Current Promising Strategies against Antibiotic-Resistant Bacterial Infections
Abstract
1. Introduction
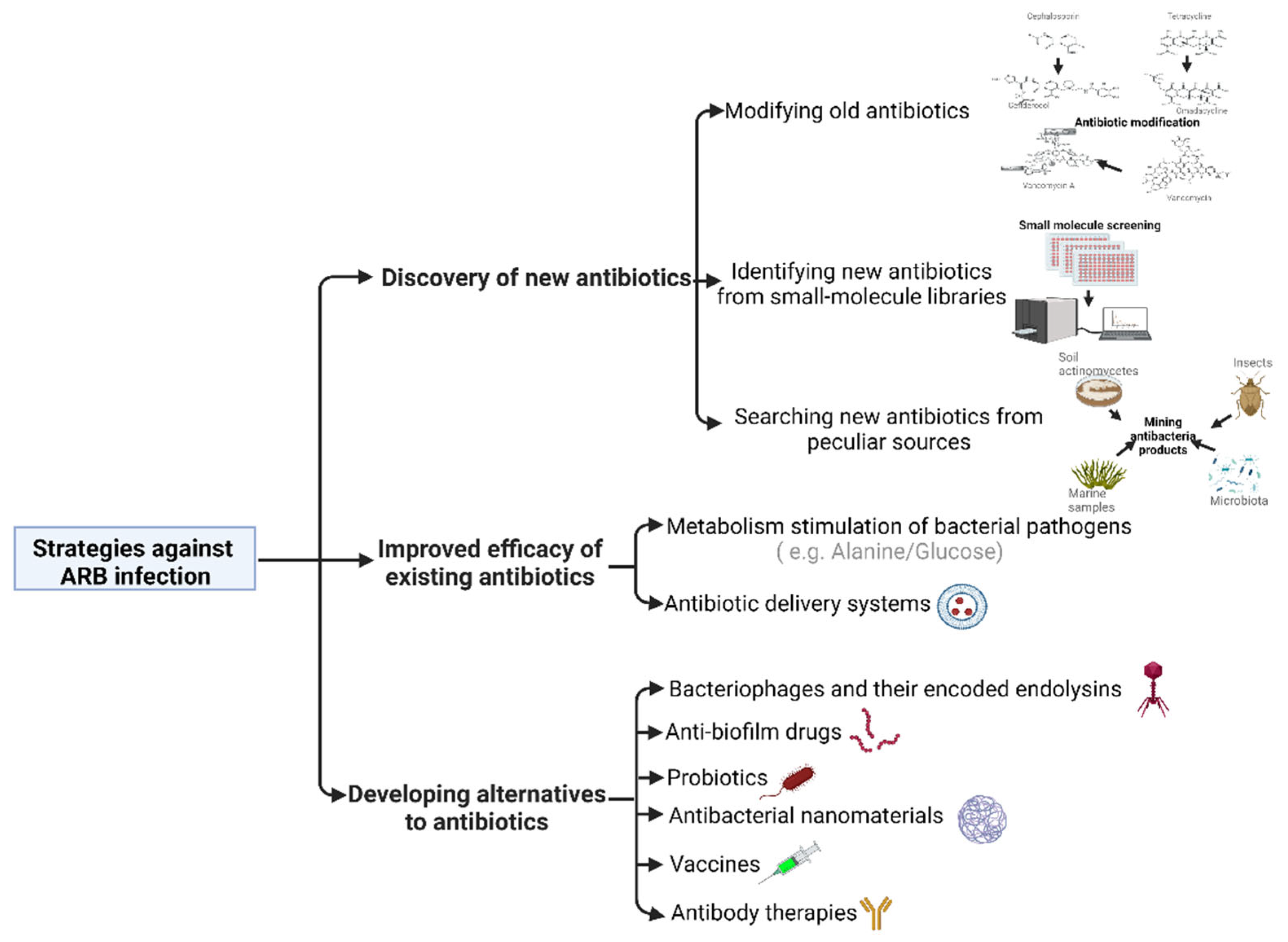
2. Discovery of New Antibiotics
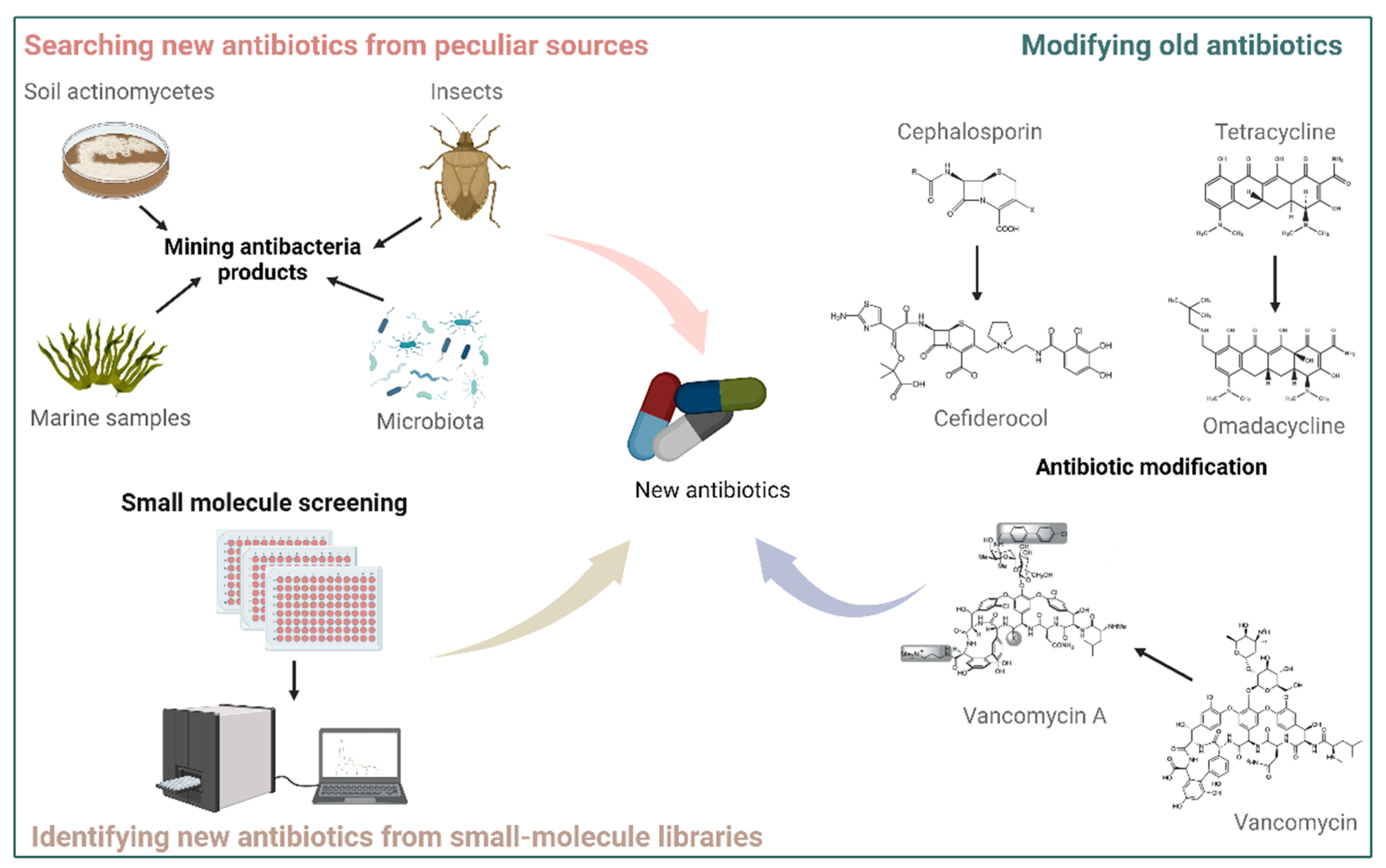
2.1. Modifying Old Antibiotics
2.2. Identifying New Antibiotics from Small-Molecule Libraries
2.3. Searching for New Antibiotics from Peculiar Sources
3. Improved Efficacy of Existing Antibiotics
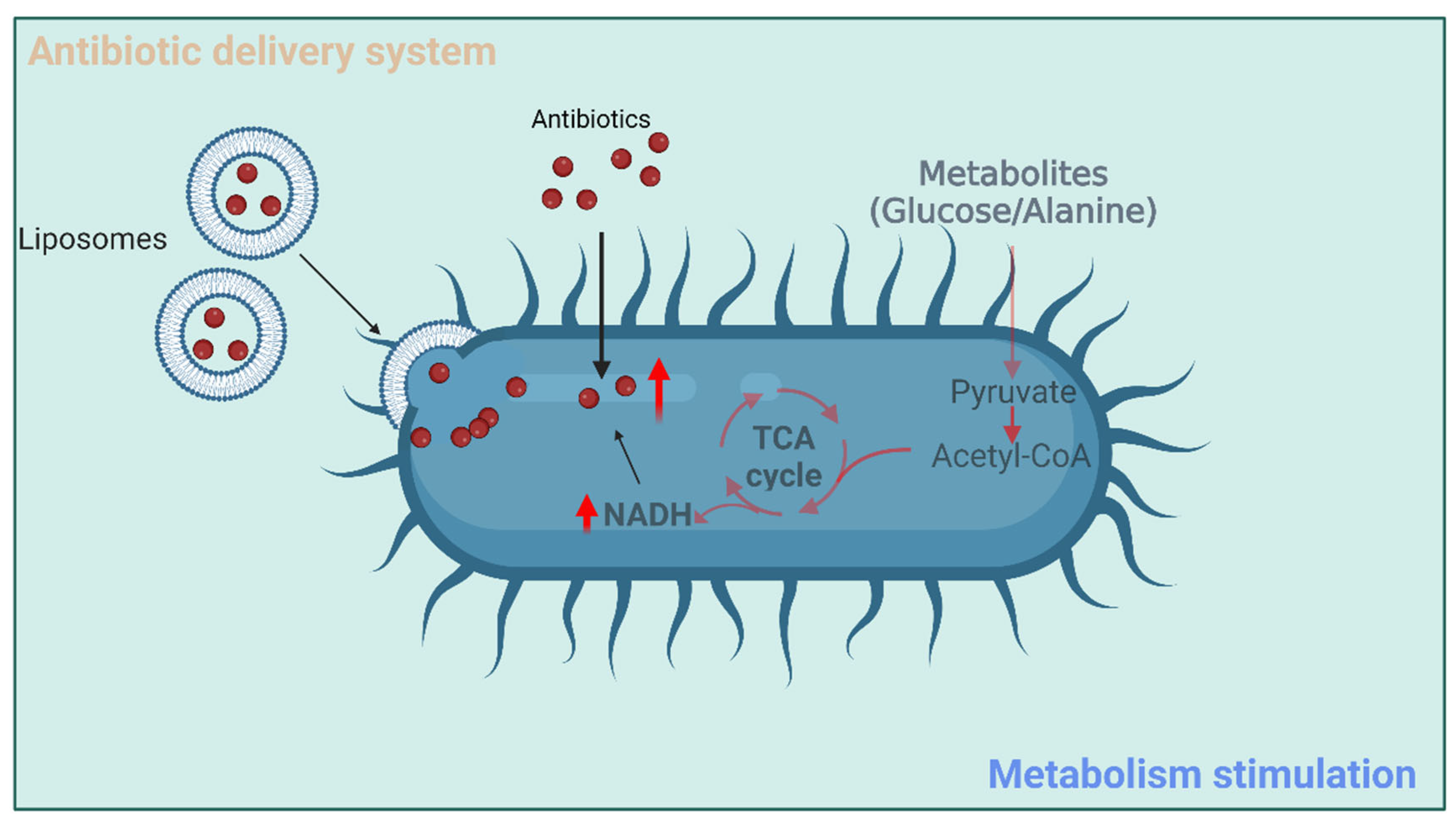
3.1. Metabolism Stimulation of Bacterial Pathogens
3.2. Antibiotic Delivery Systems
4. Developing Alternatives to Antibiotics
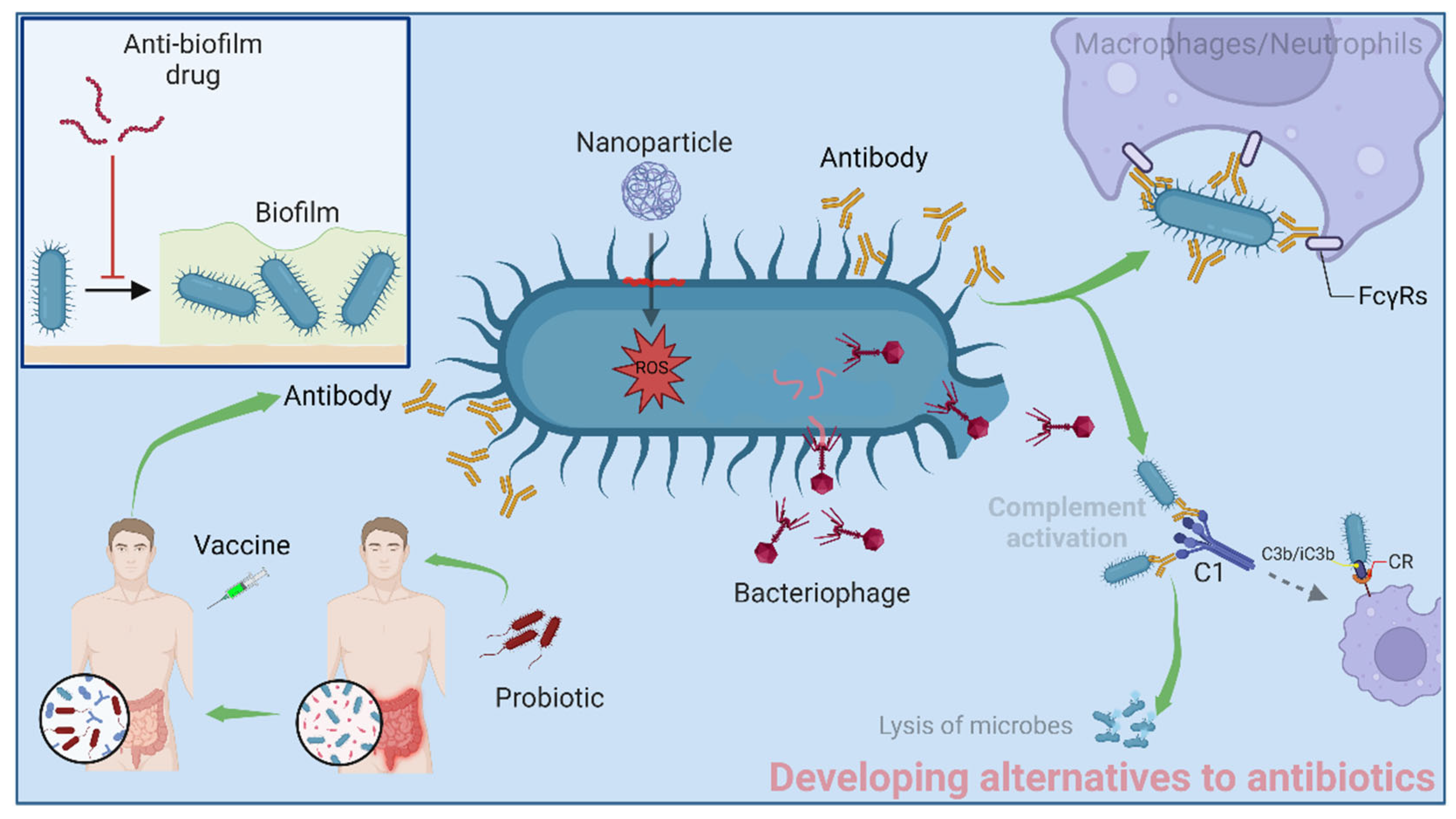
4.1. Bacteriophages and Their Encoded Endolysins
4.2. Anti-Biofilm Drugs
4.3. Probiotics
4.4. Antibacterial Nanomaterials
4.5. Vaccines
4.6. Antibody Therapies
5. Future Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J. Old and modern antibiotic structures with potential for today’s infections. ADMET DMPK 2022, 10, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Lawandi, A.; Kadri, S.S. Can financial rewards for stewardship in primary care curb antibiotic resistance? Lancet Infect. Dis. 2021, 21, 1618–1620. [Google Scholar] [CrossRef] [PubMed]
- Aliabadi, S.; Anyanwu, P.; Beech, E.; Jauneikaite, E.; Wilson, P.; Hope, R.; Majeed, A.; Muller-Pebody, B.; Costelloe, C. Effect of antibiotic stewardship interventions in primary care on antimicrobial resistance of Escherichia coli bacteraemia in England (2013–18): A quasi-experimental, ecological, data linkage study. Lancet Infect. Dis. 2021, 21, 1689–1700. [Google Scholar] [CrossRef] [PubMed]
- Pickens, C.I.; Wunderink, R.G. Principles and Practice of Antibiotic Stewardship in the ICU. Chest 2019, 156, 163–171. [Google Scholar] [CrossRef]
- Lindsay, P.J.; Rohailla, S.; Taggart, L.R.; Lightfoot, D.; Havey, T.; Daneman, N.; Lowe, C.; Muller, M.P. Antimicrobial Stewardship and Intensive Care Unit Mortality: A Systematic Review. Clin. Infect. Dis. 2019, 68, 748–756. [Google Scholar] [CrossRef]
- Karlowsky, J.A.; Steenbergen, J.; Zhanel, G.G. Microbiology and preclinical review of omadacycline. Clin. Infect. Dis. 2019, 69, S6–S15. [Google Scholar] [CrossRef]
- Wu, J.Y.; Srinivas, P.; Pogue, J.M. Cefiderocol: A Novel Agent for the Management of Multidrug-Resistant Gram-Negative Organisms. Infect. Dis. 2020, 9, 17–40. [Google Scholar] [CrossRef]
- Hsu, Y.-P.; Meng, X.; VanNieuwenhze, M. Methods for visualization of peptidoglycan biosynthesis. In Methods in Microbiology; Elsevier: Amsterdam, The Netherlands, 2016; Volume 43, pp. 3–48. [Google Scholar]
- Vollmer, W. Peptidoglycan. In Molecular Medical Microbiology; Elsevier: Amsterdam, The Netherlands, 2015; pp. 105–124. [Google Scholar]
- Okano, A.; Isley, N.A.; Boger, D.L. Peripheral modifications of [Ψ [CH2NH] Tpg4] vancomycin with added synergistic mechanisms of action provide durable and potent antibiotics. Proc. Natl. Acad. Sci. USA 2017, 114, E5052–E5061. [Google Scholar] [CrossRef]
- Wu, Z.-C.; Cameron, M.D.; Boger, D.L. Vancomycin C-Terminus Guanidine Modifications and Further Insights into an Added Mechanism of Action Imparted by a Peripheral Structural Modification. ACS Infect. Dis. 2020, 6, 2169–2180. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.-C.; Boger, D.L. Maxamycins: Durable antibiotics derived by rational redesign of vancomycin. Acc. Chem. Res. 2020, 53, 2587–2599. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.J.; Qu, S.; Tan, C.; Cai, Y.; Mogi, Y.; Jamin Keith, D.; Boger, D.L. Next-generation total synthesis of vancomycin. J. Am. Chem. Soc. 2020, 142, 16039–16050. [Google Scholar] [CrossRef]
- Flint, A.J.; Davis, A.P. Vancomycin mimicry: Towards new supramolecular antibiotics. Org. Biomol. Chem. 2022, 20, 7694–7712. [Google Scholar] [CrossRef] [PubMed]
- Lenci, E.; Trabocchi, A. Synthetic approaches toward small molecule libraries. In Small Molecule Drug Discovery; Elsevier: Amsterdam, The Netherlands, 2020; pp. 1–34. [Google Scholar]
- Richter, S.G.; Elli, D.; Kim, H.K.; Hendrickx, A.P.A.; Sorg, J.A.; Schneewind, O.; Missiakas, D. Small molecule inhibitor of lipoteichoic acid synthesis is an antibiotic for Gram-positive bacteria. Proc. Natl. Acad. Sci. USA 2013, 110, 3531–3536. [Google Scholar] [CrossRef] [PubMed]
- Opoku-Temeng, C.; Naclerio, G.A.; Mohammad, H.; Dayal, N.; Abutaleb, N.S.; Seleem, M.N.; Sintim, H.O. N-(1, 3, 4-oxadiazol-2-yl) benzamide analogs, bacteriostatic agents against methicillin-and vancomycin-resistant bacteria. Eur. J. Med. Chem. 2018, 155, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Naclerio, G.A.; Karanja, C.W.; Opoku-Temeng, C.; Sintim, H.O. Antibacterial small molecules that potently inhibit Staphylococcus aureus lipoteichoic acid biosynthesis. ChemMedChem 2019, 14, 1000–1004. [Google Scholar] [CrossRef] [PubMed]
- Naclerio, G.A.; Onyedibe, K.I.; Sintim, H.O. Lipoteichoic acid biosynthesis inhibitors as potent inhibitors of S. aureus and E. faecalis growth and biofilm formation. Molecules 2020, 25, 2277. [Google Scholar] [CrossRef]
- Brown, S.; Santa Maria, J.P., Jr.; Walker, S. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef]
- Lee, K.; Campbell, J.; Swoboda, J.G.; Cuny, G.D.; Walker, S. Development of improved inhibitors of wall teichoic acid biosynthesis with potent activity against Staphylococcus aureus. Bioorg. Med. Chem. Lett. 2010, 20, 1767–1770. [Google Scholar] [CrossRef]
- Swoboda, J.G.; Meredith, T.C.; Campbell, J.; Brown, S.; Suzuki, T.; Bollenbach, T.; Malhowski, A.J.; Kishony, R.; Gilmore, M.S.; Walker, S. Discovery of a small molecule that blocks wall teichoic acid biosynthesis in Staphylococcus aureus. ACS Chem. Biol. 2009, 4, 875–883. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gill, C.J.; Lee, S.H.; Mann, P.; Zuck, P.; Meredith, T.C.; Murgolo, N.; She, X.; Kales, S.; Liang, L. Discovery of wall teichoic acid inhibitors as potential anti-MRSA β-lactam combination agents. Chem. Biol. 2013, 20, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.; Singh, A.K.; Swoboda, J.G.; Gilmore, M.S.; Wilkinson, B.J.; Walker, S. An antibiotic that inhibits a late step in wall teichoic acid biosynthesis induces the cell wall stress stimulon in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012, 56, 1810–1820. [Google Scholar] [CrossRef] [PubMed]
- Sauvage, E.; Kerff, F.; Terrak, M.; Ayala, J.A.; Charlier, P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 234–258. [Google Scholar] [CrossRef] [PubMed]
- Fishovitz, J.; Hermoso, J.A.; Chang, M.; Mobashery, S. Penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. IUBMB Life 2014, 66, 572–577. [Google Scholar] [CrossRef]
- O’Daniel, P.I.; Peng, Z.; Pi, H.; Testero, S.A.; Ding, D.; Spink, E.; Leemans, E.; Boudreau, M.A.; Yamaguchi, T.; Schroeder, V.A. Discovery of a new class of non-β-lactam inhibitors of penicillin-binding proteins with gram-positive antibacterial activity. J. Am. Chem. Soc. 2014, 136, 3664–3672. [Google Scholar] [CrossRef]
- Bouley, R.; Kumarasiri, M.; Peng, Z.; Otero, L.H.; Song, W.; Suckow, M.A.; Schroeder, V.A.; Wolter, W.R.; Lastochkin, E.; Antunes, N.T. Discovery of antibiotic (E)-3-(3-carboxyphenyl)-2-(4-cyanostyryl) quinazolin-4 (3 H)-one. J. Am. Chem. Soc. 2015, 137, 1738–1741. [Google Scholar] [CrossRef]
- Janardhanan, J.; Bouley, R.; Martínez-Caballero, S.; Peng, Z.; Batuecas-Mordillo, M.; Meisel, J.E.; Ding, D.; Schroeder, V.A.; Wolter, W.R.; Mahasenan, K.V. The quinazolinone allosteric inhibitor of PBP 2a synergizes with piperacillin and tazobactam against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2019, 63, e02637-18. [Google Scholar] [CrossRef]
- Ghai, I.; Ghai, S. Understanding antibiotic resistance via outer membrane permeability. Infect. Drug Resist. 2018, 11, 523–530. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Srinivas, N.; Jetter, P.; Ueberbacher, B.J.; Werneburg, M.; Zerbe, K.; Steinmann, J.; Van der Meijden, B.; Bernardini, F.; Lederer, A.; Dias, R.L.A.; et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 2010, 327, 1010–1013. [Google Scholar] [CrossRef]
- Andolina, G.; Bencze, L.-C.; Zerbe, K.; Müller, M.; Steinmann, J.; Kocherla, H.; Mondal, M.; Sobek, J.; Moehle, K.; Malojčić, G.; et al. A Peptidomimetic Antibiotic Interacts with the Periplasmic Domain of LptD from Pseudomonas aeruginosa. ACS Chem. Biol. 2018, 13, 666–675. [Google Scholar] [CrossRef]
- Urfer, M.; Bogdanovic, J.; Lo Monte, F.; Moehle, K.; Zerbe, K.; Omasits, U.; Ahrens, C.H.; Pessi, G.; Eberl, L.; Robinson, J.A. A Peptidomimetic Antibiotic Targets Outer Membrane Proteins and Disrupts Selectively the Outer Membrane in Escherichia coli. J. Biol. Chem. 2016, 291, 1921–1932. [Google Scholar] [CrossRef] [PubMed]
- Hart, E.M.; Mitchell, A.M.; Konovalova, A.; Grabowicz, M.; Sheng, J.; Han, X.; Rodriguez-Rivera, F.P.; Schwaid, A.G.; Malinverni, J.C.; Balibar, C.J.; et al. A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier. Proc. Natl. Acad. Sci. USA 2019, 116, 21748–21757. [Google Scholar] [CrossRef]
- Yakushi, T.; Masuda, K.; Narita, S.; Matsuyama, S.; Tokuda, H. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2000, 2, 212–218. [Google Scholar] [CrossRef]
- McLeod, S.M.; Fleming, P.R.; MacCormack, K.; McLaughlin, R.E.; Whiteaker, J.D.; Narita, S.-I.; Mori, M.; Tokuda, H.; Miller, A.A. Small-molecule inhibitors of gram-negative lipoprotein trafficking discovered by phenotypic screening. J. Bacteriol. 2015, 197, 1075–1082. [Google Scholar] [CrossRef]
- Nayar, A.S.; Dougherty, T.J.; Ferguson, K.E.; Granger, B.A.; McWilliams, L.; Stacey, C.; Leach, L.J.; Narita, S.-I.; Tokuda, H.; Miller, A.A.; et al. Novel antibacterial targets and compounds revealed by a high-throughput cell wall reporter assay. J. Bacteriol. 2015, 197, 1726–1734. [Google Scholar] [CrossRef]
- Nickerson, N.N.; Jao, C.C.; Xu, Y.; Quinn, J.; Skippington, E.; Alexander, M.K.; Miu, A.; Skelton, N.; Hankins, J.V.; Lopez, M.S.; et al. A Novel Inhibitor of the LolCDE ABC Transporter Essential for Lipoprotein Trafficking in Gram-Negative Bacteria. Antimicrob. Agents Chemother. 2018, 62, e02151-17. [Google Scholar] [CrossRef]
- Naclerio, G.A.; Sintim, H.O. Multiple ways to kill bacteria via inhibiting novel cell wall or membrane targets. Future Med. Chem. 2020, 12, 1253–1279. [Google Scholar] [CrossRef]
- Pathania, R.; Zlitni, S.; Barker, C.; Das, R.; Gerritsma, D.A.; Lebert, J.; Awuah, E.; Melacini, G.; Capretta, F.A.; Brown, E.D. Chemical genomics in Escherichia coli identifies an inhibitor of bacterial lipoprotein targeting. Nat. Chem. Biol. 2009, 5, 849–856. [Google Scholar] [CrossRef]
- Lehman, K.M.; Smith, H.C.; Grabowicz, M. A Biological Signature for the Inhibition of Outer Membrane Lipoprotein Biogenesis. mBio 2022, 13, e0075722. [Google Scholar] [CrossRef]
- Svanberg Frisinger, F.; Pirolo, M.; Ng, D.Y.K.; Mao, X.; Nielsen, D.S.; Guardabassi, L. Impact of the Gram-Negative-Selective Inhibitor MAC13243 on In Vitro Simulated Gut Microbiota. Pharmaceuticals 2022, 15, 731. [Google Scholar] [CrossRef]
- Mori, H.; Ito, K. The Sec protein-translocation pathway. Trends Microbiol. 2001, 9, 494–500. [Google Scholar] [CrossRef]
- Chen, W.; Huang, Y.-j.; Gundala, S.R.; Yang, H.; Li, M.; Tai, P.C.; Wang, B. The first low microM SecA inhibitors. Bioorg. Med. Chem. 2010, 18, 1617–1625. [Google Scholar] [CrossRef]
- Cui, J.; Jin, J.; Chaudhary, A.S.; Hsieh, Y.-h.; Zhang, H.; Dai, C.; Damera, K.; Chen, W.; Tai, P.C.; Wang, B. Design, Synthesis and Evaluation of Triazole-Pyrimidine Analogues as SecA Inhibitors. ChemMedChem 2016, 11, 43–56. [Google Scholar] [CrossRef]
- Cui, P.; Li, X.; Zhu, M.; Wang, B.; Liu, J.; Chen, H. Design, synthesis and antimicrobial activities of thiouracil derivatives containing triazolo-thiadiazole as SecA inhibitors. Eur. J. Med. Chem. 2017, 127, 159–165. [Google Scholar] [CrossRef]
- Lewis, K. Antibiotics: Recover the lost art of drug discovery. Nature 2012, 485, 439–440. [Google Scholar] [CrossRef]
- Liu, M.; El-Hossary, E.M.; Oelschlaeger, T.A.; Donia, M.S.; Quinn, R.J.; Abdelmohsen, U.R. Potential of marine natural products against drug-resistant bacterial infections. Lancet Infect. Dis. 2019, 19, e237–e245. [Google Scholar] [CrossRef]
- Fehlbaum, P.; Bulet, P.; Chernysh, S.; Briand, J.P.; Roussel, J.P.; Letellier, L.; Hetru, C.; Hoffmann, J.A. Structure-activity analysis of thanatin, a 21-residue inducible insect defense peptide with sequence homology to frog skin antimicrobial peptides. Proc. Natl. Acad. Sci. USA 1996, 93, 1221–1225. [Google Scholar] [CrossRef]
- Ma, B.; Niu, C.; Zhou, Y.; Xue, X.; Meng, J.; Luo, X.; Hou, Z. The Disulfide Bond of the Peptide Thanatin Is Dispensible for Its Antimicrobial Activity In Vivo and In Vitro. Antimicrob. Agents Chemother. 2016, 60, 4283–4289. [Google Scholar] [CrossRef]
- Ma, B.; Fang, C.; Lu, L.; Wang, M.; Xue, X.; Zhou, Y.; Li, M.; Hu, Y.; Luo, X.; Hou, Z. The antimicrobial peptide thanatin disrupts the bacterial outer membrane and inactivates the NDM-1 metallo-β-lactamase. Nat. Commun. 2019, 10, 3517. [Google Scholar] [CrossRef]
- Wu, G.; Fan, X.; Li, L.; Wang, H.; Ding, J.; Hongbin, W.; Zhao, R.; Gou, L.; Shen, Z.; Xi, T. Interaction of antimicrobial peptide s-thanatin with lipopolysaccharide in vitro and in an experimental mouse model of septic shock caused by a multidrug-resistant clinical isolate of Escherichia coli. Int. J. Antimicrob. Agents 2010, 35, 250–254. [Google Scholar] [CrossRef]
- Wu, G.; Wu, P.; Xue, X.; Yan, X.; Liu, S.; Zhang, C.; Shen, Z.; Xi, T. Application of S-thanatin, an antimicrobial peptide derived from thanatin, in mouse model of Klebsiella pneumoniae infection. Peptides 2013, 45, 73–77. [Google Scholar] [CrossRef]
- Hou, Z.; Lu, J.; Fang, C.; Zhou, Y.; Bai, H.; Zhang, X.; Xue, X.; Chen, Y.; Luo, X. Underlying mechanism of in vivo and in vitro activity of C-terminal-amidated thanatin against clinical isolates of extended-spectrum beta-lactamase-producing Escherichia coli. J. Infect. Dis. 2011, 203, 273–282. [Google Scholar] [CrossRef]
- Dash, R.; Bhattacharjya, S. Thanatin: An Emerging Host Defense Antimicrobial Peptide with Multiple Modes of Action. Int. J. Mol. Sci. 2021, 22, 1522. [Google Scholar] [CrossRef]
- Zipperer, A.; Konnerth, M.C.; Laux, C.; Berscheid, A.; Janek, D.; Weidenmaier, C.; Burian, M.; Schilling, N.A.; Slavetinsky, C.; Marschal, M.; et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016, 535, 511–516. [Google Scholar] [CrossRef]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef]
- Shukla, R.; Lavore, F.; Maity, S.; Derks, M.G.N.; Jones, C.R.; Vermeulen, B.J.A.; Melcrová, A.; Morris, M.A.; Becker, L.M.; Wang, X.; et al. Teixobactin kills bacteria by a two-pronged attack on the cell envelope. Nature 2022, 608, 390–396. [Google Scholar] [CrossRef]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heuer, O.E.; et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef]
- Stokes, J.M.; Lopatkin, A.J.; Lobritz, M.A.; Collins, J.J. Bacterial Metabolism and Antibiotic Efficacy. Cell Metab. 2019, 30, 251–259. [Google Scholar] [CrossRef]
- Peng, B.; Su, Y.-B.; Li, H.; Han, Y.; Guo, C.; Tian, Y.-M.; Peng, X.-X. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 2015, 21, 249–262. [Google Scholar] [CrossRef]
- Allison, K.R.; Brynildsen, M.P.; Collins, J.J. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 2011, 473, 216–220. [Google Scholar] [CrossRef]
- Meylan, S.; Porter, C.B.M.; Yang, J.H.; Belenky, P.; Gutierrez, A.; Lobritz, M.A.; Park, J.; Kim, S.H.; Moskowitz, S.M.; Collins, J.J. Carbon Sources Tune Antibiotic Susceptibility in Pseudomonas aeruginosa via Tricarboxylic Acid Cycle Control. Cell Chem. Biol. 2017, 24, 195–206. [Google Scholar] [CrossRef]
- Zhao, X.-L.; Chen, Z.-G.; Yang, T.-C.; Jiang, M.; Wang, J.; Cheng, Z.-X.; Yang, M.-J.; Zhu, J.-X.; Zhang, T.-T.; Li, H.; et al. Glutamine promotes antibiotic uptake to kill multidrug-resistant uropathogenic bacteria. Sci. Transl. Med. 2021, 13, eabj0716. [Google Scholar] [CrossRef]
- Yang, J.H.; Wright, S.N.; Hamblin, M.; McCloskey, D.; Alcantar, M.A.; Schrübbers, L.; Lopatkin, A.J.; Satish, S.; Nili, A.; Palsson, B.O.; et al. A White-Box Machine Learning Approach for Revealing Antibiotic Mechanisms of Action. Cell 2019, 177, 1649–1661. [Google Scholar] [CrossRef]
- Ye, J.; Su, Y.; Peng, X.; Li, H. Reactive oxygen species-related ceftazidime resistance is caused by the pyruvate cycle perturbation and reverted by Fe3+ in Edwardsiella tarda. Front. Microbiol. 2021, 12, 654783. [Google Scholar] [CrossRef]
- Ye, J.-z.; Su, Y.-b.; Lin, X.-m.; Lai, S.-s.; Li, W.-x.; Ali, F.; Zheng, J.; Peng, B. Alanine enhances aminoglycosides-induced ROS production as revealed by proteomic analysis. Front. Microbiol. 2018, 9, 29. [Google Scholar] [CrossRef]
- Ferreira, M.; Ogren, M.; Dias, J.N.R.; Silva, M.; Gil, S.; Tavares, L.; Aires-da-Silva, F.; Gaspar, M.M.; Aguiar, S.I. Liposomes as Antibiotic Delivery Systems: A Promising Nanotechnological Strategy against Antimicrobial Resistance. Molecules 2021, 26, 2047. [Google Scholar] [CrossRef]
- Zazo, H.; Colino, C.I.; Lanao, J.M. Current applications of nanoparticles in infectious diseases. J. Control. Release 2016, 224, 86–102. [Google Scholar] [CrossRef]
- Turos, E.; Shim, J.-Y.; Wang, Y.; Greenhalgh, K.; Reddy, G.S.K.; Dickey, S.; Lim, D.V. Antibiotic-conjugated polyacrylate nanoparticles: New opportunities for development of anti-MRSA agents. Bioorg. Med. Chem. Lett. 2007, 17, 53–56. [Google Scholar] [CrossRef]
- Turos, E.; Reddy, G.S.K.; Greenhalgh, K.; Ramaraju, P.; Abeylath, S.C.; Jang, S.; Dickey, S.; Lim, D.V. Penicillin-bound polyacrylate nanoparticles: Restoring the activity of beta-lactam antibiotics against MRSA. Bioorg. Med. Chem. Lett. 2007, 17, 3468–3472. [Google Scholar] [CrossRef]
- Greenhalgh, K.; Turos, E. In vivo studies of polyacrylate nanoparticle emulsions for topical and systemic applications. Nanomedicine 2009, 5, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Lima, R.; Del Fiol, F.S.; Balcão, V.M. Prospects for the Use of New Technologies to Combat Multidrug-Resistant Bacteria. Front. Pharm. 2019, 10, 692. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez Gomez, A.; Hosseinidoust, Z. Liposomes for Antibiotic Encapsulation and Delivery. ACS Infect. Dis 2020, 6, 896–908. [Google Scholar] [CrossRef] [PubMed]
- Khan, O.; Chaudary, N. The Use of Amikacin Liposome Inhalation Suspension (Arikayce) in the Treatment of Refractory Nontuberculous Mycobacterial Lung Disease in Adults. Drug Des. Devel. 2020, 14, 2287–2294. [Google Scholar] [CrossRef]
- Alipour, M.; Halwani, M.; Omri, A.; Suntres, Z.E. Antimicrobial effectiveness of liposomal polymyxin B against resistant Gram-negative bacterial strains. Int. J. Pharm. 2008, 355, 293–298. [Google Scholar] [CrossRef]
- Mugabe, C.; Halwani, M.; Azghani, A.O.; Lafrenie, R.M.; Omri, A. Mechanism of enhanced activity of liposome-entrapped aminoglycosides against resistant strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2006, 50, 2016–2022. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, Y.; Khalil, H.; Wang, R.; Lu, T.; Zhao, W.; Zhang, Y.; Chen, J.; Chen, T. Fusion between fluid liposomes and intact bacteria: Study of driving parameters and in vitro bactericidal efficacy. Int. J. Nanomed. 2016, 11, 4025–4036. [Google Scholar] [CrossRef]
- Nacucchio, M.C.; Bellora, M.J.; Sordelli, D.O.; D’Aquino, M. Enhanced liposome-mediated activity of piperacillin against staphylococci. Antimicrob. Agents Chemother. 1985, 27, 137–139. [Google Scholar] [CrossRef]
- Gaspar, M.M.; Cruz, A.; Penha, A.F.; Reymão, J.; Sousa, A.C.; Eleutério, C.V.; Domingues, S.A.; Fraga, A.G.; Filho, A.L.; Cruz, M.E.M.; et al. Rifabutin encapsulated in liposomes exhibits increased therapeutic activity in a model of disseminated tuberculosis. Int. J. Antimicrob. Agents 2008, 31, 37–45. [Google Scholar]
- Huang, W.; Zhang, Q.; Li, W.; Yuan, M.; Zhou, J.; Hua, L.; Chen, Y.; Ye, C.; Ma, Y. Development of novel nanoantibiotics using an outer membrane vesicle-based drug efflux mechanism. J. Control. Release 2020, 317, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Huang, Y.; Yan, J.; Li, Y.; Wang, J.; Yang, Y.Y.; Yuan, P.; Ding, X. Bacterial outer membrane-coated mesoporous silica nanoparticles for targeted delivery of antibiotic rifampicin against Gram-negative bacterial infection in vivo. Adv. Funct. Mater. 2021, 31, 2103442. [Google Scholar] [CrossRef]
- Cao, X.; Li, S.; Chen, L.; Ding, H.; Xu, H.; Huang, Y.; Li, J.; Liu, N.; Cao, W.; Zhu, Y.; et al. Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Res. 2009, 37, 4621–4628. [Google Scholar] [CrossRef] [PubMed]
- Kavruk, M.; Celikbicak, O.; Ozalp, V.C.; Borsa, B.A.; Hernandez, F.J.; Bayramoglu, G.; Salih, B.; Arica, M.Y. Antibiotic loaded nanocapsules functionalized with aptamer gates for targeted destruction of pathogens. Chem. Commun. 2015, 51, 8492–8495. [Google Scholar] [CrossRef] [PubMed]
- Nikolich, M.P.; Filippov, A.A. Bacteriophage Therapy: Developments and Directions. Antibiotics 2020, 9, 135. [Google Scholar] [CrossRef]
- Jault, P.; Leclerc, T.; Jennes, S.; Pirnay, J.P.; Que, Y.-A.; Resch, G.; Rousseau, A.F.; Ravat, F.; Carsin, H.; Le Floch, R.; et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): A randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis 2019, 19, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Chen, H.; Zhang, M.; Zhao, Y.; Jiang, Y.; Liu, X.; Huang, W.; Ma, Y. Clinical Experience of Personalized Phage Therapy Against Carbapenem-Resistant Lung Infection in a Patient with Chronic Obstructive Pulmonary Disease. Front. Cell Infect. Microbiol. 2021, 11, 631585. [Google Scholar] [CrossRef]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails to Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 2017, 61, e00954-17. [Google Scholar] [CrossRef]
- LaVergne, S.; Hamilton, T.; Biswas, B.; Kumaraswamy, M.; Schooley, R.T.; Wooten, D. Phage Therapy for a Multidrug-Resistant Craniectomy Site Infection. Open Forum Infect. Dis. 2018, 5, ofy064. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Fernández, L.; Rodríguez, A.; García, P. Are Phage Lytic Proteins the Secret Weapon to Kill? mBio 2018, 9, e01923-17. [Google Scholar] [CrossRef]
- Antonova, N.P.; Vasina, D.V.; Lendel, A.M.; Usachev, E.V.; Makarov, V.V.; Gintsburg, A.L.; Tkachuk, A.P.; Gushchin, V.A. Broad Bactericidal Activity of the Bacteriophage Lysins LysAm24, LysECD7, and LysSi3 against Gram-Negative ESKAPE Pathogens. Viruses 2019, 11, 284. [Google Scholar] [CrossRef]
- Grygorcewicz, B.; Roszak, M.; Golec, P.; Śleboda-Taront, D.; Łubowska, N.; Górska, M.; Jursa-Kulesza, J.; Rakoczy, R.; Wojciuk, B.; Dołęgowska, B. Antibiotics Act with vB_AbaP_AGC01 Phage against in Human Heat-Inactivated Plasma Blood and Models. Int. J. Mol. Sci. 2020, 21, 4390. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.Y.; Jang, I.J.; Yoon, S.; Jang, K.; Yu, K.-S.; Cho, J.Y.; Seong, M.-W.; Jung, G.M.; Yoon, S.J.; Kang, S.H. Pharmacokinetics and Tolerance of the Phage Endolysin-Based Candidate Drug SAL200 after a Single Intravenous Administration among Healthy Volunteers. Antimicrob. Agents Chemother. 2017, 61, e02629-16. [Google Scholar] [CrossRef] [PubMed]
- Totté, J.E.E.; van Doorn, M.B.; Pasmans, S.G.M.A. Successful Treatment of Chronic Related Dermatoses with the Topical Endolysin Staphefekt SA.100: A Report of 3 Cases. Case Rep. Derm. 2017, 9, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Schuch, R.; Cassino, C.; Vila-Farres, X. Direct Lytic Agents: Novel, Rapidly Acting Potential Antimicrobial Treatment Modalities for Systemic Use in the Era of Rising Antibiotic Resistance. Front. Microbiol. 2022, 13, 841905. [Google Scholar] [CrossRef]
- Fowler, V.G.; Das, A.F.; Lipka-Diamond, J.; Schuch, R.; Pomerantz, R.; Jáuregui-Peredo, L.; Bressler, A.; Evans, D.; Moran, G.J.; Rupp, M.E.; et al. Exebacase for patients with Staphylococcus aureus bloodstream infection and endocarditis. J. Clin. Investig. 2020, 130, 3750–3760. [Google Scholar] [CrossRef]
- Hospenthal, M.K.; Waksman, G. The Remarkable Biomechanical Properties of the Type 1 Chaperone-Usher Pilus: A Structural and Molecular Perspective. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Cusumano, C.K.; Pinkner, J.S.; Han, Z.; Greene, S.E.; Ford, B.A.; Crowley, J.R.; Henderson, J.P.; Janetka, J.W.; Hultgren, S.J. Treatment and prevention of urinary tract infection with orally active FimH inhibitors. Sci. Transl. Med. 2011, 3, 109ra115. [Google Scholar] [CrossRef]
- Hemmati, F.; Rezaee, M.A.; Ebrahimzadeh, S.; Yousefi, L.; Nouri, R.; Kafil, H.S.; Gholizadeh, P. Novel Strategies to Combat Bacterial Biofilms. Mol. Biotechnol. 2021, 63, 569–586. [Google Scholar] [CrossRef]
- Cegelski, L.; Pinkner, J.S.; Hammer, N.D.; Cusumano, C.K.; Hung, C.S.; Chorell, E.; Aberg, V.; Walker, J.N.; Seed, P.C.; Almqvist, F.; et al. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat. Chem. Biol. 2009, 5, 913–919. [Google Scholar] [CrossRef]
- de la Fuente-Núñez, C.; Reffuveille, F.; Haney, E.F.; Straus, S.K.; Hancock, R.E.W. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 2014, 10, e1004152. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente-Núñez, C.; Reffuveille, F.; Mansour, S.C.; Reckseidler-Zenteno, S.L.; Hernández, D.; Brackman, G.; Coenye, T.; Hancock, R.E.W. D-enantiomeric peptides that eradicate wild-type and multidrug-resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chem. Biol. 2015, 22, 196–205. [Google Scholar] [CrossRef]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [PubMed]
- Shahed-Al-Mahmud, M.; Roy, R.; Sugiokto, F.G.; Islam, M.N.; Lin, M.-D.; Lin, L.-C.; Lin, N.-T. Phage φAB6-Borne Depolymerase Combats Biofilm Formation and Infection. Antibiotics 2021, 10, 279. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Cheng, L.; Ling, M.; Feng, X.; Chen, L.; Wu, M.; Deng, L. Efficient suppression of biofilm formation by a nucleic acid aptamer. Pathog. Dis. 2015, 73, ftv034. [Google Scholar] [CrossRef]
- Ommen, P.; Hansen, L.; Hansen, B.K.; Vu-Quang, H.; Kjems, J.; Meyer, R.L. Aptamer-Targeted Drug Delivery for Biofilm. Front. Cell Infect. Microbiol. 2022, 12, 814340. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Cummings, J.H. Probiotics, infection and immunity. Curr. Opin. Infect. Dis. 2002, 15, 501–506. [Google Scholar] [CrossRef]
- Piewngam, P.; Otto, M. Probiotics to prevent Staphylococcus aureus disease? Gut Microbes 2020, 11, 94–101. [Google Scholar] [CrossRef]
- Soto, N.E.; Vaghjimal, A.; Stahl-Avicolli, A.; Protic, J.R.; Lutwick, L.I.; Chapnick, E.K. Bacitracin versus mupirocin for Staphylococcus aureus nasal colonization. Infect. Control. Hosp. Epidemiol. 1999, 20, 351–353. [Google Scholar] [CrossRef]
- Piewngam, P.; Zheng, Y.; Nguyen, T.H.; Dickey, S.W.; Joo, H.-S.; Villaruz, A.E.; Glose, K.A.; Fisher, E.L.; Hunt, R.L.; Li, B.; et al. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 2018, 562, 532–537. [Google Scholar] [CrossRef]
- Saeidi, N.; Wong, C.K.; Lo, T.M.; Nguyen, H.X.; Ling, H.; Leong, S.S.; Poh, C.L.; Chang, M.W. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol. Syst. Biol. 2011, 7, 521. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.Y.; Koh, E.; Wong, A.; March, J.C.; Bentley, W.E.; Lee, Y.S.; Chang, M.W. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat. Commun. 2017, 8, 15028. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Hong, M.; Xie, W.; Chen, Z.; Chen, D.; Xie, S. Progress and prospects of nanomaterials against resistant bacteria. J. Control. Release 2022, 351, 301–323. [Google Scholar] [CrossRef] [PubMed]
- Linklater, D.P.; Baulin, V.A.; Le Guével, X.; Fleury, J.-B.; Hanssen, E.; Nguyen, T.H.P.; Juodkazis, S.; Bryant, G.; Crawford, R.J.; Stoodley, P.; et al. Antibacterial Action of Nanoparticles by Lethal Stretching of Bacterial Cell Membranes. Adv. Mater. 2020, 32, e2005679. [Google Scholar] [CrossRef]
- Tang, S.; Zheng, J. Antibacterial Activity of Silver Nanoparticles: Structural Effects. Adv. Heal. Mater. 2018, 7, e1701503. [Google Scholar] [CrossRef]
- Mathews, S.; Kumar, R.; Solioz, M. Copper Reduction and Contact Killing of Bacteria by Iron Surfaces. Appl. Environ. Microbiol. 2015, 81, 6399–6403. [Google Scholar] [CrossRef]
- Gunawan, C.; Faiz, M.B.; Mann, R.; Ting, S.R.S.; Sotiriou, G.A.; Marquis, C.P.; Amal, R. Nanosilver Targets the Bacterial Cell Envelope: The Link with Generation of Reactive Oxygen Radicals. ACS Appl. Mater. Interfaces 2020, 12, 5557–5568. [Google Scholar] [CrossRef]
- Su, Y.; Zheng, X.; Chen, Y.; Li, M.; Liu, K. Alteration of intracellular protein expressions as a key mechanism of the deterioration of bacterial denitrification caused by copper oxide nanoparticles. Sci. Rep. 2015, 5, 15824. [Google Scholar] [CrossRef]
- Hoque, J.; Akkapeddi, P.; Yarlagadda, V.; Uppu, D.S.S.M.; Kumar, P.; Haldar, J. Cleavable cationic antibacterial amphiphiles: Synthesis, mechanism of action, and cytotoxicities. Langmuir 2012, 28, 12225–12234. [Google Scholar] [CrossRef]
- Hu, D.; Li, H.; Wang, B.; Ye, Z.; Lei, W.; Jia, F.; Jin, Q.; Ren, K.-F.; Ji, J. Surface-Adaptive Gold Nanoparticles with Effective Adherence and Enhanced Photothermal Ablation of Methicillin-Resistant Staphylococcus aureus Biofilm. ACS Nano 2017, 11, 9330–9339. [Google Scholar] [CrossRef]
- de Moraes, A.C.M.; Lima, B.A.; de Faria, A.F.; Brocchi, M.; Alves, O.L. Graphene oxide-silver nanocomposite as a promising biocidal agent against methicillin-resistant Staphylococcus aureus. Int. J. Nanomed. 2015, 10, 6847–6861. [Google Scholar] [CrossRef] [PubMed]
- Rytel, M.W.; Ferstenfeld, J.E.; Rose, H.D.; Balay, J.; Pierce, W.E.; Lynch, K.L. Efficacy of a “mixed bacterial vaccine” in prophylaxis of acute respiratory infections: Possible role of interferon. Am. J. Epidemiol. 1974, 99, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Lauener, P.A. Bacterial lysates and ribosomes as inducers of specific immune responses: A comparative study. Scand. J. Immunol. 1994, 40, 466–467. [Google Scholar] [CrossRef]
- Fattom, A.; Matalon, A.; Buerkert, J.; Taylor, K.; Damaso, S.; Boutriau, D. Efficacy profile of a bivalent Staphylococcus aureus glycoconjugated vaccine in adults on hemodialysis: Phase III randomized study. Hum. Vaccin. Immunother. 2015, 11, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Fowler, V.G.; Allen, K.B.; Moreira, E.D.; Moustafa, M.; Isgro, F.; Boucher, H.W.; Corey, G.R.; Carmeli, Y.; Betts, R.; Hartzel, J.S.; et al. Effect of an investigational vaccine for preventing Staphylococcus aureus infections after cardiothoracic surgery: A randomized trial. JAMA 2013, 309, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Frenck, R.W.; Creech, C.B.; Sheldon, E.A.; Seiden, D.J.; Kankam, M.K.; Baber, J.; Zito, E.; Hubler, R.; Eiden, J.; Severs, J.M.; et al. Safety, tolerability, and immunogenicity of a 4-antigen Staphylococcus aureus vaccine (SA4Ag): Results from a first-in-human randomised, placebo-controlled phase 1/2 study. Vaccine 2017, 35, 375–384. [Google Scholar] [CrossRef]
- Langford, D.T.; Hiller, J. Prospective, controlled study of a polyvalent pseudomonas vaccine in cystic fibrosis—Three year results. Arch. Dis. Child. 1984, 59, 1131–1134. [Google Scholar] [CrossRef]
- Sharma, A.; Krause, A.; Worgall, S. Recent developments for Pseudomonas vaccines. Hum. Vaccin. 2011, 7, 999–1011. [Google Scholar] [CrossRef]
- Adlbrecht, C.; Wurm, R.; Depuydt, P.; Spapen, H.; Lorente, J.A.; Staudinger, T.; Creteur, J.; Zauner, C.; Meier-Hellmann, A.; Eller, P.; et al. Efficacy, immunogenicity, and safety of IC43 recombinant Pseudomonas aeruginosa vaccine in mechanically ventilated intensive care patients-a randomized clinical trial. Crit. Care 2020, 24, 74. [Google Scholar] [CrossRef]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti Infect. 2013, 11, 297–308. [Google Scholar] [CrossRef]
- Lindstedt, K.; Buczek, D.; Pedersen, T.; Hjerde, E.; Raffelsberger, N.; Suzuki, Y.; Brisse, S.; Holt, K.; Samuelsen, Ø.; Sundsfjord, A. Detection of human gut carriage: A comparison of culture, qPCR, and whole metagenomic sequencing methods. Gut Microbes 2022, 14, 2118500. [Google Scholar] [CrossRef] [PubMed]
- Swierstra, J.; Debets, S.; de Vogel, C.; Lemmens-den Toom, N.; Verkaik, N.; Ramdani-Bouguessa, N.; Jonkman, M.F.; van Dijl, J.M.; Fahal, A.; van Belkum, A.; et al. IgG4 subclass-specific responses to Staphylococcus aureus antigens shed new light on host-pathogen interaction. Infect. Immun. 2015, 83, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Leech, J.M.; Dhariwala, M.O.; Lowe, M.M.; Chu, K.; Merana, G.R.; Cornuot, C.; Weckel, A.; Ma, J.M.; Leitner, E.G.; Gonzalez, J.R.; et al. Toxin-Triggered Interleukin-1 Receptor Signaling Enables Early-Life Discrimination of Pathogenic versus Commensal Skin Bacteria. Cell Host Microbe 2019, 26, 795–809. [Google Scholar] [CrossRef] [PubMed]
- Si, Y.; Zhao, F.; Beesetty, P.; Weiskopf, D.; Li, Z.; Tian, Q.; Alegre, M.-L.; Sette, A.; Chong, A.S.; Montgomery, C.P. Inhibition of protective immunity against infection by MHC-restricted immunodominance is overcome by vaccination. Sci. Adv. 2020, 6, eaaw7713. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-M.; Caldera, J.R.; Hajam, I.A.; Chiang, A.W.T.; Tsai, C.-H.; Li, H.; Díez, M.L.; Gonzalez, C.; Trieu, D.; Martins, G.A.; et al. Non-protective immune imprint underlies failure of Staphylococcus aureus IsdB vaccine. Cell Host Microbe 2022, 30, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Pauli, N.T.; Kim, H.K.; Falugi, F.; Huang, M.; Dulac, J.; Henry Dunand, C.; Zheng, N.-Y.; Kaur, K.; Andrews, S.F.; Huang, Y.; et al. Staphylococcus aureus infection induces protein A-mediated immune evasion in humans. J. Exp. Med. 2014, 211, 2331–2339. [Google Scholar] [CrossRef]
- Radke, E.E.; Li, Z.; Hernandez, D.N.; El Bannoudi, H.; Kosakovsky Pond, S.L.; Shopsin, B.; Lopez, P.; Fenyö, D.; Silverman, G.J. Diversity of Functionally Distinct Clonal Sets of Human Conventional Memory B Cells That Bind Staphylococcal Protein A. Front. Immunol. 2021, 12, 662782. [Google Scholar] [CrossRef]
- Shi, M.; Chen, X.; Sun, Y.; Kim, H.K.; Schneewind, O.; Missiakas, D. A protein A based Staphylococcus aureus vaccine with improved safety. Vaccine 2021, 39, 3907–3915. [Google Scholar] [CrossRef]
- Lu, R.-M.; Hwang, Y.-C.; Liu, I.J.; Lee, C.-C.; Tsai, H.-Z.; Li, H.-J.; Wu, H.-C. Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 2020, 27, 1. [Google Scholar] [CrossRef]
- Chen, X.; Shi, M.; Tong, X.; Kim, H.K.; Wang, L.-X.; Schneewind, O.; Missiakas, D. Glycosylation-dependent opsonophagocytic activity of staphylococcal protein A antibodies. Proc. Natl. Acad. Sci. USA 2020, 117, 22992–23000. [Google Scholar] [CrossRef]
- Falugi, F.; Kim, H.K.; Missiakas, D.M.; Schneewind, O. Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. mBio 2013, 4, e00575-13. [Google Scholar] [CrossRef] [PubMed]
- Pietrocola, G.; Nobile, G.; Rindi, S.; Speziale, P. Manipulates Innate Immunity through Own and Host-Expressed Proteases. Front. Cell Infect. Microbiol. 2017, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Schneewind, O.; Missiakas, D. Engineered human antibodies for the opsonization and killing of Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2022, 119, e2114478119. [Google Scholar] [CrossRef] [PubMed]
- Leo, J.C.; Goldman, A. The immunoglobulin-binding Eib proteins from Escherichia coli are receptors for IgG Fc. Mol. Immunol. 2009, 46, 1860–1866. [Google Scholar] [CrossRef] [PubMed]
- Sandt, C.H.; Hill, C.W. Four different genes responsible for nonimmune immunoglobulin-binding activities within a single strain of Escherichia coli. Infect. Immun 2000, 68, 2205–2214. [Google Scholar] [CrossRef] [PubMed]
- Rubin, D.; Zhang, W.; Karch, H.; Kuczius, T. Distinct Expression of Immunoglobulin-Binding Proteins in Shiga Toxin-Producing Escherichia coli Implicates High Protein Stability and a Characteristic Phenotype. Toxins 2017, 9, 153. [Google Scholar] [CrossRef]
- Mills, J.O.; Ghosh, P. Nonimmune antibody interactions of Group A Streptococcus M and M-like proteins. PLoS Pathog. 2021, 17, e1009248. [Google Scholar] [CrossRef]
- Brezski, R.J.; Jordan, R.E. Cleavage of IgGs by proteases associated with invasive diseases: An evasion tactic against host immunity? MAbs 2010, 2, 212–220. [Google Scholar] [CrossRef]
- Naegeli, A.; Bratanis, E.; Karlsson, C.; Shannon, O.; Kalluru, R.; Linder, A.; Malmström, J.; Collin, M. Streptococcus pyogenes evades adaptive immunity through specific IgG glycan hydrolysis. J. Exp. Med. 2019, 216, 1615–1629. [Google Scholar] [CrossRef]
- Nordenfelt, P.; Waldemarson, S.; Linder, A.; Mörgelin, M.; Karlsson, C.; Malmström, J.; Björck, L. Antibody orientation at bacterial surfaces is related to invasive infection. J. Exp. Med. 2012, 209, 2367–2381. [Google Scholar] [CrossRef]
- Lehar, S.M.; Pillow, T.; Xu, M.; Staben, L.; Kajihara, K.K.; Vandlen, R.; DePalatis, L.; Raab, H.; Hazenbos, W.L.; Morisaki, J.H.; et al. Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature 2015, 527, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Chastre, J.; François, B.; Bourgeois, M.; Komnos, A.; Ferrer, R.; Rahav, G.; De Schryver, N.; Lepape, A.; Koksal, I.; Luyt, C.-E.; et al. Safety, efficacy, and pharmacokinetics of gremubamab (MEDI3902), an anti-Pseudomonas aeruginosa bispecific human monoclonal antibody, in P. aeruginosa-colonised, mechanically ventilated intensive care unit patients: A randomised controlled trial. Crit. Care 2022, 26, 355. [Google Scholar] [CrossRef]
- Goydel, R.S.; Rader, C. Antibody-based cancer therapy. Oncogene 2021, 40, 3655–3664. [Google Scholar] [CrossRef] [PubMed]
- Slavetinsky, C.J.; Hauser, J.N.; Gekeler, C.; Slavetinsky, J.; Geyer, A.; Kraus, A.; Heilingbrunner, D.; Wagner, S.; Tesar, M.; Krismer, B.; et al. Sensitizing Staphylococcus aureus to antibacterial agents by decoding and blocking the lipid flippase MprF. Elife 2022, 11, e66376. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, J.; Chen, X. Current Promising Strategies against Antibiotic-Resistant Bacterial Infections. Antibiotics 2023, 12, 67. https://doi.org/10.3390/antibiotics12010067
Ye J, Chen X. Current Promising Strategies against Antibiotic-Resistant Bacterial Infections. Antibiotics. 2023; 12(1):67. https://doi.org/10.3390/antibiotics12010067
Chicago/Turabian StyleYe, Jinzhou, and Xinhai Chen. 2023. "Current Promising Strategies against Antibiotic-Resistant Bacterial Infections" Antibiotics 12, no. 1: 67. https://doi.org/10.3390/antibiotics12010067
APA StyleYe, J., & Chen, X. (2023). Current Promising Strategies against Antibiotic-Resistant Bacterial Infections. Antibiotics, 12(1), 67. https://doi.org/10.3390/antibiotics12010067





