Abstract
Infectious disease is one of the greatest causes of morbidity and mortality worldwide, and with the emergence of antimicrobial resistance, the situation is worsening. In order to prevent this crisis, antimicrobial resistance needs to be monitored carefully to control the spread of multidrug-resistant bacteria. Therefore, in this study, we aimed to determine the prevalence of infection caused by Klebsiella pneumoniae and investigate the antimicrobial profile pattern of K. pneumoniae in the last eleven years. This retrospective study was conducted in a tertiary hospital in Makkah, Saudi Arabia. Data were collected from January 2011 to December 2021. From 2011 to 2021, a total of 61,027 bacterial isolates were collected from clinical samples, among which 14.7% (n = 9014) were K. pneumoniae. The antibiotic susceptibility pattern of K. pneumoniae revealed a significant increase in the resistance rate in most tested antibiotics during the study period. A marked jump in the resistance rate was seen in amoxicillin/clavulanate and piperacillin/tazobactam, from 33.6% and 13.6% in 2011 to 71.4% and 84.9% in 2021, respectively. Ceftazidime, cefotaxime, and cefepime resistance rates increased from 29.9%, 26.2%, and 53.9%, respectively, in 2011 to become 84.9%, 85.1%, and 85.8% in 2021. Moreover, a significant increase in the resistance rate was seen in both imipenem and amikacin, with an average resistance rate rise from 6.6% for imipenem and 11.9% for amikacin in 2011 to 59.9% and 62.2% in 2021, respectively. The present study showed that the prevalence and drug resistance of K. pneumoniae increased over the study period. Thus, preventing hospital-acquired infection and the reasonable use of antibiotics must be implemented to control and reduce antimicrobial resistance.
Keywords:
Klebsiella pneumoniae; antibiotic resistance; MDR; ESBL; Saudi Arabia; Makkah; antibiogram pattern 1. Introduction
Klebsiella pneumoniae (K. pneumoniae) is a Gram-negative encapsulated bacterium that colonizes multiple sites of the human body, including the gastrointestinal tract, respiratory tract, oral cavities, and skin [1]. K. pneumoniae is an opportunistic pathogen and a major cause of hospital-acquired infections, including bloodstream infections, urinary tract infections, and pneumonia [2]. To survive the lethal effects of antibiotics, K. pneumoniae developed multiple resistance mechanisms, such as modifying the target site, drug inactivation, reduced cell permeability, and efflux pump activation [3,4]. For example, some strains of K. pneumoniae can survive and overcome the effect of β-lactam antibiotics by producing extended-spectrum beta-lactamase (ESBL) enzymes. ESBLs can hydrolyze and inactivate β-lactam antibiotics, including penicillins, cephalosporins (first-, second-, and third-generation), and aztreonam, but not cephamycins or carbapenems [4]. However, the effect of ESBL enzymes can be overcome by β-lactams inhibitors, such as clavulanic acid [3]. ESBLs are encoded by transferable plasmid-mediated genes, such as TEM, SHV, and CTX-M [5]. Since the identification of the first ESBL-producing bacteria in 1980, the incidence of ESBL-producing organisms has increased drastically to become a significant public health threat [6,7]. Recently, it has been noticed that ESBL-producing bacteria exhibit a high resistance rate to other antibiotic classes, such as fluoroquinolones, sulfonamides, and aminoglycosides [8]. In addition to ESBL, K. pneumoniae acquired resistance to carbapenems through three major mechanisms: enzyme production, efflux pumps, and porin mutations [9]. Among these, enzyme production (carbapenemases) is the most dominant resistance mechanism [9]. Furthermore, colistin resistance was also observed in K. pneumoniae and other Gram-negative bacteria. The mechanism of colistin resistance is generally through the modification of the lipopolysaccharide (LPS) outer membrane structure, which would lower the binding affinity of colistin to the LPS [10]. Due to this increase in antibiotic resistance, multiple studies were conducted worldwide to monitor the antibiotic resistance of K. pneumoniae [11].
In Saudi Arabia, the surveillance of antibiotic resistance of K. pneumoniae revealed an increase in the resistance rate among different antibiotic classes. For example, in Aseer (south), several studies conducted between 2015 and 2019 showed an increase in the resistance rate against penicillin and cephalosporin. Nearly all the isolates were resistant to ceftazidime, piperacillin, and ampicillin [12,13]. Similarly, a previous study in Riyadh (center) showed that 55% of K. pneumoniae were capable of producing ESBL and exhibited a high resistance rate to ceftazidime (95%) and cefotaxime (97%) [5]. In addition, during a four-year surveillance of K. pneumoniae in Al-Medina (north), Saudi Arabia, over half of the isolates were found to be resistant to different classes of antibiotics (with an average of 61.7%). Among carbapenems, 38.7% and 46.1% resistance rates were found towards imipenem and meropenem, respectively [14]. Moreover, the study revealed a high resistance to colistin and tigecycline. Although the result of colistin resistance was not reliable due to the low sample size, this finding is worrying because of the limited options for treating infection caused by multidrug-resistant K. pneumoniae [14,15]. The aforementioned studies indicated a continuous increase in the antibiotic resistance rate of K. pneumoniae, which requires urgent and continuous monitoring. Therefore, this study aimed to determine the prevalence of K. pneumoniae infections and the resistance trend over eleven years in Makkah, Saudi Arabia.
2. Results
2.1. Patient and Samples Demographics
This retrospective study was carried out to determine the burden of infections caused by K. pneumoniae over eleven years, from January 2011 to December 2021. Out of 61,027 isolates found during the study period, 9014 (14.7%) were K. pneumoniae. The results showed an increase in K. pneumoniae isolates over the years, with an annual number of isolates ranging from 622 to 1131 and an average of 819.5 ± 205.7. The detection rates of K. pneumoniae increased from 7.7% in 2011 to 25.9% in 2020. However, the results revealed that the rates of K. pneumoniae isolates drastically reduced from 25.9% in 2020 to 17.9% in 2022, which could be due to the strong implementation of infection control policies following the COVID-19 pandemic (Table 1).

Table 1.
The demographic distribution of K. pneumoniae.
Throughout the study period, the results showed a significantly higher rate of K. pneumoniae isolated from inpatients (mean: 672.5 ± 193.6) compared to outpatients (mean: 147 ± 51.9) (Table 1 and Figure 1). In addition, higher rates of K. pneumoniae isolates were obtained from male subjects compared to females (p value ≤ 0.0001) (Table 1). The results also showed a direct correlation between age and the rates of K. pneumoniae isolates (Figure 1). Collectively, these results may suggest that prolonged hospitalization and weakened immunity due to aging are potential risk factors for K. pneumoniae infection.
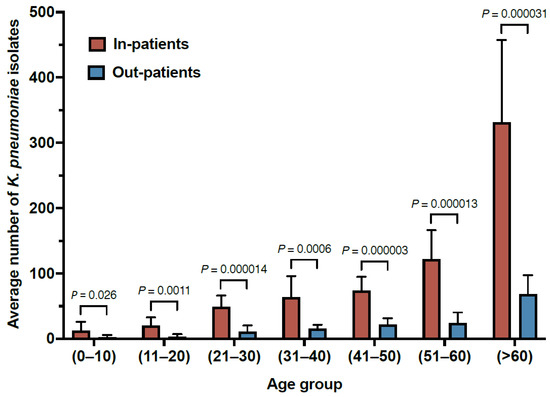
Figure 1.
The distribution of K. pneumoniae isolates from inpatient and outpatient departments from 2011–2021.
During the study period, K. pneumoniae isolates were reported mostly from blood samples (28.4%), followed by sputum (21.8%), wounds (21.3%), urine (14.7), and tip and catheter (1.5%) (Figure 2). Overall, the results showed a gradual increase in the percentage of detection of K. pneumoniae isolates from different samples over the years (Table 2). Regardless of the sample origin, K. pneumoniae isolates were significantly higher from inpatient specimens (mean: 82.55 ± 6.3) compared to outpatients (mean: 17.45 ± 6.4) (Figure 3). For example, 83.65% of K. pneumoniae isolated from blood samples were obtained from inpatient specimens, and the remaining 16.35% were from outpatients (Figure 3).
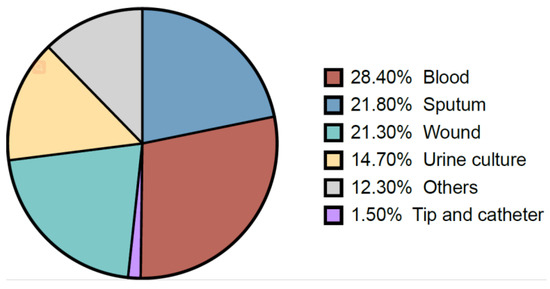
Figure 2.
The proportion of different specimen types of K. pneumoniae.

Table 2.
The proportion of different specimen types from which K. pneumoniae was obtained.
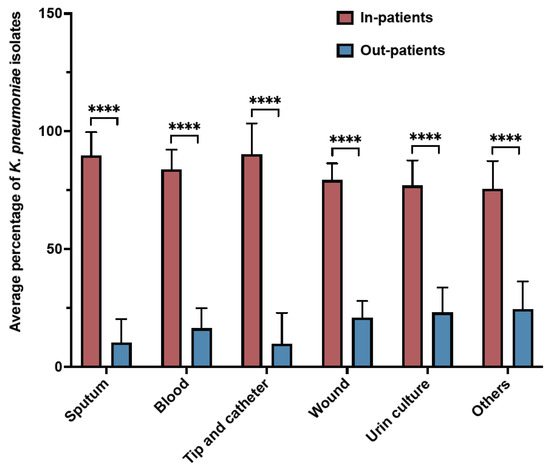
Figure 3.
Samples distribution of K. pneumoniae isolates from inpatients and outpatients over 11 years. **** p ≤ 0.000001.
2.2. Antimicrobial Resistance Profile of K. pneumoniae
The analysis of the antimicrobial susceptibility pattern of K. pneumoniae revealed a sharp increase in the resistance rate in most of the tested antibiotics (Table 3, Figure 4A–C). Throughout the duration of the study, the highest resistance rate was observed in ampicillin (97.6%), and the lowest resistance rate was for tigecycline (15.7%) (Table 3). The complex of β-lactam/β-lactamase-inhibitor antibiotics (amoxicillin/clavulanate and piperacillin/tazobactam) showed a significant gradual increase in the resistance rate over the eleven years (Table 3 and Figure 4A). Similarly, a sharp increase in resistance was observed in the third- and fourth-generation cephalosporins (ceftazidime, cefotaxime, and cefepime) (Table 3 and Figure 4A). Among the cephalosporin antibiotics, cefotaxime showed the lowest resistance rate (68.6%), followed by ceftazidime (74.9%), and cefepime (81.0%) (Table 3). Notably, imipenem was the most sensitive antibiotic among the β-lactam antibiotic family in the first two years of the study; however, the resistance rate of imipenem escalated progressively to reach 60% in 2021 (Table 3 and Figure 4A). Like β-lactam antibiotics, aminoglycosides (amikacin and gentamicin) showed a significant increase in the resistance rate over the study period. Over the eleven years, the average resistance rate of amikacin was 47.8%, whereas that of gentamicin was 54.9% (Table 3 and Figure 4B). Similarly, ciprofloxacin and cotrimoxazole (trimethoprim-sulfamethoxazole) showed a significant increase in the resistance rate over time, with average resistance rates of 71.6% and 74.9%, respectively (Table 3 and Figure 4B). In addition, the results showed that the overall resistance rates of colistin and tigecycline for the study period were 25.4% and 15.7%, respectively. However, the results for both antibiotics were not statistically significant due to the low sample size (Table 3).

Table 3.
Antibiogram pattern of K. pneumoniae.
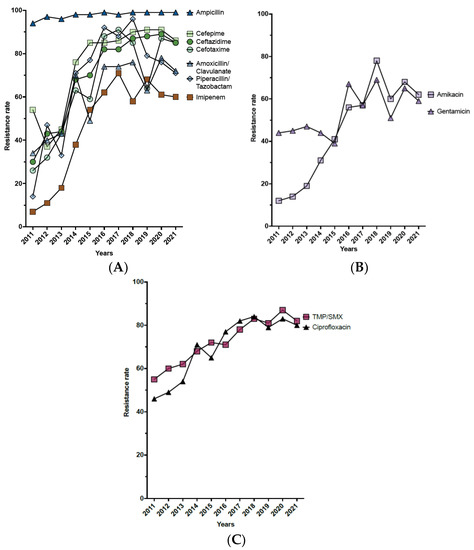
Figure 4.
Schematic demonstration of the antimicrobial resistance trends of K. pneumoniae. (A). Evolution of resistance to β-lactam antibiotics. (B). Evolution of resistance to aminoglycoside. (C). Evolution of resistance to ciprofloxacin and cotrimoxazole (trimethoprim-sulfamethoxazole).
Next, we examined the prevalence and antimicrobial susceptibility pattern of the extended-spectrum β-lactamase (ESBL) K. pneumoniae (Figure 5). The results showed that the average prevalence of ESBL throughout the study period was 21.3%. In addition, our result showed that ampicillin and cephalosporin exhibited low activity against ESBL K. pneumoniae, with an average resistance rate of more than 90%. In contrast, the most effective antibiotics against ESBL K. pneumoniae were imipenem, with an average resistance rate of 14.63 ± 18.20, followed by tigecycline (19.67 ± 22.97), amikacin (21.31 ± 17.44), and colistin (21.52 ± 35.01) (Figure 5).
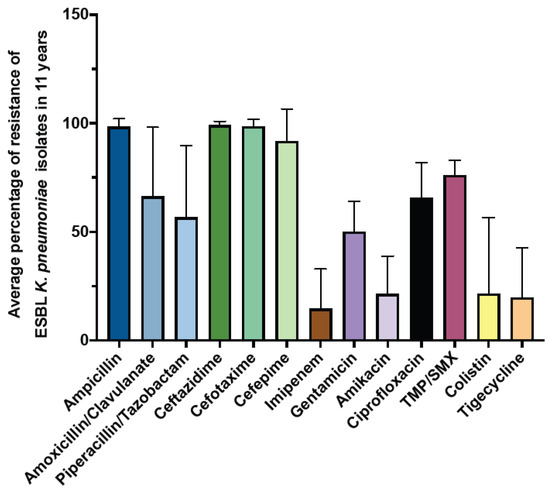
Figure 5.
The average percentage of resistance of ESBL K. pneumoniae over 11 years.
3. Discussion
Antimicrobial resistance (AMR) is a global threat to human health. More than 1 million deaths could have been avoided in 2019 if sensitive drugs had been replaced by resistant drugs in the world [16]. The misuse and overuse of antibiotics, together with the ability of resistant microbes to transmit from person to person, have significantly amplified the problem of AMR [17,18]. For instance, the emergence of multidrug-resistant K. pneumoniae was directly linked to the misuse and overuse of antibiotics when treating hospitalized patients [19]. Therefore, active surveillance of AMR trends is essential worldwide. In Saudi Arabia, only a limited number of studies were conducted to monitor the AMR trends of K. pneumoniae. Thus, in this study, we helped to fill the gap by investigating the prevalence and trends of AMR of K. pneumoniae.
Our results showed a gradual increase in the rate of infections with K. pneumoniae over the years. The prevalence of K. pneumoniae dramatically increased from 7.7% in 2011 to a peak in 2020 of 25.9%. The average rate of infections with K. pneumoniae in this study was 14.7% (n = 9014) over 11 years. This rate of infections with K. pneumoniae is lower than that reported previously in other regions in Saudi Arabia. For example, the prevalence of infections with K. pneumoniae was 39% and 18.6% in Asser and Bisha, respectively [13,20]. Nevertheless, the prevalence of infections with K. pneumoniae in Saudi Arabia is similar to that reported worldwide (i.e., 18.8 to 87.7% in Asia and 5 to 35% in Western countries) [21]. In this study, the majority of K. pneumoniae isolates were found to be from male patients, with an average of 58.5% ± 4.9. However, it is crucial to notice that the majority of total isolates were also obtained from male patients with an average of 59.3 ± 2.8. This result is in agreement with the previous studies, where the majority of K. pneumoniae isolates were obtained from male patients [20,22].
In addition, a direct correlation between the rate of infections with K. pneumoniae and age was observed. A similar observation was reported in Aseer, Saudi Arabia, where 42% of K. pneumoniae infections were in patients >60 years old [13]. This is possibly due to the decline of the immune system with aging, which allows the bacteria to multiply and cause infection [23]. Moreover, this study showed a significantly higher rate of K. pneumoniae isolated from inpatients compared to outpatients. Taken together, these findings may suggest that weakened immunity due to aging and extended hospitalization are potential risk factors for K. pneumoniae infection.
With regard to K. pneumoniae detection from different samples, the results showed that the highest rates were in blood samples, followed by sputum. This finding is consistent with the previous reports from Saudi Arabia, in which the majority of K. pneumoniae were isolated from blood and respiratory specimens [12]. The present finding relies on the fact that K. pneumoniae is one of the main causative agents of bloodstream infection and respiratory tract infection, and it should be taken into consideration during the diagnosis of respiratory infection [24,25].
In agreement with national and international reports, our results revealed the emergence of antimicrobial resistance, where K. pneumoniae is resistant to the most commonly used antibiotics [11,20,26]. Throughout the study period, ampicillin showed the highest resistance rate among the tested antibiotics, with a 97.6% average resistance rate. Similar results were reported in previous studies in Saudi Arabia; for example, in Aseer and Medina, ampicillin showed a 100% and 99.9% resistance rate, respectively [13,14]. Similarly, the results revealed that the resistance rate of K. pneumoniae to third- and fourth-generation cephalosporin rose significantly with 74.9% (n = 4451), 68.6% (n = 2244), and 81.0% (n = 4451) for ceftazidime, cefotaxime, and cefepime, respectively. This finding is in agreement with previous reports from Saudi Arabia, which indicated an increase in the resistance rate of K. pneumoniae to cephalosporin antibiotics [13,14]. The same finding was also reported in China and other countries worldwide, which suggests a global rise in resistance to third- and fourth-generation cephalosporins by K. pneumoniae [11,27]. A similar increase in the resistance rate of K. pneumoniae was also detected for amikacin. Interestingly, during the earlier years of the study, amikacin was one of the most effective antibiotics, with a resistance rate of 11.9%. Unfortunately, the rate of resistance started to increase aggressively over the period of study to reach its peak of 77.7% in 2018. A similar finding was reported in Medina, Saudi Arabia, where the resistance rate of K. pneumoniae to amikacin increased from 28.9% to 39.7% over the four-year duration of the study [14].
Carbapenem is a potent antibiotic to treat infections caused by Gram-negative bacteria. It has been used for a long time as a first choice to treat infections caused by ESBL-producing bacteria. However, resistance to carbapenems is alarmingly increasing, resulting in a future global crisis [28,29]. In this study, we assessed the resistance profile to imipenem, which showed a remarkable increase in the resistance rate over the study period from only 6.6% in 2011 to 59.9% in 2021. The rapid increase in carbapenem-resistant Enterobacteriaceae (CRE) has been reported both locally and globally [12,13,14,30,31]. This increase in CRE is linked to the increased use and consumption of β-lactamase inhibitors [32,33].
Colistin was an antibiotic commonly used to treat infections caused by Gram-negative bacteria in the 1960s and 1970s [34]. However, as a result of colistin renal- and neurotoxicity, the drug was discontinued [35]. Nevertheless, due to the emergence of CRE, the use of colistin has become a necessity since colistin is the last resort to treat CRE infections [15]. Consequently, even a low resistant rate could result in a threatening situation in the future. In this study, no significant change in the resistance rate of colistin was observed over the years, which could be due to the low number of tested isolates. Nevertheless, the resistance pattern showed some fluctuation during the study period. Multiple studies revealed a notable rise in colistin resistance both in the Kingdom of Saudi Arabia and globally. For example, the prevalence of colistin resistance in Medina, Ha’il, and Asser (Saudi Arabia) was reported to be 40.7%, 26.80%, and 35.10%, respectively [13,14,36]. Internationally, the prevalence of colistin resistance varies widely, with 13% in the USA and 73% in Italy [37,38].
In conclusion, our study revealed that the prevalence of K. pneumoniae increased significantly during the study period. In addition, we observed a significant increase in the resistance rate to nearly all of the tested antibiotics (except colistin and tigecycline) over the study years. Unfortunately, no significant decline in the resistance pattern was found in any of the tested antibiotics. Therefore, the reasonable use of antibiotics, an active antimicrobial stewardship program, and intensive educational programs for physicians as well as clinical pharmacists must be implemented to control and reduce antimicrobial resistance among K. pneumoniae and other multidrug-resistant bacteria. This study was limited to a single center from Makkah in Saudi Arabia. This investigation was conducted with a total sample size of 61,027 isolates collected over 11 years. Thus, we can comfortably state that this is a representative sample of Makkah. However, this does not exclude the need to perform similar studies in other regions and include more cities and centers to obtain a representative sample of the country. Nevertheless, such a study will need more funding and time to perform. Due to the unavailability of antimicrobial prescription patterns over the study period, we were unable to analyze the impact of prescription practices in Makkah on the emergence of antimicrobial resistance. Thus, we recommend further studies in this field that include more cities and centers and with more details on prescription patterns and patient characteristics such as comorbidity and mortality.
4. Materials and Methods
4.1. Study Design
This retrospective study was conducted to explore antibiotic susceptibility patterns among all K. pneumoniae isolates from a tertiary hospital for a duration of eleven years. Samples from January 2011 to December 2021 were included in the study. During that period, the total number of isolates was 61,027. Samples were collected from different wards and included different sample types, such as urine, blood, pus, sputum, swabs, and body fluid. Standard microbiology techniques were performed, and all samples were cultured at 37 °C for 24–48 h in two different media: blood sheep agar and McConkey agar, except for urine samples which were cultured in cystine–lactose–electrolyte-deficient agar (CLED). After 24–48 h, bacterial identification and antibiotic susceptibility patterns were determined using the Vitek-2 (bioMérieux, Marcy-l’Étoile, France) automated system (GN-21341 cards were used for identification, whereas the N291, N292, and N204 cards were used for antibiotic susceptibility) and Microscan walkaway (Beckman Coulter, Brea, CA, USA) automated system (Negative Breakpoint Combo 50, NBC 50). Antibiotics investigated in this study included ampicillin, amoxicillin/clavulanate, piperacillin/tazobactam, ceftazidime, cefotaxime, cefepime, imipenem, gentamicin, amikacin, ciprofloxacin, colistin, tigecycline, and cotrimoxazole. The interpretation of the MIC results relied on Clinical Laboratory Standard Institute guidelines [39]. Identification of ESBL was performed using a simultaneous examination of the inhibitory effects of ceftazidime, cefotaxime, and cefepime, with and without clavulanate.
4.2. Statistical Analysis
The total number of patients, specimen type, and antibiograms were entered into a database. Descriptive analysis was conducted to identify the frequency and distribution of all variables. A comparison of antibiotic susceptibility from different years was made using the chi-square test for trends. A p-value less than 0.05 was considered statistically significant. Statistical analysis was performed using GraphPad Prism 9.3.0 (GraphPad Software Inc., San Diego, CA, USA).
Author Contributions
Conceptualization, N.A.J.; methodology, A.M.A.-G., A.M.M. and F.S.B.; software, N.A.J. and A.M.A.-G.; validation, S.S.A., F.B., S.H.H. and A.K.J.; formal analysis, A.M.A.-G. and A.M.M.; investigation, A.M.M., A.A.B. and H.M.A.-S.; resources, N.A.J.; data curation, A.M.A.-G., A.M.M. and N.A.J.; writing—original draft preparation, N.A.J.; writing—review and editing, N.A.J., H.F., A.M.M. and S.S.A.; visualization, N.A.J.; supervision, N.A.J.; project administration, N.A.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Research Ethics and Advisory Committees of the College of Medicine, Umm Al-Qura University (reference no HAPO-02-K-012-2022-02-946).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Badger-Emeka, L.I.; Al-Sultan, A.A.; Bohol, M.F.F.; Al-Anazi, M.R.; Al-Qahtani, A.A. Genetic Analysis, Population Structure, and Characterisation of Multidrug-Resistant Klebsiella pneumoniae from the Al-Hofuf Region of Saudi Arabia. Pathogens 2021, 10, 1097. [Google Scholar] [CrossRef] [PubMed]
- Medrzycka-Dabrowska, W.; Lange, S.; Zorena, K.; Dabrowski, S.; Ozga, D.; Tomaszek, L. Carbapenem-Resistant Klebsiella pneumoniae Infections in ICU COVID-19 Patients-A Scoping Review. J Clin. Med. 2021, 10, 2067. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.L.; da Silva, B.C.M.; Rezende, G.S.; Nakamura-Silva, R.; Pitondo-Silva, A.; Campanini, E.B.; Brito, M.C.A.; da Silva, E.M.L.; Freire, C.C.d.M.; Cunha, A.F.d.; et al. High Prevalence of Multidrug-Resistant Klebsiella pneumoniae Harboring Several Virulence and β-Lactamase Encoding Genes in a Brazilian Intensive Care Unit. Front. Microbiol. 2019, 9, 3198. [Google Scholar] [CrossRef] [PubMed]
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. Biomed. Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef] [PubMed]
- Al-Agamy, M.H.; Shibl, A.M.; Tawfik, A.F. Prevalence and molecular characterization of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in Riyadh, Saudi Arabia. Ann. Saudi Med. 2009, 29, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Alasmary, M.Y. Antimicrobial Resistance Patterns and ESBL of Uropathogens Isolated from Adult Females in Najran Region of Saudi Arabia. Clin. Pract. 2021, 11, 650–658. [Google Scholar] [CrossRef]
- Knothe, H.; Shah, P.; Krcmery, V.; Antal, M.; Mitsuhashi, S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 1983, 11, 315–317. [Google Scholar] [CrossRef]
- Teklu, D.S.; Negeri, A.A.; Legese, M.H.; Bedada, T.L.; Woldemariam, H.K.; Tullu, K.D. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob. Resist. Infect. Control 2019, 8, 39. [Google Scholar] [CrossRef]
- Armstrong, T.; Fenn, S.J.; Hardie, K.R. JMM Profile: Carbapenems: A broad-spectrum antibiotic. J. Med. Microbiol. 2021, 70, 1462. [Google Scholar] [CrossRef]
- Loho, T.; Dharmayanti, A. Colistin: An antibiotic and its role in multiresistant Gram-negative infections. Acta Med. Indones. 2015, 47, 157–168. [Google Scholar]
- Mohd Asri, N.A.; Ahmad, S.; Mohamud, R.; Mohd Hanafi, N.; Mohd Zaidi, N.F.; Irekeola, A.A.; Shueb, R.H.; Yee, L.C.; Mohd Noor, N.; Mustafa, F.H.; et al. Global Prevalence of Nosocomial Multidrug-Resistant Klebsiella pneumoniae: A Systematic Review and Meta-Analysis. Antibiotics 2021, 10, 1508. [Google Scholar] [CrossRef] [PubMed]
- Al-Zahrani, I.A.; Alsiri, B.A. The emergence of carbapenem-resistant Klebsiella pneumoniae isolates producing OXA-48 and NDM in the Southern (Asir) province, Saudi Arabia. Saudi Med. J. 2018, 39, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Al Bshabshe, A.; Al-Hakami, A.; Alshehri, B.; Al-Shahrani, K.A.; Alshehri, A.A.; Al Shahrani, M.B.; Assiry, I.; Joseph, M.R.; Alkahtani, A.; Hamid, M.E. Rising Klebsiella pneumoniae Infections and Its Expanding Drug Resistance in the Intensive Care Unit of a Tertiary Healthcare Hospital, Saudi Arabia. Cureus 2020, 12, e10060. [Google Scholar] [CrossRef] [PubMed]
- Al-Zalabani, A.; AlThobyane, O.A.; Alshehri, A.H.; Alrehaili, A.O.; Namankani, M.O.; Aljafri, O.H. Prevalence of Klebsiella pneumoniae Antibiotic Resistance in Medina, Saudi Arabia, 2014–2018. Cureus 2020, 12, e9714. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed Ahmed, M.A.E.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef]
- Antimicrobial Resistance, C. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Gil-Gil, T.; Laborda, P.; Sanz-Garcia, F.; Hernando-Amado, S.; Blanco, P.; Martinez, J.L. Antimicrobial resistance: A multifaceted problem with multipronged solutions. Microbiologyopen 2019, 8, e945. [Google Scholar] [CrossRef]
- Brinkac, L.; Voorhies, A.; Gomez, A.; Nelson, K.E. The Threat of Antimicrobial Resistance on the Human Microbiome. Microb. Ecol. 2017, 74, 1001–1008. [Google Scholar] [CrossRef]
- Anes, J.; Hurley, D.; Martins, M.; Fanning, S. Exploring the Genome and Phenotype of Multi-Drug Resistant Klebsiella pneumoniae of Clinical Origin. Front. Microbiol. 2017, 8, 1913. [Google Scholar] [CrossRef]
- Ibrahim, M.E. High antimicrobial resistant rates among Gram-negative pathogens in intensive care units. A retrospective study at a tertiary care hospital in Southwest Saudi Arabia. Saudi Med. J. 2018, 39, 1035–1043. [Google Scholar] [CrossRef]
- Chang; Sharma, L.; Dela Cruz, C.S.; Zhang, D. Clinical Epidemiology, Risk Factors, and Control Strategies of Klebsiella pneumoniae Infection. Front. Microbiol. 2021, 12, 750662. [Google Scholar] [CrossRef] [PubMed]
- Nirwati, H.; Sinanjung, K.; Fahrunissa, F.; Wijaya, F.; Napitupulu, S.; Hati, V.P.; Hakim, M.S.; Meliala, A.; Aman, A.T.; Nuryastuti, T. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proc. 2019, 13, 20. [Google Scholar] [CrossRef] [PubMed]
- Montecino-Rodriguez, E.; Berent-Maoz, B.; Dorshkind, K. Causes, consequences, and reversal of immune system aging. J. Clin. Investig. 2013, 123, 958–965. [Google Scholar] [CrossRef] [PubMed]
- Claassen-Weitz, S.; Lim, K.Y.L.; Mullally, C.; Zar, H.J.; Nicol, M.P. The association between bacteria colonizing the upper respiratory tract and lower respiratory tract infection in young children: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2021, 27, 1262–1270. [Google Scholar] [CrossRef]
- Kern, W.V.; Rieg, S. Burden of bacterial bloodstream infection-a brief update on epidemiology and significance of multidrug-resistant pathogens. Clin. Microbiol. Infect. 2020, 26, 151–157. [Google Scholar] [CrossRef]
- Yezli, S.; Shibl, A.M.; Livermore, D.M.; Memish, Z.A. Prevalence and antimicrobial resistance among Gram-negative pathogens in Saudi Arabia. J. Chemother. 2014, 26, 257–272. [Google Scholar] [CrossRef]
- Pei, N.; Liu, Q.; Cheng, X.; Liang, T.; Jian, Z.; Wang, S.; Zhong, Y.; He, J.; Zhou, M.; Kristiansen, K.; et al. Longitudinal Study of the Drug Resistance in Klebsiella pneumoniae of a Tertiary Hospital, China: Phenotypic Epidemiology Analysis (2013–2018). Infect. Drug Resist. 2021, 14, 613–626. [Google Scholar] [CrossRef]
- Hansen, G.T. Continuous Evolution: Perspective on the Epidemiology of Carbapenemase Resistance Among Enterobacterales and Other Gram-Negative Bacteria. Infect. Dis. Ther. 2021, 10, 75–92. [Google Scholar] [CrossRef]
- Potter, R.F.; D’Souza, A.W.; Dantas, G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist. Updates 2016, 29, 30–46. [Google Scholar] [CrossRef]
- Li, Y.; Sun, Q.L.; Shen, Y.; Zhang, Y.; Yang, J.W.; Shu, L.B.; Zhou, H.W.; Wang, Y.; Wang, B.; Zhang, R.; et al. Rapid Increase in Prevalence of Carbapenem-Resistant Enterobacteriaceae (CRE) and Emergence of Colistin Resistance Gene mcr-1 in CRE in a Hospital in Henan, China. J. Clin. Microbiol. 2018, 56, e01932-17. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Q.; Yin, Y.; Chen, H.; Jin, L.; Gu, B.; Xie, L.; Yang, C.; Ma, X.; Li, H.; et al. Epidemiology of Carbapenem-Resistant Enterobacteriaceae Infections: Report from the China CRE Network. Antimicrob. Agents Chemother. 2018, 62, e01882-17. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Wang, H.; Chen, C.; Tao, Z.; Yin, C.; Yin, A.; Ma, C.; Idris, A. Surveillance of carbapenem-resistant Klebsiella pneumoniae in Chinese hospitals—A five-year retrospective study. J Infect. Dev. Ctries. 2019, 13, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Federico, M.P.; Furtado, G.H. Immediate and later impacts of antimicrobial consumption on carbapenem-resistant Acinetobacter spp., Pseudomonas aeruginosa, and Klebsiella spp. in a teaching hospital in Brazil: A 10-year trend study. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 2153–2158. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.M.; Bachman, M.A. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Kasiakou, S.K. Toxicity of polymyxins: A systematic review of the evidence from old and recent studies. Crit. Care 2006, 10, R27. [Google Scholar] [CrossRef]
- Said, K.B.; Alsolami, A.; Khalifa, A.M.; Khalil, N.A.; Moursi, S.; Osman, A.; Fahad, D.; Rakha, E.; Rashidi, M.; Moussa, S.; et al. A Multi-Point Surveillance for Antimicrobial Resistance Profiles among Clinical Isolates of Gram-Negative Bacteria Recovered from Major Ha’il Hospitals, Saudi Arabia. Microorganisms 2021, 9, 2024. [Google Scholar] [CrossRef] [PubMed]
- Rojas, L.J.; Salim, M.; Cober, E.; Richter, S.S.; Perez, F.; Salata, R.A.; Kalayjian, R.C.; Watkins, R.R.; Marshall, S.; Rudin, S.D.; et al. Colistin Resistance in Carbapenem-Resistant Klebsiella pneumoniae: Laboratory Detection and Impact on Mortality. Clin. Infect. Dis. 2017, 64, 711–718. [Google Scholar] [CrossRef]
- Di Tella, D.; Tamburro, M.; Guerrizio, G.; Fanelli, I.; Sammarco, M.L.; Ripabelli, G. Molecular Epidemiological Insights into Colistin-Resistant and Carbapenemases-Producing Clinical Klebsiella pneumoniae Isolates. Infect. Drug Resist. 2019, 12, 3783–3795. [Google Scholar] [CrossRef]
- Ferraro, M.J. Performance Standards for Antimicrobial Susceptibility Testing; NCCLS: Orlando, FL, USA, 2001. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).