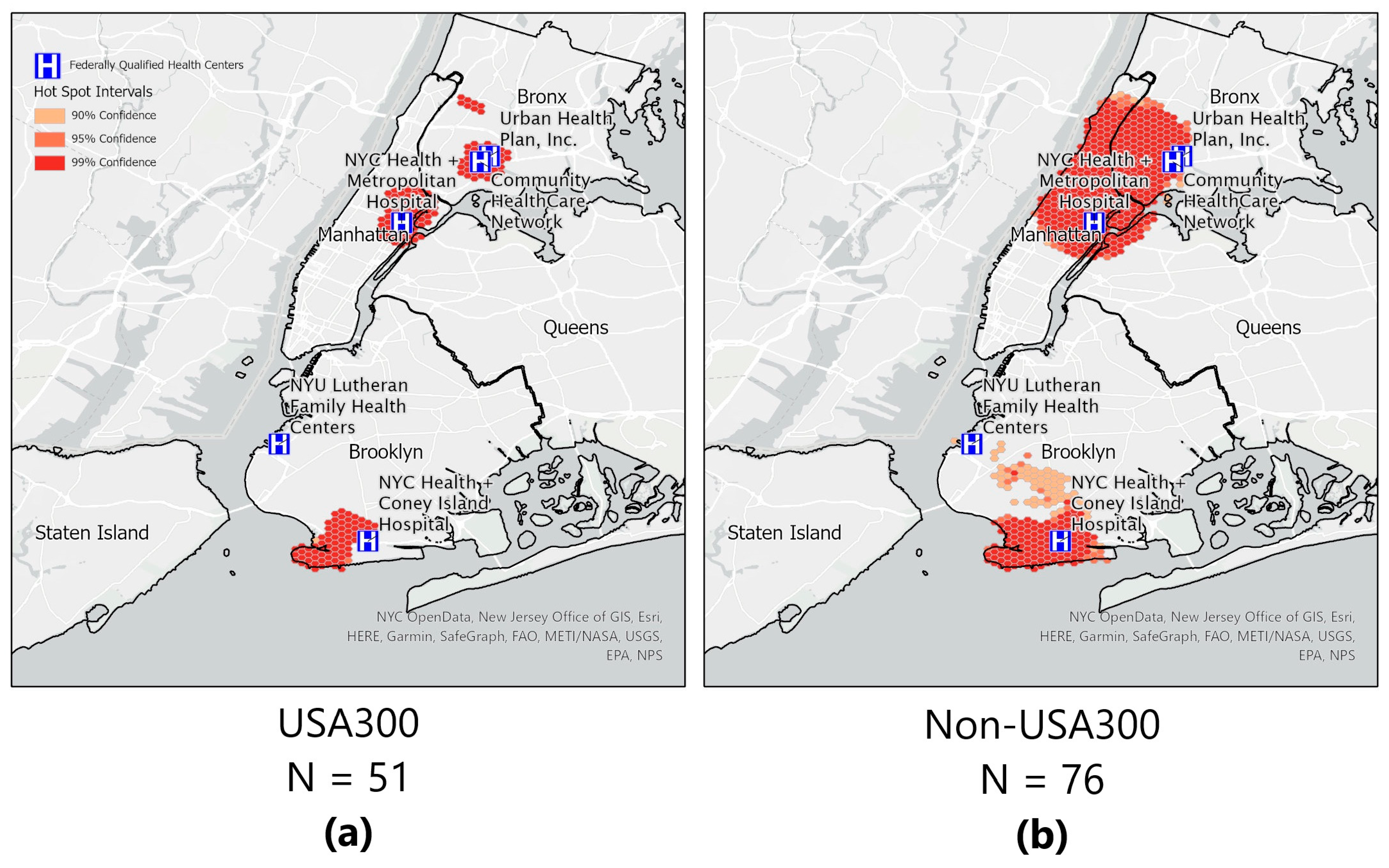
2.8. Hot/Cold Spot Analyses for S. aureus and Genotype-USA300
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Altman, S.A.; Bastian, P. DHL Global Connectedness Index; Deutsche Post DHL Group: Bonn, Germany, 2019; pp. 1–56. [Google Scholar]
- Itani, K.M.; Merchant, S.; Lin, S.J.; Akhras, K.; Alandete, J.C.; Hatoum, H.T. Outcomes and management costs in patients hospitalized for skin and skin-structure infections. Am. J. Infect. Control 2011, 39, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Planet, P.J. Life After USA300: The Rise and Fall of a Superbug. J. Infect. Dis. 2017, 215, S71–S77. [Google Scholar] [CrossRef] [PubMed]
- David, M.Z.; Daum, R.S. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010, 23, 616–687. [Google Scholar] [CrossRef] [PubMed]
- Mistry, R.D.; Shapiro, D.J.; Goyal, M.K.; Zaoutis, T.E.; Gerber, J.S.; Liu, C.; Hersh, A.L. Clinical management of skin and soft tissue infections in the US Emergency Departments. West. J. Emerg. Med. 2014, 15, 491–498. [Google Scholar] [CrossRef]
- Ray, G.T.; Suaya, J.A.; Baxter, R. Trends and characteristics of culture-confirmed Staphylococcus aureus infections in a large US integrated health care organization. J. Clin. Microbiol. 2012, 50, 1950–1957. [Google Scholar] [CrossRef] [PubMed]
- Casey, J.A.; Cosgrove, S.E.; Stewart, W.F.; Pollak, J.; Schwartz, B.S. A population-based study of the epidemiology and clinical features of methicillin-resistant Staphylococcus aureus infection in Pennsylvania, 2001–2010. Epidemiol. Infect. 2013, 141, 1166–1179. [Google Scholar] [CrossRef]
- Talan, D.A.; Krishnadasan, A.; Gorwitz, R.J.; Fosheim, G.E.; Limbago, B.; Albrecht, V.; Moran, G.J. Comparison of Staphylococcus aureus from skin and soft-tissue infections in US emergency department patients, 2004 and 2008. Clin. Infect. Dis. 2011, 53, 144–149. [Google Scholar] [CrossRef]
- Nurjadi, D.; Friedrich-Janicke, B.; Schafer, J.; Van Genderen, P.J.; Goorhuis, A.; Perignon, A.; Neumayr, A.; Mueller, A.; Kantele, A.; Schunk, M.; et al. Skin and soft tissue infections in intercontinental travelers and the import of multi-resistant Staphylococcus aureus to Europe. Clin. Microbiol. Infect. 2015, 21, 567.e1–567.e10. [Google Scholar] [CrossRef]
- Ray, G.T.; Suaya, J.A.; Baxter, R. Incidence, microbiology, and patient characteristics of skin and soft-tissue infections in a US population: A retrospective population-based study. BMC Infect. Dis. 2013, 13, 252. [Google Scholar] [CrossRef]
- Tenover, F.C.; Goering, R.V. Methicillin-resistant Staphylococcus aureus strain USA300: Origin and epidemiology. J. Antimicrob. Chemother. 2009, 64, 441–446. [Google Scholar] [CrossRef]
- Gill, V.C.; Ma, I.; Guo, M.; Gregson, D.B.; Naugler, C.; Church, D.L. Sociodemographic and geospatial associations with community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) infections in a large Canadian city: An 11 year retrospective study. BMC Public Health 2019, 19, 914. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.; Rincon, S.; Diaz, L.; Panesso, D.; Contreras, G.A.; Zurita, J.; Carrillo, C.; Rizzi, A.; Guzman, M.; Adachi, J.; et al. Dissemination of methicillin-resistant Staphylococcus aureus USA300 sequence type 8 lineage in Latin America. Clin. Infect. Dis. 2009, 49, 1861–1867. [Google Scholar] [CrossRef] [PubMed]
- Shallcross, L.J.; Fragaszy, E.; Johnson, A.M.; Hayward, A.C. The role of the Panton-Valentine leucocidin toxin in staphylococcal disease: A systematic review and meta-analysis. Lancet Infect. Dis. 2013, 13, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Blanco, R.; Tristan, A.; Ezpeleta, G.; Larsen, A.R.; Bes, M.; Etienne, J.; Cisterna, R.; Laurent, F. Molecular epidemiology of Panton-Valentine Leukocidin-positive Staphylococcus aureus in Spain: Emergence of the USA300 clone in an autochthonous population. J. Clin. Microbiol. 2011, 49, 433–436. [Google Scholar] [CrossRef]
- Tristan, A.; Bes, M.B.; Meugnier, H.; Lina, G.L.; Bozdogan, B.; Courvalin, P.; Reverdy, M.-E.; Enright, M.C.; Vandenesch, F.; Etienne, J. Global Distribution of Panton-Valentine Leukocidin–positive Methicillin-resistant Staphylococcus aureus, 2006. Emerg. Infect. Dis. 2007, 13, 594–600. [Google Scholar] [CrossRef]
- Planet, P.J.; Diaz, L.; Kolokotronis, S.O.; Narechania, A.; Reyes, J.; Xing, G.; Rincon, S.; Smith, H.; Panesso, D.; Ryan, C.; et al. Parallel Epidemics of Community-Associated Methicillin-Resistant Staphylococcus aureus USA300 Infection in North and South America. J. Infect. Dis. 2015, 212, 1874–1882. [Google Scholar] [CrossRef]
- What Is a Health Center? Available online: https://bphc.hrsa.gov/about/what-is-a-health-center/index.html (accessed on 7 October 2020).
- Piper Jenks, N.; Pardos de la Gandara, M.; D’Orazio, B.M.; Correa da Rosa, J.; Kost, R.G.; Khalida, C.; Vasquez, K.S.; Coffran, C.; Pastagia, M.; Evering, T.H.; et al. Differences in prevalence of community-associated MRSA and MSSA among US and non-US born populations in six New York Community Health Centers. Travel Med. Infect. Dis. 2016, 14, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Tobin, J.N.; Hower, S.; D’Orazio, B.M.; Pardos de la Gandara, M.; Evering, T.H.; Khalida, C.; Ramachandran, J.; Gonzalez, L.J.; Kost, R.G.; Vasquez, K.S.; et al. Comparative Effectiveness Study of Home-Based Interventions to Prevent CA-MRSA Infection Recurrence. Antibiotics 2021, 10, 1105. [Google Scholar] [CrossRef]
- Fessenden, F.; Roberts, S. Then as Now—New York’s Shifting Ethnic Mosaic. The New York Times. 22 January 2011. Available online: https://archive.nytimes.com/query.nytimes.com/gst/fullpage-9B05E4D7123EF930A15752C0A9679D8B63.html#:~:text=’’New%20York%20has%20evolved%20into,in%20the%20city’s%20foreign%2Dborn (accessed on 26 October 2020).
- Esri. Esri Tapestry Segmentation. Available online: https://doc.arcgis.com/en/esri-demographics/latest/regional-data/tapestry-segmentation.htm (accessed on 15 April 2023).
- Esri. LifeModeGroup: Next Wave High-Rise Renters. Available online: https://downloads.esri.com/esri_content_doc/dbl/us/tapestry/segment64.pdf (accessed on 15 April 2023).
- Di Ruscio, F.; Bjornholt, J.V.; Leegaard, T.M.; Moen, A.E.F.; de Blasio, B.F. MRSA infections in Norway: A study of the temporal evolution, 2006–2015. PLoS ONE 2017, 12, e0179771. [Google Scholar] [CrossRef]
- Hagmann, S.H.F. When less is best-Why non-US born patients could have less MRSA. Travel Med. Infect. Dis. 2016, 14, 546–547. [Google Scholar] [CrossRef]
- Baptiste, K.E.; Williams, K.; Williams, N.J.; Wattret, A.; Clegg, P.D.; Dawson, S.; Corkill, J.E.; O’Neill, T.; Hart, C.A. Methicillin-resistant Staphylococci in Companion Animals. Emerg. Infect. Dis. 2005, 11, 1942–1944. [Google Scholar] [CrossRef] [PubMed]
- Mama, O.M.; Gomez-Sanz, E.; Ruiz-Ripa, L.; Gomez, P.; Torres, C. Diversity of staphylococcal species in food producing animals in Spain, with detection of PVL-positive MRSA ST8 (USA300). Vet. Microbiol. 2019, 233, 5–10. [Google Scholar] [CrossRef]
- Haenni, M.; Saras, E.; Chatre, P.; Medaille, C.; Bes, M.; Madec, J.Y.; Laurent, F. A USA300 variant and other human-related methicillin-resistant Staphylococcus aureus strains infecting cats and dogs in France. J. Antimicrob. Chemother. 2012, 67, 326–329. [Google Scholar] [CrossRef]
- Lin, Y.; Barker, E.; Kislow, J.; Kaldhone, P.; Stemper, M.E.; Pantrangi, M.; Moore, F.M.; Hall, M.; Fritsche, T.R.; Novicki, T.; et al. Evidence of multiple virulence subtypes in nosocomial and community-associated MRSA genotypes in companion animals from the upper midwestern and northeastern United States. Clin. Med. Res. 2011, 9, 7–16. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Loeffler, A.; Lloyd, D.H. Companion animals: A reservoir for methicillin-resistant Staphylococcus aureus in the community? Epidemiol. Infect. 2010, 138, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Kadariya, J.; Thapaliya, D.; Bhatta, S.; Mahatara, R.L.; Bempah, S.; Dhakal, N.; Smith, T.C. Multidrug-resistant Staphylococcus aureus Colonization in Healthy Adults Is More Common in Bhutanese Refugees in Nepal than Those Resettled in Ohio. BioMed Res. Int. 2019, 2019, 5739247. [Google Scholar] [CrossRef] [PubMed]
- Iverson, S.A.; Brazil, A.M.; Ferguson, J.M.; Nelson, K.; Lautenbach, E.; Rankin, S.C.; Morris, D.O.; Davis, M.F. Anatomical patterns of colonization of pets with staphylococcal species in homes of people with methicillin-resistant Staphylococcus aureus (MRSA) skin or soft tissue infection (SSTI). Vet. Microbiol. 2015, 176, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Cotter, C.J.; Ferradas, C.; Ludwig, S.; Dalton, K.; Larsen, J.; Laucks, D.; Iverson, S.A.; Baron, P.; Tolomeo, P.C.; Brazil, A.M.; et al. Risk factors for meticillin-resistant Staphylococcus aureus (MRSA) carriage in MRSA-exposed household pets. Vet. Dermatol. 2023, 34, 22–27. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.L.; Park, M.; Program, N.C.S.; Kong, H.H.; Segre, J.A. Temporal Stability of the Human Skin Microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef]
- Frost, I.; Van Boeckel, T.P.; Pires, J.; Craig, J.; Laxminarayan, R. Global geographic trends in antimicrobial resistance: The role of international travel. J. Travel. Med. 2019, 26, taz036. [Google Scholar] [CrossRef]
- Steinig, E.; Aglua, I.; Duchene, S.; Meehan, M.T.; Yoannes, M.; Firth, C.; Jaworski, J.; Drekore, J.; Urakoko, B.; Poka, H.; et al. Phylodynamic signatures in the emergence of community-associated MRSA. Proc. Natl. Acad. Sci. USA 2022, 119, e2204993119. [Google Scholar] [CrossRef] [PubMed]
- van Tonder, A.J.; McCullagh, F.; McKeand, H.; Thaw, S.; Bellis, K.; Raisen, C.; Lay, L.; Aggarwal, D.; Holmes, M.; Parkhill, J.; et al. Colonization and transmission of Staphylococcus aureus in schools: A citizen science project. Microb. Genom. 2023, 9, mgen000993. [Google Scholar] [CrossRef] [PubMed]
- Almeida, S.T.; Paulo, A.C.; Lencastre, H.d.; Sá-Leão, R. Evaluation of Methicillin-Resistant Staphylococcus aureus Carriage in the Elderly in Portugal Using Selective Enrichment Followed by Quantitative Real-Time PCR. Microb. Drug Resist. 2022, 28, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Earls, M.R.; Steinig, E.J.; Monecke, S.; Samaniego Castruita, J.A.; Simbeck, A.; Schneider-Brachert, W.; Vremera, T.; Dorneanu, O.S.; Loncaric, I.; Bes, M.; et al. Exploring the evolution and epidemiology of European CC1-MRSA-IV: Tracking a multidrug-resistant community-associated meticillin-resistant Staphylococcus aureus clone. Microb. Genom. 2021, 7, 000601. [Google Scholar] [CrossRef]
- Ita, T.; Luvsansharav, U.O.; Smith, R.M.; Mugoh, R.; Ayodo, C.; Oduor, B.; Jepleting, M.; Oguta, W.; Ouma, C.; Juma, J.; et al. Prevalence of colonization with multidrug-resistant bacteria in communities and hospitals in Kenya. Sci. Rep. 2022, 12, 22290. [Google Scholar] [CrossRef] [PubMed]
- Morgan Bustamante, B.L.; May, L.; Fejerman, L.; Martinez-Lopez, B. A Bayesian multilevel analysis exploring population-level effects mediating the relationship between area-level poverty and community-acquired Methicillin-resistant Staphylococcus aureus (CA-MRSA) infection across California communities. Health Place 2023, 83, 103094. [Google Scholar] [CrossRef]
- Russakoff, B.; Wood, C.; Lininger, M.R.; Barger, S.D.; Trotter, R.T.; Maltinsky, S.; Mbegbu, M.; Coyne, B.; Panisello Yague, D.; Kyman, S.; et al. A Quantitative Assessment of Staphylococcus aureus Community Carriage in Yuma, Arizona. J. Infect. Dis. 2023, 227, 1031–1041. [Google Scholar] [CrossRef]
- Fisher, T.L.; Burnet, D.L.; Huang, E.S.; Chin, M.H.; Cagney, K.A. Cultural Leverage: Interventions Using Culture to Narrow Racial Disparities in Health Care. Med. Care Res. Rev. 2007, 64, 243S–282S. [Google Scholar] [CrossRef]
- Stevens, D.L.; Bisno, A.L.; Chambers, H.F.; Patchen Dellinger, E.; Goldstein, E.J.C.; Gorbach, S.L.; Hirschmann, J.V.; Kaplan, S.L.; Montoya, J.G.; Wade, J.C. Practice guidelines for the diagnosis and management of skin and soft tissue infections: 2014 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2014, 59, e10–e52. [Google Scholar] [CrossRef]
- Tobin, J.; de la Gándara, M.P.; D’Orazio, B.; Hower, S.; Vaughan, R.; Corrêa da Rosa, J.; Kost, R.; Vasquez, K.; Evering, T.; Vasquez, K.; et al. Testing a Way to Keep Staph Infections from Recurring. 2020. Available online: https://www.pcori.org/research-results/2014/testing-way-keep-staph-infections-recurring (accessed on 6 October 2023). [CrossRef]
- Balachandra, S.; Pardos de la Gandara, M.; Salvato, S.; Urban, T.; Parola, C.; Khalida, C.; Kost, R.G.; Evering, T.H.; Pastagia, M.; D’Orazio, B.M.; et al. Recurrent furunculosis caused by a community-acquired Staphylococcus aureus strain belonging to the USA300 clone. Microb. Drug Resist. 2015, 21, 237–243. [Google Scholar] [CrossRef]
- Aires-de-Sousa, M.; Boye, K.; de Lencastre, H.; Deplano, A.; Enright, M.C.; Etienne, J.; Friedrich, A.; Harmsen, D.; Holmes, A.; Huijsdens, X.W.; et al. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J. Clin. Microbiol. 2006, 44, 619–621. [Google Scholar] [CrossRef]
- Crisostomo, M.I.; Westh, H.; Tomasz, A.; Chung, M.; Oliveira, D.C.; Lencastre, H.D. The evolution of methicillin resistance in Staphylococcus aureus: Similarity of genetic backgrounds in historically early methicillin susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 2001, 98, 9865–9870. [Google Scholar] [CrossRef] [PubMed]
- Enright, M.C.; Day, N.P.J.; Davies, C.E.; Spratt, B.G. Multilocus Sequence Typing for Characterization of Methicillin Resistant and Methicillin-Susceptible Clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.; Lencastre, H.D.; Matthews, P.; Tomasz, A.; Aadamsson, M.P.C.I.; Sousa, M.A.D.; Camou, T.; Cocuzza, C.; Corso, A.; Couto, I.; et al. Molecular Typing of Methicillin-Resistant Staphylococcus aureus by Pulsed-Field Gel Electrophoresis: Comparison of Results Obtained in a Multilaboratory Effort Using Identical Protocols and MRSA Strains. Microb. Drug Resist. 2000, 6, 189–198. [Google Scholar] [CrossRef]
- McDougal, L.K.; Steward, C.D.; Killgore, G.E.; Chaitram, J.M.; McAllister, S.K.; Tenover, F.C. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: Establishing a national database. J. Clin. Microbiol. 2003, 41, 5113–5120. [Google Scholar] [CrossRef] [PubMed]
- Milheirico, C.; Oliveira, D.C.; de Lencastre, H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob. Agents Chemother. 2007, 51, 3374–3377. [Google Scholar] [CrossRef]
- Milheirico, C.; Oliveira, D.C.; de Lencastre, H. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’. J. Antimicrob. Chemother. 2007, 60, 42–48. [Google Scholar] [CrossRef]
- Diep, B.A.; Stone, G.G.; Basuino, L.; Graber, C.J.; Miller, A.; des Etages, S.A.; Jones, A.; Palazzolo-Ballance, A.M.; Perdreau-Remington, F.; Sensabaugh, G.F.; et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: Convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 2008, 197, 1523–1530. [Google Scholar] [CrossRef]
- Vandenesch, F.; Naimi, T.; Enright, M.C.; Lina, G.; Nimmo, G.R.; Heffernan, H.; Liassine, N.; Bes, M.; Greenland, T.; Reverdy, M.-E.; et al. Community-Acquired Methicillin Resistant Staphylococcus aureus Carrying Panton-Valentine Leukocidin Genes: Worldwide Emergence. Emerg. Infect. Dis. 2003, 9, 978–984. [Google Scholar] [CrossRef]
- Pardos de la Gandara, M.; Raygoza Garay, J.A.; Mwangi, M.; Tobin, J.N.; Tsang, A.; Khalida, C.; D’Orazio, B.; Kost, R.G.; Leinberger-Jabari, A.; Coffran, C.; et al. Molecular Types of Methicillin-Resistant Staphylococcus aureus and Methicillin-Sensitive Staphylococcus aureus Strains Causing Skin and Soft Tissue Infections and Nasal Colonization, Identified in Community Health Centers in New York City. J. Clin. Microbiol. 2015, 53, 2648–2658. [Google Scholar] [CrossRef]
- Oliveira, D.C.; de Lencastre, H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2002, 46, 2155–2161. [Google Scholar] [CrossRef] [PubMed]
- City of New York. NYC Open Data. Available online: https://opendata.cityofnewyork.us/ (accessed on 6 November 2019).
- U.S. Census Bureau. Explore Census Data. Available online: https://data.census.gov/ (accessed on 26 April 2020).
- Esri. ArcGIS Pro; Esri, Inc.: Redlands, CA, USA, 2022. [Google Scholar]
- Esri Data Development. 2022 Esri Tapestry Segmentation. Available online: https://storymaps.arcgis.com/stories/6e8f2d8c08d8427892e816d1aeb373f8 (accessed on 19 April 2023).


| US-Born (n = 81) | Non-US-Born (n = 46) | Total (n = 127) | p-Value | |
|---|---|---|---|---|
| Gender, n (%) | 0.75 1 | |||
| Female | 34 (42.0%) | 18 (39.1%) | 52 (40.9%) | |
| Male | 47 (58.0%) | 28 (60.9%) | 75 (59.1%) | |
| Race, n (%) | 0.008 2 | |||
| Black | 27 (45.8%) | 3 (13.6%) | 30 (37.0%) | |
| White | 17 (28.8%) | 6 (27.3%) | 23 (28.4%) | |
| Other Race | 15 (25.4%) | 13 (59.1%) | 28 (34.6%) | |
| Missing | 22 | 24 | 46 | |
| Ethnicity, n (%) | 0.007 1 | |||
| Hispanic | 43 (57.3%) | 33 (82.5%) | 76 (66.1%) | |
| Non-Hispanic | 32 (42.7%) | 7 (17.5%) | 39 (33.9%) | |
| Missing | 6 | 6 | 12 | |
| Age (years), n (%) | 0.01 2 | |||
| <19 | 10 (12.3%) | 0 (0.0%) | 10 (7.9%) | |
| 19–45 | 43 (53.1%) | 31 (67.4%) | 74 (58.3%) | |
| 45–65 | 23 (28.4%) | 15 (32.6%) | 38 (29.9%) | |
| >65 | 5 (6.2%) | 0 (0.0%) | 5 (3.9%) | |
| Marital Status, n (%) | <0.0001 1 | |||
| Couple | 16 (19.8%) | 28 (63.6%) | 44 (35.2%) | |
| Single | 65 (80.2%) | 16 (36.4%) | 81 (64.8%) | |
| Missing | 0 | 2 | 2 | |
| Education, n (%) | 0.001 2 | |||
| High School or lower | 54 (67.5%) | 30 (66.7%) | 84 (67.2%) | |
| College | 21 (26.3%) | 3 (6.7%) | 24 (19.2%) | |
| Bachelor or higher | 5 (6.3%) | 12 (26.7%) | 17 (13.6%) | |
| Missing | 1 | 1 | 2 | |
| Health Insurance, n (%) | <0.0001 1 | |||
| Private or Other | 14 (17.3%) | 10 (22.7%) | 24 (19.2%) | |
| Public (Medicare or Medicaid) | 59 (72.8%) | 14 (31.8%) | 73 (58.4%) | |
| None | 8 (9.9%) | 20 (45.5%) | 28 (22.4%) | |
| Missing | 0 | 2 | 2 | |
| Health Quality, n (%) | 0.77 2 | |||
| Good | 50 (64.9%) | 32 (71.1%) | 82 (67.2%) | |
| Fair | 22 (28.6%) | 11 (24.4%) | 33 (27.0%) | |
| Poor | 5 (6.5%) | 2 (4.4%) | 7 (5.7%) | |
| Missing | 4 | 1 | 5 | |
| Income, n (%) | 0.50 2 | |||
| <USD 40,000 | 56 (90.3%) | 27 (84.4%) | 83 (88.3%) | |
| ≥USD 40,000 | 6 (9.7%) | 5 (15.6%) | 11 (11.7%) | |
| Missing | 19 | 14 | 33 | |
| First Time Infection, n (%) | 0.22 1 | |||
| Yes | 53 (69.7%) | 36 (80.0%) | 89 (73.6%) | |
| No | 23 (30.3%) | 9 (20.0%) | 32 (26.4%) | |
| Missing | 5 | 1 | 6 | |
| Crowding Life Environment, n (%) | 0.27 1 | |||
| Yes | 47 (63.5%) | 33 (73.3%) | 80 (67.2%) | |
| No | 27 (36.5%) | 12 (26.7%) | 39 (32.8%) | |
| Missing | 7 | 1 | 8 | |
| Healthcare Exposure, n (%) | 0.24 1 | |||
| Yes | 30 (39.5%) | 13 (28.9%) | 43 (35.5%) | |
| No | 46 (60.5%) | 32 (71.1%) | 78 (64.5%) | |
| Missing | 5 | 1 | 6 | |
| Animal Contact, n (%) | 0.17 1 | |||
| Yes | 30 (39.0%) | 12 (26.7%) | 42 (34.4%) | |
| No | 47 (61.0%) | 33 (73.3%) | 80 (65.6%) | |
| Missing | 4 | 1 | 5 | |
| Had Wounds, n (%) | 0.15 1 | |||
| Yes | 29 (37.2%) | 11 (24.4%) | 40 (32.5%) | |
| No | 49 (62.8%) | 34 (75.6%) | 83 (67.5%) | |
| Missing | 3 | 1 | 4 | |
| Social Network, n (%) | 0.76 1 | |||
| Yes | 17 (22.4%) | 9 (20.0%) | 26 (21.5%) | |
| No | 59 (77.6%) | 36 (80.0%) | 95 (78.5%) | |
| Missing | 5 | 1 | 6 | |
| Household Crowding, n (%) | 0.11 1 | |||
| <2 People | 25 (35.7%) | 8 (21.1%) | 33 (30.6%) | |
| >2 People | 45 (64.3%) | 30 (78.9%) | 75 (69.4%) | |
| Missing | 11 | 8 | 19 | |
| Personal Hygiene, n (%) | 0.35 1 | |||
| Not Sharing | 28 (40.6%) | 19 (50.0%) | 47 (43.9%) | |
| Sharing | 41 (59.4%) | 19 (50.0%) | 60 (56.1%) | |
| Missing | 12 | 8 | 20 | |
| Hand Washing, n (%) | 0.53 1 | |||
| <10 Times/Day | 53 (67.9%) | 33 (73.3%) | 86 (69.9%) | |
| >10 Times/Day | 25 (32.1%) | 12 (26.7%) | 37 (30.1%) | |
| Missing | 3 | 1 | 4 |
| US-Born (n = 81) | Non-US-Born (n = 46) | Total (n = 127) | p-Value | |
|---|---|---|---|---|
| S. aureus, n (%) | 0.11 1 | |||
| MRSA | 47 (58.0%) | 20 (43.5%) | 67 (52.8%) | |
| MSSA | 34 (42.0%) | 26 (56.5%) | 60 (47.2%) | |
| S. aureus Genotype, n (%) | 0.04 1 | |||
| USA300 | 38 (46.9%) | 13 (28.3%) | 51 (40.2%) | |
| Non-USA300 | 43 (53.1%) | 33 (71.7%) | 76 (59.8%) | |
| S. aureus + Genotype, n (%) | 0.17 1 | |||
| MRSA, USA300 | 30 (37.0%) | 9 (19.6%) | 39 (30.7%) | |
| MRSA, non-USA300 | 17 (21.0%) | 11 (23.9%) | 28 (22.0%) | |
| MSSA, USA300 | 8 (9.9%) | 4 (8.7%) | 12 (9.4%) | |
| MSSA, non-USA300 | 26 (32.1%) | 22 (47.8%) | 48 (37.8%) | |
| mecA Gene from S. aureus Wound, n (%) | 0.16 2 | |||
| Positive | 42 (54.5%) | 18 (39.1%) | 60 (48.8%) | |
| Negative | 32 (41.6%) | 27 (58.7%) | 59 (48.0%) | |
| Not Determined | 3 (3.9%) | 1 (2.2%) | 4 (3.3%) | |
| Missing | 4 | 0 | 4 | |
| ACME Gene from S. aureus Wound, n (%) | 0.04 2 | |||
| Negative | 42 (54.5%) | 36 (78.3%) | 78 (63.4%) | |
| Type I | 31 (40.3%) | 9 (19.6%) | 40 (32.5%) | |
| Type III | 1 (1.3%) | 0 (0.0%) | 1 (0.8%) | |
| Not Determined | 3 (3.9%) | 1 (2.2%) | 4 (3.3%) | |
| Missing | 4 | 0 | 4 | |
| SCCmec Gene from S. aureus Wound, n (%) | 0.82 2 | |||
| Negative | 1 (1.3%) | 1 (2.2%) | 2 (1.6%) | |
| IVa | 35 (45.5%) | 17 (37.0%) | 52 (42.3%) | |
| IVb | 1 (1.3%) | 0 (0.0%) | 1 (0.8%) | |
| IVc | 2 (2.6%) | 1 (2.2%) | 3 (2.4%) | |
| IVg | 1 (1.3%) | 0 (0.0%) | 1 (0.8%) | |
| IVh | 1 (1.3%) | 0 (0.0%) | 1 (0.8%) | |
| Not Determined | 34 (44.2%) | 27 (58.7%) | 61 (49.6%) | |
| Novel Type | 2 (2.6%) | 0 (0.0%) | 2 (1.6%) | |
| Missing | 4 | 0 | 4 | |
| PVL Gene from S. aureus Wound, n (%) | 0.36 1 | |||
| Positive | 47 (61.0%) | 23 (50.0%) | 70 (56.9%) | |
| Negative | 27 (35.1%) | 22 (47.8%) | 49 (39.8%) | |
| Not Determined | 3 (3.9%) | 1 (2.2%) | 4 (3.3%) | |
| Missing | 4 | 0 | 4 |
| US-Born | Non-US-Born | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MRSA USA300 (n = 30) | MRSA Non-USA300 (n = 17) | MSSA USA300 (n = 8) | MSSA Non-USA300 (n = 26) | p-Value | MRSA USA300 (n = 9) | MRSA Non-USA300 (n = 11) | MSSA USA300 (n = 4) | MSSA Non-USA300 (n = 22) | p-Value | |
| Gender, n (%) | 0.47 1 | 0.89 2 | ||||||||
| Female | 14 (46.7%) | 9 (52.9%) | 3 (37.5%) | 8 (30.8%) | 4 (44.4%) | 5 (45.5%) | 1 (25.0%) | 8 (36.4%) | ||
| Male | 16 (53.3%) | 8 (47.1%) | 5 (62.5%) | 18 (69.2%) | 5 (55.6%) | 6 (54.5%) | 3 (75.0%) | 14 (63.6%) | ||
| Race, n (%) | 0.98 2 | 0.41 2 | ||||||||
| Black | 10 (50.0%) | 5 (38.5%) | 3 (50.0%) | 9 (45.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 3 (25.0%) | ||
| White | 5 (25.0%) | 5 (38.5%) | 2 (33.3%) | 5 (25.0%) | 2 (50.0%) | 0 (0.0%) | 0 (0.0%) | 4 (33.3%) | ||
| Other Race | 5 (25.0%) | 3 (23.1%) | 1 (16.7%) | 6 (30.0%) | 2 (50.0%) | 4 (100.0%) | 2 (100.0%) | 5 (41.7%) | ||
| Missing | 10 | 4 | 2 | 6 | 5 | 7 | 2 | 10 | ||
| Ethnicity, n (%) | 0.87 1 | 0.19 2 | ||||||||
| Hispanic | 17 (60.7%) | 10 (58.8%) | 5 (62.5%) | 11 (50.0%) | 6 (66.7%) | 10 (100.0%) | 4 (100.0%) | 13 (76.5%) | ||
| Non-Hispanic | 11 (39.3%) | 7 (41.2%) | 3 (37.5%) | 11 (50.0%) | 3 (33.3%) | 0 (0.0%) | 0 (0.0%) | 4 (23.5%) | ||
| Missing | 2 | 0 | 0 | 4 | 0 | 1 | 0 | 5 | ||
| Age (years), n (%) | 0.46 2 | 0.08 2 | ||||||||
| <19 | 2 (6.7%) | 4 (23.5%) | 1 (12.5%) | 3 (11.5%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | ||
| 19–45 | 20 (66.7%) | 8 (47.1%) | 2 (25.0%) | 13 (50.0%) | 5 (55.6%) | 10 (90.9%) | 4 (100.0%) | 12 (54.5%) | ||
| 45–65 | 7 (23.3%) | 4 (23.5%) | 4 (50.0%) | 8 (30.8%) | 4 (44.4%) | 1 (9.1%) | 0 (0.0%) | 10 (45.5%) | ||
| >65 | 1 (3.3%) | 1 (5.9%) | 1 (12.5%) | 2 (7.7%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | ||
| Marital Status, n (%) | 0.50 2 | 0.85 2 | ||||||||
| Couple | 6 (20.0%) | 5 (29.4%) | 2 (25.0%) | 3 (11.5%) | 5 (62.5%) | 8 (72.7%) | 3 (75.0%) | 12 (57.1%) | ||
| Single | 24 (80.0%) | 12 (70.6%) | 6 (75.0%) | 23 (88.5%) | 3 (37.5%) | 3 (27.3%) | 1 (25.0%) | 9 (42.9%) | ||
| Missing | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | ||
| Education, n (%) | 0.71 2 | 0.77 2 | ||||||||
| High School or lower | 19 (63.3%) | 11 (64.7%) | 5 (62.5%) | 19 (76.0%) | 5 (62.5%) | 6 (54.5%) | 4 (100.0%) | 15 (68.2%) | ||
| College | 10 (33.3%) | 4 (23.5%) | 2 (25.0%) | 5 (20.0%) | 0 (0.0%) | 1 (9.1%) | 0 (0.0%) | 2 (9.1%) | ||
| Bachelor or higher | 1 (3.3%) | 2 (11.8%) | 1 (12.5%) | 1 (4.0%) | 3 (37.5%) | 4 (36.4%) | 0 (0.0%) | 5 (22.7%) | ||
| Missing | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | ||
| Health Insurance, n (%) | 0.79 2 | 0.86 2 | ||||||||
| Private/Other | 8 (26.7%) | 2 (11.8%) | 1 (12.5%) | 3 (11.5%) | 2 (25.0%) | 2 (18.2%) | 1 (25.0%) | 5 (23.8%) | ||
| Public (Medicare or Medicaid) | 20 (66.7%) | 13 (76.5%) | 6 (75.0%) | 20 (76.9%) | 2 (25.0%) | 4 (36.4%) | 0 (0.0%) | 8 (38.1%) | ||
| None | 2 (6.7%) | 2 (11.8%) | 1 (12.5%) | 3 (11.5%) | 4 (50.0%) | 5 (45.5%) | 3 (75.0%) | 8 (38.1%) | ||
| Missing | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | ||
| Health Quality, n (%) | 0.31 2 | 0.68 2 | ||||||||
| Good | 17 (58.6%) | 11 (73.3%) | 5 (62.5%) | 17 (68.0%) | 7 (87.5%) | 9 (81.8%) | 2 (50.0%) | 14 (63.6%) | ||
| Fair | 10 (34.5%) | 3 (20.0%) | 1 (12.5%) | 8 (32.0%) | 1 (12.5%) | 2 (18.2%) | 2 (50.0%) | 6 (27.3%) | ||
| Poor | 2 (6.9%) | 1 (6.7%) | 2 (25.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 2 (9.1%) | ||
| Missing | 1 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | ||
| Income, n (%) | 0.35 2 | 0.88 2 | ||||||||
| <USD 40,000 | 25 (96.2%) | 10 (83.3%) | 4 (80.0%) | 17 (89.5%) | 5 (83.3%) | 5 (83.3%) | 2 (66.7%) | 15 (88.2%) | ||
| ≥USD 40,000 | 1 (3.8%) | 2 (16.7%) | 1 (20.0%) | 2 (10.5%) | 1 (16.7%) | 1 (16.7%) | 1 (33.3%) | 2 (11.8%) | ||
| Missing | 4 | 5 | 3 | 7 | 3 | 5 | 1 | 5 | ||
| Years in the US, Average (SD) | 36.1 (13.2) | 31.9 (17.9) | 42.4 (15.1) | 33.8 (16.8) | 0.44 3 | 11.7 (6.0) | 7.8 (8.3) | 7.3 (1.2) | 17.8 (11.9) | 0.08 3 |
| First Time Infection, n (%) | 0.27 1 | 0.42 2 | ||||||||
| Yes | 18 (62.1%) | 13 (86.7) | 7 (87.5%) | 15 (62.5%) | 7 (87.5%) | 7 (63.6%) | 3 (75.0%) | 19 (86.4%) | ||
| No | 11 (37.9%) | 2 (13.3%) | 1 (12.5%) | 9 (37.5%) | 1 (12.5%) | 4 (36.4%) | 1 (25.0%) | 3 (13.6%) | ||
| Missing | 1 | 2 | 0 | 2 | 1 | 0 | 0 | 0 | ||
| Crowded Life Environment, n (%) | 0.08 1 | 0.82 2 | ||||||||
| Yes | 20 (76.9%) | 11 (73.3%) | 5 (62.5%) | 11 (44.0%) | 7 (87.5%) | 8 (72.7%) | 3 (75.0%) | 15 (68.2%) | ||
| No | 6 (23.1%) | 4 (26.7%) | 3 (37.5%) | 14 (56.0%) | 1 (12.5%) | 3 (27.3%) | 1 (25.0%) | 7 (31.8%) | ||
| Missing | 4 | 2 | 9 | 1 | 1 | 0 | 0 | 0 | ||
| Healthcare Exposure, n (%) | 0.84 1 | 0.46 2 | ||||||||
| Yes | 9 (33.3%) | 7 (46.7%) | 3 (37.5%) | 11 (42.3%) | 3 (37.5%) | 2 (18.2%) | 0 (0.0%) | 8 (36.4%) | ||
| No | 18 (66.7%) | 8 (53.3%) | 6 (62.5%) | 15 (57.7%) | 5 (62.5%) | 9 (81.8%) | 4 (100.0%) | 14 (63.6%) | ||
| Missing | 3 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | ||
| Animal Contact, n (%) | 0.42 1 | 0.06 2 | ||||||||
| Yes | 14 (50.0%) | 6 (40.0%) | 2 (25.0%) | 8 (30.8%) | 3 (37.5%) | 3 (27.3%) | 3 (75.0%) | 3 (13.6%) | ||
| No | 14 (50.0%) | 9 (60.0%) | 6 (75.0%) | 18 (69.2%) | 5 (62.5%) | 8 (72.7%) | 1 (25.0%) | 19 (86.4%) | ||
| Missing | 2 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | ||
| Previous Wounds, n (%) | 0.19 1 | 0.80 2 | ||||||||
| Yes | 9 (31.0%) | 4 (26.7%) | 2 (25.0%) | 14 (53.8%) | 1 (12.5%) | 2 (18.2%) | 1 (25.0%) | 7 (31.8%) | ||
| No | 20 (69.0%) | 11 (73.3%) | 6 (75.0%) | 12 (46.2%) | 7 (87.5%) | 9 (81.8%) | 3 (75.0%) | 15 (68.2%) | ||
| Missing | 1 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | ||
| Social Network, n (%) | 0.14 2 | 1.0 2 | ||||||||
| Yes | 6 (21.4%) | 6 (42.9%) | 0 (0.0%) | 5 (19.2%) | 1 (12.5%) | 2 (18.2%) | 1 (25.0%) | 5 (22.7%) | ||
| No | 22 (78.6%) | 8 (57.1%) | 8 (100%) | 21 (80.8%) | 7 (87.5%) | 9 (81.8%) | 3 (75.0%) | 17 (77.3%) | ||
| Missing | 2 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | ||
| Household Crowding, n (%) | 0.39 1 | 0.55 2 | ||||||||
| <2 People/Room | 8 (30.8%) | 7 (50.0%) | 1 (14.3%) | 9 (39.1%) | 1 (16.7%) | 1 (11.1%) | 0 (0.0%) | 6 (31.6%) | ||
| >2 People/Room | 18 (69.2%) | 7 (50.0%) | 6 (85.7%) | 14 (60.9%) | 5 (83.3%) | 8 (88.9%) | 4 (100.0%) | 13 (68.4%) | ||
| Missing | 4 | 3 | 1 | 3 | 3 | 2 | 0 | 3 | ||
| Personal Hygiene, n (%) | 0.38 2 | 0.10 2 | ||||||||
| Sharing | 12 (46.2%) | 10 (71.4%) | 5 (71.4%) | 14 (63.6%) | 1 (16.7%) | 3 (33.3%) | 2 (50.0%) | 13 (68.4%) | ||
| Not Sharing | 14 (53.8%) | 4 (28.6%) | 2 (28.6%) | 8 (36.4%) | 5 (83.3%) | 6 (66.7%) | 2 (50.0%) | 6 (31.6%) | ||
| Missing | 4 | 3 | 1 | 4 | 3 | 2 | 0 | 3 | ||
| Hand Washing, n (%) | 0.81 1 | 0.12 2 | ||||||||
| <10 times/day | 18 (62.1%) | 10 (66.7%) | 6 (75.0%) | 19 (73.1%) | 7 (87.5%) | 5 (45.5%) | 3 (75.0%) | 18 (81.8%) | ||
| >10 times/day | 11 (37.9%) | 5 (33.3%) | 32 (25.0%) | 7 (26.9%) | 1 (12.5%) | 6 (54.5%) | 1 (25.0%) | 4 (18.2%) | ||
| Missing | 1 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | ||
| US-Born | Non-US-Born | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MRSA USA300 (n = 30) | MRSA Non-USA300 (n = 17) | MSSA USA300 (n = 8) | MSSA Non-USA300 (n = 26) | p-Value | MRSA USA300 (n = 9) | MRSA Non-USA300 (n = 11) | MSSA USA300 (n = 4) | MSSA Non-USA300 (n = 22) | p-Value | |
| mecA Gene from S. aureus Wound, n (%) | <0.0001 | <0.0001 | ||||||||
| Positive | 30 (100.0%) | 12 (75.0%) | 0 (0.0%) | 0 (0.0%) | 9 (100.0%) | 9 (81.8%) | 0 (0.0%) | 0 (0.0%) | ||
| Negative | 0 (0.0%) | 4 (25.0%) | 7 (100.0%) | 21 (87.5%) | 0 (0.0%) | 2 (18.2%) | 4 (100.0%) | 21 (95.5%) | ||
| ND | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 3 (12.5%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (4.5%) | ||
| Missing | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | ||
| ACME Gene from S. aureus Wound, n (%) | <0.0001 | <0.0001 | ||||||||
| Type I | 26 (86.7%) | 3 (18.8%) | 2 (28.6%) | 0 (0.0%) | 7 (77.8%) | 2 (18.2%) | 0 (0.0%) | 0 (0.0%) | ||
| Type III | 1 (3.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | ||
| Negative | 3 (10.0%) | 13 (81.3%) | 5 (71.4%) | 21 (87.5%) | 2 (25.0%) | 9 (81.8%) | 4 (100.0%) | 21 (95.5%) | ||
| ND | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 3 (12.5%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (4.5%) | ||
| Missing | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | ||
| SCCmec Gene from S. aureus Wound, n (%) | <0.0001 | <0.0001 | ||||||||
| IVa | 29 (96.7%) | 6 (37.5%) | 0 (0.0%) | 0 (0.0%) | 9 (100.0%) | 8 (72.7%) | 0 (0.0%) | 0 (0.0%) | ||
| IVb | 0 (0.0%) | 1 (6.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | ||
| IVc | 1 (3.3%) | 1 (6.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (9.1%) | 0 (0.0%) | 0 (0.0%) | ||
| IVg | 0 (0.0%) | 1 (6.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | ||
| IVh | 0 (0.0%) | 1 (6.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | ||
| Novel Type | 0 (0.0%) | 2 (12.5%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | ||
| Negative | 0 (0.0%) | 0 (0.0%) | 1 (14.3%) | 0 (0.0%) | 0 (0.0%) | 1 (9.1%) | 0 (0.0%) | 0 (0.0%) | ||
| Not Determined | 0 (0.0%) | 4 (25.0%) | 6 (85.7%) | 24 (100.0%) | 0 (0.0%) | 1 (9.1%) | 4 (100.0%) | 22 (100.0%) | ||
| PVL Gene from S. aureus Wound, n (%) | <0.0001 | 0.08 | ||||||||
| Positive | 27 (90.0%) | 9 (56.3%) | 5 (71.4%) | 6 (25.0%) | 8 (88.9%) | 6 (54.5%) | 1 (25.0%) | 8 (36.4%) | ||
| Negative | 3 (10.0%) | 7 (43.8%) | 2 (28.6%) | 15 (62.5%) | 1 (11.1%) | 5 (45.5%) | 3 (75.0%) | 13 (59.1%) | ||
| ND | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 3 (12.5%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (4.5%) | ||
| Missing | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | ||
| Unadjusted | Adjusted | |||||
|---|---|---|---|---|---|---|
| OR | 95% CI | p-Value | OR | 95% CI | p-Value | |
| Birthplace | 0.06 | 0.03 | ||||
| US-born | 2.55 | 0.99–7.20 | 3.20 | 1.15–9.89 | ||
| Non-US-born | ref | ref | ||||
| Crowded Living | 0.16 | 0.07 | ||||
| ≥2 People/Room | 1.89 | 0.79–4.75 | 3.04 | 0.93–10.95 | ||
| <2 People/Room | ref | ref | ||||
| Contact Animal | 0.03 | 0.10 | ||||
| Yes | 2.53 | 1.08–6.06 | 2.13 | 0.87–5.31 | ||
| No | ref | ref | ||||
| Age, Unit = 1 Year | 1.00 | 0.98–1.03 | 0.88 | 1.03 | 0.99–1.07 | 0.14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Immergluck, L.C.; Lin, X.; Geng, R.; Edelson, M.; Ali, F.; Li, C.; Lin, T.; Khalida, C.; Piper-Jenks, N.; Pardos de la Gandara, M.; et al. Molecular Epidemiologic and Geo-Spatial Characterization of Staphylococcus aureus Cultured from Skin and Soft Tissue Infections from United States-Born and Immigrant Patients Living in New York City. Antibiotics 2023, 12, 1541. https://doi.org/10.3390/antibiotics12101541
Immergluck LC, Lin X, Geng R, Edelson M, Ali F, Li C, Lin T, Khalida C, Piper-Jenks N, Pardos de la Gandara M, et al. Molecular Epidemiologic and Geo-Spatial Characterization of Staphylococcus aureus Cultured from Skin and Soft Tissue Infections from United States-Born and Immigrant Patients Living in New York City. Antibiotics. 2023; 12(10):1541. https://doi.org/10.3390/antibiotics12101541
Chicago/Turabian StyleImmergluck, Lilly Cheng, Xiting Lin, Ruijin Geng, Mike Edelson, Fatima Ali, Chaohua Li, TJ Lin, Chamanara Khalida, Nancy Piper-Jenks, Maria Pardos de la Gandara, and et al. 2023. "Molecular Epidemiologic and Geo-Spatial Characterization of Staphylococcus aureus Cultured from Skin and Soft Tissue Infections from United States-Born and Immigrant Patients Living in New York City" Antibiotics 12, no. 10: 1541. https://doi.org/10.3390/antibiotics12101541
APA StyleImmergluck, L. C., Lin, X., Geng, R., Edelson, M., Ali, F., Li, C., Lin, T., Khalida, C., Piper-Jenks, N., Pardos de la Gandara, M., de Lencastre, H., Tomasz, A., Evering, T. H., Kost, R. G., Vaughan, R., & Tobin, J. N. (2023). Molecular Epidemiologic and Geo-Spatial Characterization of Staphylococcus aureus Cultured from Skin and Soft Tissue Infections from United States-Born and Immigrant Patients Living in New York City. Antibiotics, 12(10), 1541. https://doi.org/10.3390/antibiotics12101541








