A Review of Biofilm Formation of Staphylococcus aureus and Its Regulation Mechanism
Abstract
1. Introduction
2. Biofilm Formation Process
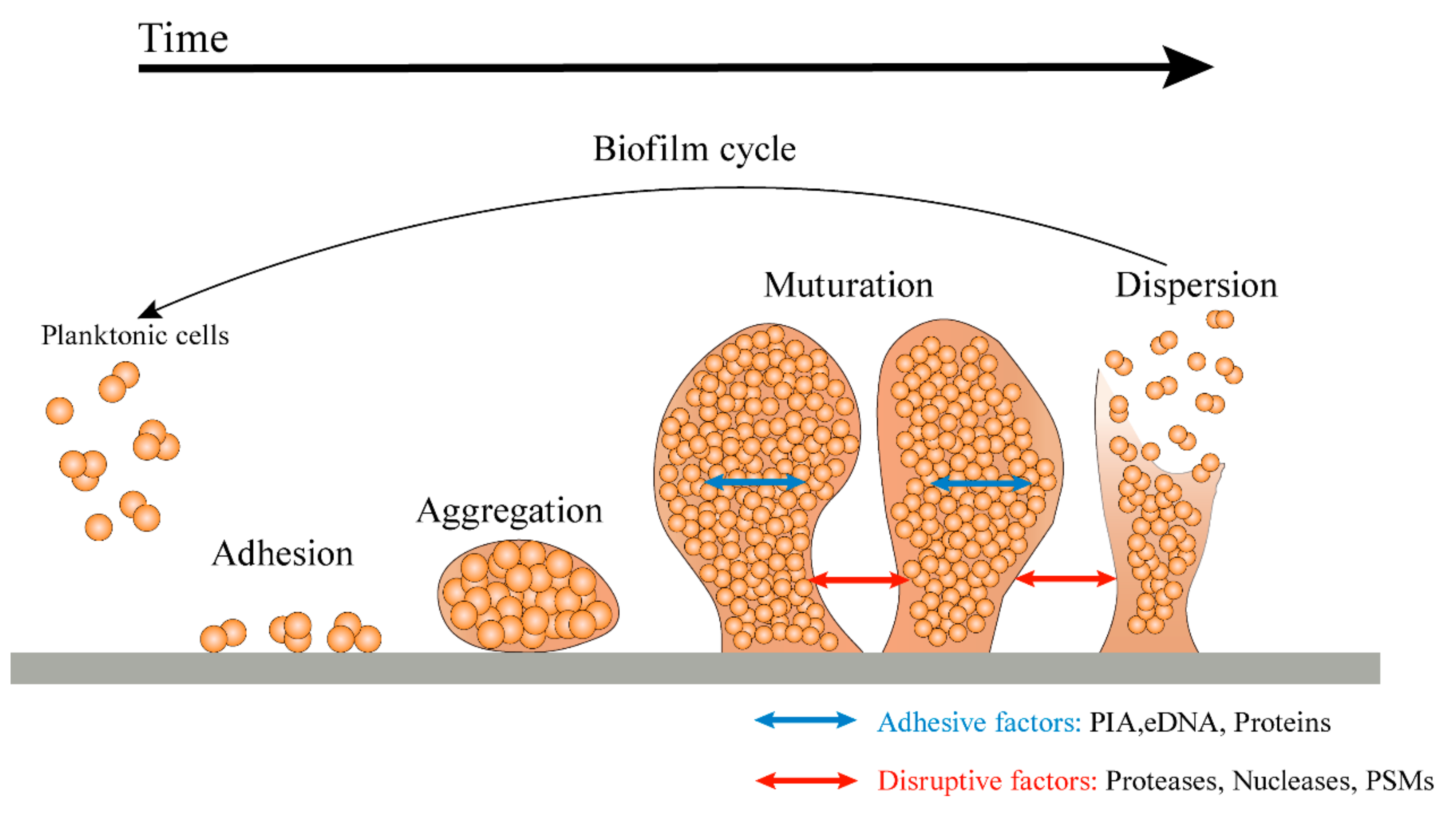
3. Biofilm Formation Mechanism
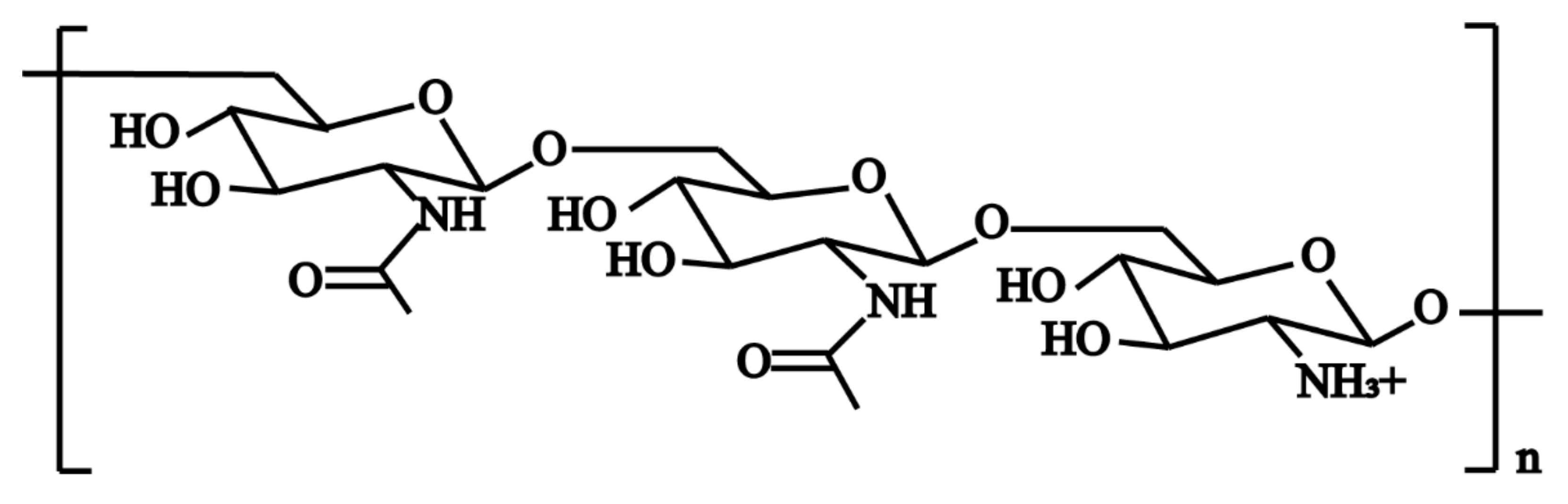
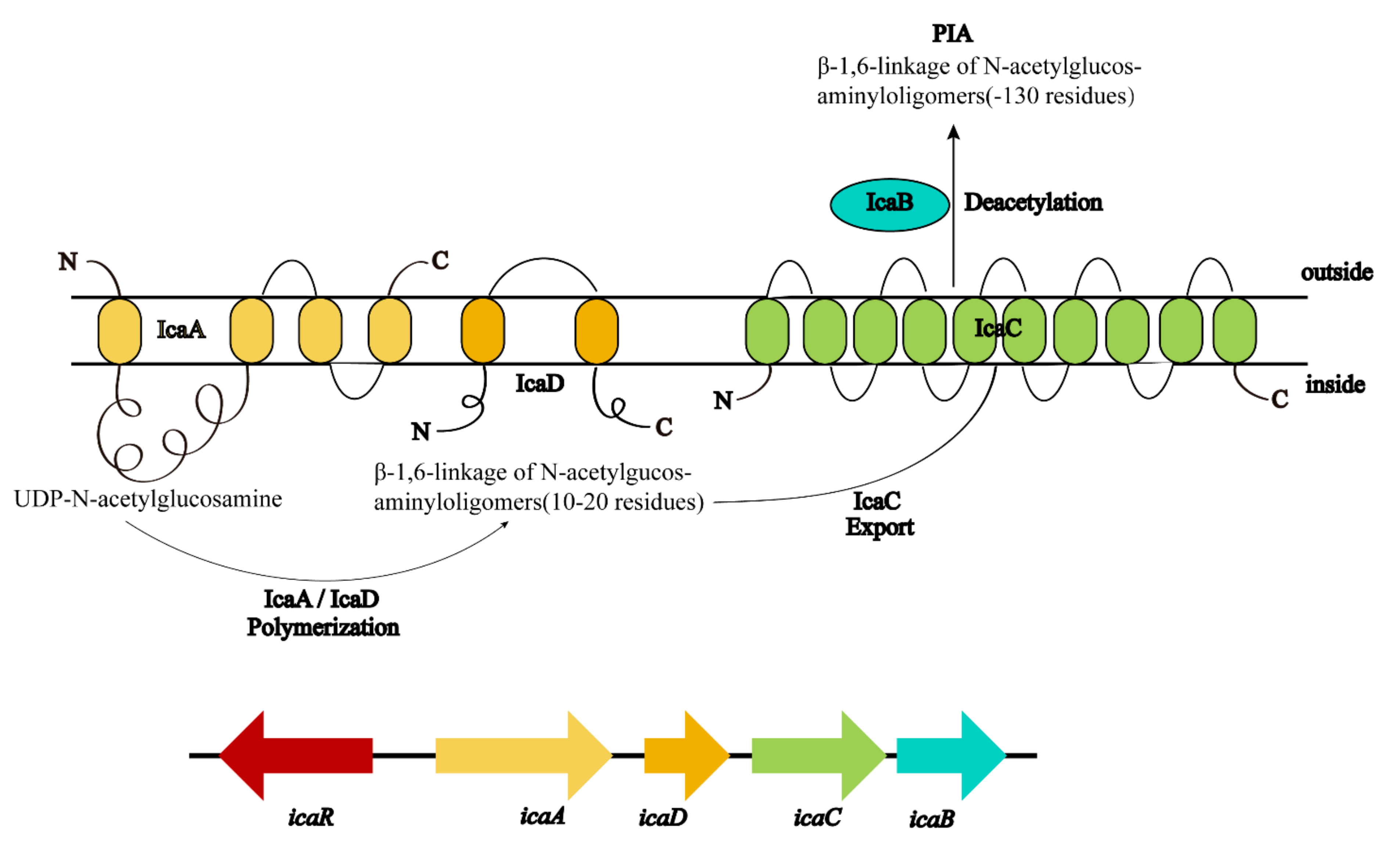
3.1. Polysaccharide Intercellular Adhesion(PIA)-Dependent Mechanism
3.2. Protein-Dependent Mechanisms
3.3. Extracellular DNA (eDNA)-Dependent Mechanism
4. Regulation Mechanism of Biofilm Formation
4.1. Regulation of Quorum-Sensing System-Mediated Biofilm Formation
4.2. The Global Response Regulator
4.3. ica Operon
4.4. Two-Component Regulatory System
4.5. The Second Messenger
4.6. sRNAs
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm Matrixome: Extracellular Components in Structured Microbial Communities. Trends Microbiol. 2020, 28, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, P. Extracellular polymeric substances, a key element in understanding biofilm phenotype. AIMS Microbiol. 2018, 4, 274–288. [Google Scholar] [CrossRef] [PubMed]
- Guilhen, C.; Forestier, C.; Balestrino, D. Biofilm dispersal: Multiple elaborate strategies for dissemination of bacteria with unique properties. Mol. Microbiol. 2017, 105, 188–210. [Google Scholar] [CrossRef] [PubMed]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.-F.; Alarcon, E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, e01067. [Google Scholar] [CrossRef]
- Lowy, F.D. Staphylococcus aureus Infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef]
- Zheng, Y.; He, L.; Asiamah, T.K.; Otto, M. Colonization of medical devices by staphylococci. Environ. Microbiol. 2018, 20, 3141–3153. [Google Scholar] [CrossRef]
- Lee, J.; Zilm, P.S.; Kidd, S.P. Novel Research Models for Staphylococcus aureus Small Colony Variants (SCV) Development: Co-pathogenesis and Growth Rate. Front. Microbiol. 2020, 11, 321. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the Natural environment to infectious diseases. Nat. Rev. Genet. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- Guo, H.; Tong, Y.; Cheng, J.; Abbas, Z.; Li, Z.; Wang, J.; Zhou, Y.; Si, D.; Zhang, R. Biofilm and Small Colony Variants—An Update on Staphylococcus aureus Strategies toward Drug Resistance. Int. J. Mol. Sci. 2022, 23, 1241. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 2002, 292, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Lee, J.-H.; Beyenal, H.; Lee, J. Fatty Acids as Antibiofilm and Antivirulence Agents. Trends Microbiol. 2020, 28, 753–768. [Google Scholar] [CrossRef] [PubMed]
- Beloin, C.; Renard, S.; Ghigo, J.-M.; Lebeaux, D. Novel approaches to combat bacterial biofilms. Curr. Opin. Pharmacol. 2014, 18, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Girish, V.M.; Liang, H.; Aguilan, J.T.; Nosanchuk, J.D.; Friedman, J.M.; Nacharaju, P. Anti-biofilm activity of garlic extract loaded nanoparticles. Nanomedicine 2019, 20, 102009. [Google Scholar] [CrossRef] [PubMed]
- Donne, J.; Dewilde, S. The Challenging World of Biofilm Physiology. In Recent Advances in Microbial Oxygen-Binding Proteins; Poole, R.K., Ed.; Advances in Microbial Physiology; Academic Press: London, UK, 2015; Volume 67, pp. 235–292. [Google Scholar]
- Ribeiro, S.M.; Felício, M.R.; Boas, E.V.; Gonçalves, S.; Costa, F.F.; Samy, R.P.; Santos, N.C.; Franco, O.L. New frontiers for anti-biofilm drug development. Pharmacol. Ther. 2016, 160, 133–144. [Google Scholar] [CrossRef]
- Idrees, M.; Sawant, S.; Karodia, N.; Rahman, A. Staphylococcus aureus Biofilm: Morphology, Genetics, Pathogenesis and Treatment Strategies. Int. J. Environ. Res. Public Health 2021, 18, 7602. [Google Scholar] [CrossRef]
- Moormeier, D.E.; Bayles, K.W. Staphylococcus aureus biofilm: A complex developmental organism. Mol. Microbiol. 2017, 104, 365–376. [Google Scholar] [CrossRef]
- Schilcher, K.; Horswill, A.R. Staphylococcal Biofilm Development: Structure, Regulation, and Treatment Strategies. Microbiol. Mol. Biol. Rev. 2020, 84, e00026-19. [Google Scholar] [CrossRef]
- O’Neill, E.; Pozzi, C.; Houston, P.; Humphreys, H.; Robinson, D.A.; Loughman, A.; Foster, T.J.; O’Gara, J.P. A Novel Staphylococcus aureus Biofilm Phenotype Mediated by the Fibronectin-Binding Proteins, FnBPA and FnBPB. J. Bacteriol. 2008, 190, 3835–3850. [Google Scholar] [CrossRef]
- Wolz, C.; Goerke, C.; Landmann, R.; Zimmerli, W.; Fluckiger, U. Transcription of Clumping Factor A in Attached and Unattached Staphylococcus aureus In Vitro and during Device-Related Infection. Infect. Immun. 2002, 70, 2758–2762. [Google Scholar] [CrossRef] [PubMed]
- Sabat, A.; Melles, D.C.; Martirosian, G.; Grundmann, H.; van Belkum, A.; Hryniewicz, W. Distribution of the Serine-Aspartate Repeat Protein-Encoding sdr Genes among Nasal-Carriage and Invasive Staphylococcus aureus Strains. J. Clin. Microbiol. 2006, 44, 1135–1138. [Google Scholar] [CrossRef] [PubMed]
- Beaussart, A.; Feuillie, C.; El-Kirat-Chatel, S. The microbial adhesive arsenal deciphered by atomic force microscopy. Nanoscale 2020, 12, 23885–23896. [Google Scholar] [CrossRef] [PubMed]
- Mangwani, N.; Kumari, S.; Das, S. Bacterial biofilms and quorum sensing: Fidelity in bioremediation technology. In Biotechnology and Genetic Engineering Reviews; Mayes, S., Harding, S.E., Eds.; Biotechnology & Genetic Engineering Reviews; Taylor & Francis Ltd.: Abingdon, UK, 2016; Volume 32, pp. 43–73. [Google Scholar]
- Otto, M. Staphylococcal biofilms. In Bacterial Biofilms; Romeo, T., Ed.; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2008; Volume 322, p. 207. [Google Scholar]
- Herman-Bausier, P.; El-Kirat-Chatel, S.; Foster, T.J.; Geoghegan, J.A.; Dufrêne, Y.F. Staphylococcus aureus Fibronectin-Binding Protein A Mediates Cell-Cell Adhesion through Low-Affinity Homophilic Bonds. mBio 2015, 6, e00413-15. [Google Scholar] [CrossRef] [PubMed]
- Rasamiravaka, T.; Labtani, Q.; Duez, P.; El Jaziri, M. The Formation of Biofilms by Pseudomonas aeruginosa: A Review of the Natural and Synthetic Compounds Interfering with Control Mechanisms. BioMed Res. Int. 2015, 2015, 759348. [Google Scholar] [CrossRef]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm Formation as Microbial Development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef]
- Gupta, P.; Sarkar, S.; Das, B.; Bhattacharjee, S.; Tribedi, P. Biofilm, pathogenesis and prevention—A journey to break the wall: A review. Arch. Microbiol. 2016, 198, 1–15. [Google Scholar] [CrossRef]
- Costerton, J. Introduction to biofilm. Int. J. Antimicrob. Agents 1999, 11, 217–221; discussion 237–239. [Google Scholar] [CrossRef]
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef]
- Boles, B.R.; Horswill, A.R. Staphylococcal biofilm disassembly. Trends Microbiol. 2011, 19, 449–455. [Google Scholar] [CrossRef]
- Vandana; Das, S. Genetic regulation, biosynthesis and applications of extracellular polysaccharides of the biofilm matrix of bacteria. Carbohydr. Polym. 2022, 291, 119536. [Google Scholar] [CrossRef] [PubMed]
- Periasamy, S.; Joo, H.-S.; Duong, A.C.; Bach, T.-H.L.; Tan, V.Y.; Chatterjee, S.S.; Cheung, G.Y.C.; Otto, M. How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Cheung, G.Y.; Joo, H.-S.; Chatterjee, S.S.; Otto, M. Phenol-soluble modulins—Critical determinants of staphylococcal virulence. FEMS Microbiol. Rev. 2014, 38, 698–719. [Google Scholar] [CrossRef]
- Le, K.Y.; Dastgheyb, S.; Ho, T.V.; Otto, M. Molecular determinants of staphylococcal biofilm dispersal and structuring. Front. Cell. Infect. Microbiol. 2014, 4, 167. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, K.; Syed, A.K.; Stephenson, R.E.; Rickard, A.H.; Boles, B.R. Functional Amyloids Composed of Phenol Soluble Modulins Stabilize Staphylococcus aureus Biofilms. PLoS Pathog. 2012, 8, e1002744. [Google Scholar] [CrossRef] [PubMed]
- Marinelli, P.; Pallares, I.; Navarro, S.; Ventura, S. Dissecting the contribution of Staphylococcus aureus α-phenol-soluble modulins to biofilm amyloid structure. Sci. Rep. 2016, 6, 34552. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal Infections: Mechanisms of Biofilm Maturation and Detachment as Critical Determinants of Pathogenicity. Annu. Rev. Med. 2013, 64, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Nguyen, T.H.; Otto, M. The staphylococcal exopolysaccharide PIA—Biosynthesis and role in biofilm formation, colonization, and infection. Comput. Struct. Biotechnol. J. 2020, 18, 3324–3334. [Google Scholar] [CrossRef]
- Mack, D.; Fischer, W.; Krokotsch, A.; Leopold, K.; Hartmann, R.; Egge, H.; Laufs, R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: Purification and structural analysis. J. Bacteriol. 1996, 178, 175–183. [Google Scholar] [CrossRef]
- Cramton, S.E.; Gerke, C.; Schnell, N.F.; Nichols, W.W.; Götz, F. The Intercellular Adhesion (ica) Locus Is Present in Staphylococcus aureus and Is Required for Biofilm Formation. Infect. Immun. 1999, 67, 5427–5433. [Google Scholar] [CrossRef] [PubMed]
- O’Gara, J.P. ica and beyond: Biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 2007, 270, 179–188. [Google Scholar] [CrossRef]
- Gerke, C.; Kraft, A.; Süßmuth, R.; Schweitzer, O.; Götz, F. Characterization of the N-Acetylglucosaminyltransferase Activity Involved in the Biosynthesis of the Staphylococcus epidermidis Polysaccharide Intercellular Adhesin. J. Biol. Chem. 1998, 273, 18586–18593. [Google Scholar] [CrossRef] [PubMed]
- Atkin, K.E.; MacDonald, S.J.; Brentnall, A.S.; Potts, J.R.; Thomas, G.H. A different path: Revealing the function of staphylococcal proteins in biofilm formation. FEBS Lett. 2014, 588, 1869–1872. [Google Scholar] [CrossRef]
- Vuong, C.; Kocianova, S.; Voyich, J.M.; Yao, Y.; Fischer, E.R.; DeLeo, F.R.; Otto, M. A Crucial Role for Exopolysaccharide Modification in Bacterial Biofilm Formation, Immune Evasion, and Virulence. J. Biol. Chem. 2004, 279, 54881–54886. [Google Scholar] [CrossRef] [PubMed]
- Sedarat, Z.; Taylor-Robinson, A.W. Biofilm Formation by Pathogenic Bacteria: Applying a Staphylococcus aureus Model to Appraise Potential Targets for Therapeutic Intervention. Pathogens 2022, 11, 388. [Google Scholar] [CrossRef] [PubMed]
- Marques, V.F.; Santos, H.A.; Santos, T.H.; Melo, D.A.; Coelho, S.M.; Coelho, I.S.; Souza, M.M. Expression of icaA and icaD genes in biofilm formation in Staphylococcus aureus isolates from bovine subclinical mastitis. Pesquisa Veterinária Brasileira 2021, 41, e06645. [Google Scholar] [CrossRef]
- Brooks, J.L.; Jefferson, K.K. Phase Variation of Poly-N-Acetylglucosamine Expression in Staphylococcus aureus. PLoS Pathog. 2014, 10, e1004292. [Google Scholar] [CrossRef]
- O’Neill, E.; Pozzi, C.; Houston, P.; Smyth, D.; Humphreys, H.; Robinson, D.A.; O’Gara, J.P. Association between Methicillin Susceptibility and Biofilm Regulation in Staphylococcus aureus Isolates from Device-Related Infections. J. Clin. Microbiol. 2007, 45, 1379–1388. [Google Scholar] [CrossRef] [PubMed]
- Cucarella, C.; Tormo, M.A.; Ubeda, C.; Trotonda, M.P.; Monzón, M.; Peris, C.; Amorena, B.; Lasa, I.; Penadés, J.R. Role of Biofilm-Associated Protein Bap in the Pathogenesis of Bovine Staphylococcus aureus. Infect. Immun. 2004, 72, 2177–2185. [Google Scholar] [CrossRef]
- Taglialegna, A.; Navarro, S.; Ventura, S.; Garnett, J.A.; Matthews, S.; Penades, J.R.; Lasa, I.; Valle, J. Staphylococcal Bap Proteins Build Amyloid Scaffold Biofilm Matrices in Response to Environmental Signals. PLoS Pathog. 2016, 12, e1005711. [Google Scholar] [CrossRef]
- Arrizubieta, M.J.; Toledo-Arana, A.; Amorena, B.; Penadés, J.R.; Lasa, I. Calcium Inhibits Bap-Dependent Multicellular Behavior in Staphylococcus aureus. J. Bacteriol. 2004, 186, 7490–7498. [Google Scholar] [CrossRef] [PubMed]
- Pietrocola, G.; Campoccia, D.; Motta, C.; Montanaro, L.; Arciola, C.R.; Speziale, P. Colonization and Infection of Indwelling Medical Devices by Staphylococcus aureus with an Emphasis on Orthopedic Implants. Int. J. Mol. Sci. 2022, 23, 5958. [Google Scholar] [CrossRef] [PubMed]
- McCourt, J.; O’Halloran, D.P.; McCarthy, H.; O’Gara, J.P.; Geoghegan, J.A. Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS Microbiol. Lett. 2014, 353, 157–164. [Google Scholar] [CrossRef]
- Herman-Bausier, P.; Labate, C.; Towell, A.M.; Derclaye, S.; Geoghegan, J.A.; Dufrêne, Y.F. Staphylococcus aureus clumping factor A is a force-sensitive molecular switch that activates bacterial adhesion. Proc. Natl. Acad. Sci. USA 2018, 115, 5564–5569. [Google Scholar] [CrossRef]
- Abraham, N.M.; Jefferson, K.K. Staphylococcus aureus clumping factor B mediates biofilm formation in the absence of calcium. Microbiology 2012, 158, 1504–1512. [Google Scholar] [CrossRef] [PubMed]
- Formosa-Dague, C.; Speziale, P.; Foster, T.J.; Geoghegan, J.A.; Dufrêne, Y.F. Zinc-dependent mechanical properties of Staphylococcus aureus biofilm-forming surface protein SasG. Proc. Natl. Acad. Sci. USA 2016, 113, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Corrigan, R.M.; Rigby, D.; Handley, P.; Foster, T.J. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 2007, 153, 2435–2446. [Google Scholar] [CrossRef]
- Schroeder, K.; Jularic, M.; Horsburgh, S.M.; Hirschhausen, N.; Neumann, C.; Bertling, A.; Schulte, A.; Foster, S.; Kehrel, B.E.; Peters, G.; et al. Molecular Characterization of a Novel Staphylococcus Aureus Surface Protein (SasC) Involved in Cell Aggregation and Biofilm Accumulation. PLoS ONE 2009, 4, e7567. [Google Scholar] [CrossRef]
- Feuillie, C.; Formosa-Dague, C.; Hays, L.M.C.; Vervaeck, O.; Derclaye, S.; Brennan, M.P.; Foster, T.J.; Geoghegan, J.A.; Dufrêne, Y.F. Molecular interactions and inhibition of the staphylococcal biofilm-forming protein SdrC. Proc. Natl. Acad. Sci. USA 2017, 114, 3738–3743. [Google Scholar] [CrossRef]
- Corrigan, R.M.; Miajlovic, H.; Foster, T.J. Surface proteins that promote adherence of Staphylococcus aureusto human desquamated nasal epithelial cells. BMC Microbiol. 2009, 9, 22. [Google Scholar] [CrossRef]
- Askarian, F.; Ajayi, C.; Hanssen, A.-M.; van Sorge, N.; Pettersen, I.; Diep, D.B.; Sollid, J.U.E.; Johannessen, M. The interaction between Staphylococcus aureus SdrD and desmoglein 1 is important for adhesion to host cells. Sci. Rep. 2016, 6, 22134. [Google Scholar] [CrossRef] [PubMed]
- Askarian, F.; Uchiyama, S.; Valderrama, J.A.; Ajayi, C.; Sollid, J.U.E.; van Sorge, N.M.; Nizet, V.; van Strijp, J.A.G.; Johannessen, M. Serine-Aspartate Repeat Protein D Increases Staphylococcus aureus Virulence and Survival in Blood. Infect. Immun. 2017, 85, e00559-16. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, M.; Hang, T.; Wang, C.; Yang, Y.; Pan, W.; Zang, J.; Zhang, M.; Zhang, X. Staphylococcus aureus SdrE captures complement factor H’s C-terminus via a novel ‘close, dock, lock and latch’ mechanism for complement evasion. Biochem. J. 2017, 474, 1619–1631. [Google Scholar] [CrossRef]
- Patti, J.M.; Boles, J.O.; Hook, M. Identification and biochemical characterization of the ligand binding domain of the collagen adhesin from Staphylococcus aureus. Biochemistry 1993, 32, 11428–11435. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.V.; Nallapareddy, S.R.; Sillanpää, J.; Murray, B.E. Importance of the Collagen Adhesin Ace in Pathogenesis and Protection against Enterococcus faecalis Experimental Endocarditis. PLoS Pathog. 2010, 6, e1000716. [Google Scholar] [CrossRef]
- Nakano, K.; Hokamura, K.; Taniguchi, N.; Wada, K.; Kudo, C.; Nomura, R.; Kojima, A.; Naka, S.; Muranaka, Y.; Thura, M.; et al. The collagen-binding protein of Streptococcus mutans is involved in haemorrhagic stroke. Nat. Commun. 2011, 2, 485. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Ko, Y.-P.; Liang, X.; Ross, C.L.; Liu, Q.; Murray, B.E.; Höök, M. Collagen-binding Microbial Surface Components Recognizing Adhesive Matrix Molecule (MSCRAMM) of Gram-positive Bacteria Inhibit Complement Activation via the Classical Pathway. J. Biol. Chem. 2013, 288, 20520–20531. [Google Scholar] [CrossRef]
- Mann, E.E.; Rice, K.C.; Boles, B.R.; Endres, J.L.; Ranjit, D.; Chandramohan, L.; Tsang, L.H.; Smeltzer, M.S.; Horswill, A.R.; Bayles, K.W. Modulation of eDNA Release and Degradation Affects Staphylococcus aureus Biofilm Maturation. PLoS ONE 2009, 4, e5822. [Google Scholar] [CrossRef]
- Campoccia, D.; Montanaro, L.; Arciola, C.R. Extracellular DNA (eDNA). A Major Ubiquitous Element of the Bacterial Biofilm Architecture. Int. J. Mol. Sci. 2021, 22, 9100. [Google Scholar] [CrossRef] [PubMed]
- Blakeman, J.T.; Morales-García, A.L.; Mukherjee, J.; Gori, K.; Hayward, A.S.; Lant, N.J.; Geoghegan, M. Extracellular DNA Provides Structural Integrity to a Micrococcus luteus Biofilm. Langmuir 2019, 35, 6468–6475. [Google Scholar] [CrossRef] [PubMed]
- Payne, D.E.; Boles, B.R. Emerging interactions between matrix components during biofilm development. Curr. Genet. 2016, 62, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Sharma, P.; Busscher, H.J.; van der Mei, H.C.; Krom, B.P. Role of Extracellular DNA in Initial Bacterial Adhesion and Surface Aggregation. Appl. Environ. Microbiol. 2010, 76, 3405–3408. [Google Scholar] [CrossRef]
- Wilton, M.; Charron-Mazenod, L.; Moore, R.; Lewenza, S. Extracellular DNA Acidifies Biofilms and Induces Aminoglycoside Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 544–553. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Batoni, G.; Maisetta, G.; Esin, S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim. Biophys. Acta 2016, 1858, 1044–1060. [Google Scholar] [CrossRef]
- Doroshenko, N.; Tseng, B.S.; Howlin, R.P.; Deacon, J.; Wharton, J.A.; Thurner, P.J.; Gilmore, B.F.; Parsek, M.R.; Stoodley, P. Extracellular DNA Impedes the Transport of Vancomycin in Staphylococcus epidermidis Biofilms Preexposed to Subinhibitory Concentrations of Vancomycin. Antimicrob. Agents Chemother. 2014, 58, 7273–7282. [Google Scholar] [CrossRef] [PubMed]
- Rice, K.C.; Firek, B.A.; Nelson, J.B.; Yang, S.-J.; Patton, T.G.; Bayles, K.W. The Staphylococcus aureus cidAB Operon: Evaluation of Its Role in Regulation of Murein Hydrolase Activity and Penicillin Tolerance. J. Bacteriol. 2003, 185, 2635–2643. [Google Scholar] [CrossRef]
- Groicher, K.H.; Firek, B.A.; Fujimoto, D.F.; Bayles, K.W. The Staphylococcus aureus lrgAB Operon Modulates Murein Hydrolase Activity and Penicillin Tolerance. J. Bacteriol. 2000, 182, 1794–1801. [Google Scholar] [CrossRef] [PubMed]
- Parsek, M.R.; Greenberg, E. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005, 13, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Hu, W.; Tian, Z.; Yuan, D.; Yi, G.; Zhou, Y.; Cheng, Q.; Zhu, J.; Li, M. Developing natural products as potential anti-biofilm agents. Chin. Med. 2019, 14, 11. [Google Scholar] [CrossRef] [PubMed]
- Sionov, R.V.; Steinberg, D. Targeting the Holy Triangle of Quorum Sensing, Biofilm Formation, and Antibiotic Resistance in Pathogenic Bacteria. Microorganisms 2022, 10, 1239. [Google Scholar] [CrossRef]
- Le, K.Y.; Otto, M. Quorum-sensing regulation in staphylococci—An overview. Front. Microbiol. 2015, 6, 1174. [Google Scholar] [CrossRef]
- Dunman, P.M.; Murphy, E.; Haney, S.; Palacios, D.; Tucker-Kellogg, G.; Wu, S.; Brown, E.L.; Zagursky, R.J.; Shlaes, D.; Projan, S.J. Transcription Profiling-Based Identification of Staphylococcus aureus Genes Regulated by the agr and/or sarA Loci. J. Bacteriol. 2001, 183, 7341–7353. [Google Scholar] [CrossRef]
- Harraghy, N.; Kerdudou, S.; Herrmann, M. Quorum-sensing systems in staphylococci as therapeutic targets. Anal. Bioanal. Chem. 2006, 387, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Ji, G.; Beavis, R.; Novick, R.P. Bacterial Interference Caused by Autoinducing Peptide Variants. Science 1997, 276, 2027–2030. [Google Scholar] [CrossRef] [PubMed]
- Gordon, C.P.; Olson, S.D.; Lister, J.L.; Kavanaugh, J.S.; Horswill, A.R. Truncated Autoinducing Peptides as Antagonists of Staphylococcus lugdunensis Quorum Sensing. J. Med. Chem. 2016, 59, 8879–8888. [Google Scholar] [CrossRef]
- Novick, R.P.; Geisinger, E. Quorum Sensing in Staphylococci. Annu. Rev. Genet. 2008, 42, 541–564. [Google Scholar] [CrossRef] [PubMed]
- Mootz, J.M.; Benson, M.A.; Heim, C.E.; Crosby, H.A.; Kavanaugh, J.S.; Dunman, P.M.; Kielian, T.; Torres, V.J.; Horswill, A.R. Rot is a key regulator of Staphylococcus aureus biofilm formation. Mol. Microbiol. 2015, 96, 388–404. [Google Scholar] [CrossRef] [PubMed]
- Yarwood, J.M.; Bartels, D.J.; Volper, E.M.; Greenberg, E.P. Quorum Sensing in Staphylococcus aureus Biofilms. J. Bacteriol. 2004, 186, 1838–1850. [Google Scholar] [CrossRef] [PubMed]
- Boles, B.R.; Horswill, A.R. agr-Mediated Dispersal of Staphylococcus aureus Biofilms. PLoS Pathog. 2008, 4, e1000052. [Google Scholar] [CrossRef] [PubMed]
- Queck, S.Y.; Jameson-Lee, M.; Villaruz, A.E.; Bach, T.-H.L.; Khan, B.A.; Sturdevant, D.E.; Ricklefs, S.M.; Li, M.; Otto, M. RNAIII-Independent Target Gene Control by the agr Quorum-Sensing System: Insight into the Evolution of Virulence Regulation in Staphylococcus aureus. Mol. Cell 2008, 32, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Le, K.Y.; Villaruz, A.E.; Zheng, Y.; He, L.; Fisher, E.L.; Nguyen, T.H.; Ho, T.V.; Yeh, A.J.; Joo, H.-S.; Cheung, G.Y.; et al. Role of Phenol-Soluble Modulins in Staphylococcus epidermidis Biofilm Formation and Infection of Indwelling Medical Devices. J. Mol. Biol. 2019, 431, 3015–3027. [Google Scholar] [CrossRef] [PubMed]
- Schauder, S.; Shokat, K.; Surette, M.G.; Bassler, B.L. The LuxS family of bacterial autoinducers: Biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 2001, 41, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Li, H.; Vuong, C.; Vadyvaloo, V.; Wang, J.; Yao, Y.; Otto, M.; Gao, Q. Role of the luxS Quorum-Sensing System in Biofilm Formation and Virulence of Staphylococcus epidermidis. Infect. Immun. 2006, 74, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Zhao, L.; Xue, T.; Sun, B. Staphylococcus aureus autoinducer-2 quorum sensing decreases biofilm formation in an icaR-dependent manner. BMC Microbiol. 2012, 12, 288. [Google Scholar] [CrossRef] [PubMed]
- Cue, D.; Lei, M.G.; Lee, C.Y. Activation of sarX by Rbf Is Required for Biofilm Formation and icaADBC Expression in Staphylococcus aureus. J. Bacteriol. 2013, 195, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Qiu, S.; Jiang, Q.; Sun, H.; Xue, T.; Cai, G.; Sun, B. AI-2 quorum sensing negatively regulates rbf expression and biofilm formation in Staphylococcus aureus. Int. J. Med. Microbiol. 2017, 307, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Ni, J.; Shang, F.; Chen, X.; Zhang, M. Autoinducer-2 increases biofilm formation via an ica- and bhp-dependent manner in Staphylococcus epidermidis RP62A. Microbes Infect. 2015, 17, 345–352. [Google Scholar] [CrossRef]
- Selvaraj, A.; Jayasree, T.; Valliammai, A.; Pandian, S.K. Myrtenol Attenuates MRSA Biofilm and Virulence by Suppressing sarA Expression Dynamism. Front. Microbiol. 2019, 10, 2027. [Google Scholar] [CrossRef]
- Balamurugan, P.; Krishna, V.P.; Bharath, D.; Lavanya, R.; Vairaprakash, P.; Princy, S.A. Staphylococcus aureus Quorum Regulator SarA Targeted Compound, 2-[(Methylamino)methyl]phenol Inhibits Biofilm and Down-Regulates Virulence Genes. Front. Microbiol. 2017, 8, 1290. [Google Scholar] [CrossRef]
- Valle, J.; Toledo-Arana, A.; Berasain, C.; Ghigo, J.-M.; Amorena, B.; Penadés, J.R.; Lasa, I. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 2003, 48, 1075–1087. [Google Scholar] [CrossRef] [PubMed]
- Chien, Y.-T.; Cheung, A.L. Molecular Interactions between Two Global Regulators, sar and agr, in Staphylococcus aureus. J. Biol. Chem. 1998, 273, 2645–2652. [Google Scholar] [CrossRef]
- Cheung, A.L.; Nishina, K.; Manna, A.C. SarA of Staphylococcus aureus Binds to the sarA Promoter To Regulate Gene Expression. J. Bacteriol. 2008, 190, 2239–2243. [Google Scholar] [CrossRef]
- Reyes, D.; Andrey, D.O.; Monod, A.; Kelley, W.L.; Zhang, G.; Cheung, A.L. Coordinated Regulation by AgrA, SarA, and SarR To Control agr Expression in Staphylococcus aureus. J. Bacteriol. 2011, 193, 6020–6031. [Google Scholar] [CrossRef]
- Chien, Y.-T.; Manna, A.C.; Projan, S.J.; Cheung, A.L. SarA, a Global Regulator of Virulence Determinants in Staphylococcus aureus, Binds to a Conserved Motif Essential for sar-dependent Gene Regulation. J. Biol. Chem. 1999, 274, 37169–37176. [Google Scholar] [CrossRef]
- Xiong, Y.Q.; Willard, J.; Yeaman, M.R.; Cheung, A.L.; Bayer, A.S. Regulation of Staphylococcus aureus α-Toxin Gene (hla) Expression by agr, sarA, and sae In Vitro and in Experimental Infective Endocarditis. J. Infect. Dis. 2006, 194, 1267–1275. [Google Scholar] [CrossRef]
- Zielinska, A.K.; Beenken, K.E.; Mrak, L.N.; Spencer, H.J.; Post, G.R.; Skinner, R.A.; Tackett, A.J.; Horswill, A.R.; Smeltzer, M.S. sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol. Microbiol. 2012, 86, 1183–1196. [Google Scholar] [CrossRef] [PubMed]
- Loughran, A.J.; Atwood, D.N.; Anthony, A.C.; Harik, N.S.; Spencer, H.J.; Beenken, K.E.; Smeltzer, M.S. Impact of individual extracellular proteases on Staphylococcus aureus biofilm formation in diverse clinical isolates and their isogenic sarA mutants. MicrobiologyOpen 2014, 3, 897–909. [Google Scholar] [CrossRef]
- Kullik, I.; Giachino, P.; Fuchs, T. Deletion of the Alternative Sigma Factor ς B in Staphylococcus aureus Reveals Its Function as a Global Regulator of Virulence Genes. J. Bacteriol. 1998, 180, 4814–4820. [Google Scholar] [CrossRef]
- Rachid, S.; Ohlsen, K.; Wallner, U.; Hacker, J.; Hecker, M.; Ziebuhr, W. Alternative Transcription Factor ς B Is Involved in Regulation of Biofilm Expression in a Staphylococcus aureus Mucosal Isolate. J. Bacteriol. 2000, 182, 6824–6826. [Google Scholar] [CrossRef] [PubMed]
- Valle, J.; Echeverz, M.; Lasa, I. σB Inhibits Poly-N-Acetylglucosamine Exopolysaccharide Synthesis and Biofilm Formation in Staphylococcus aureus. J. Bacteriol. 2019, 201, e00098-19. [Google Scholar] [CrossRef]
- Bischoff, M.; Entenza, J.M.; Giachino, P. Influence of a Functional sigB Operon on the Global Regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 2001, 183, 5171–5179. [Google Scholar] [CrossRef]
- Entenza, J.-M.; Moreillon, P.; Senn, M.M.; Kormanec, J.; Dunman, P.M.; Berger-Bächi, B.; Projan, S.; Bischoff, M.; Aguilar-Be, I.; Zardo, R.D.S.; et al. Role of σ B in the Expression of Staphylococcus aureus Cell Wall Adhesins ClfA and FnbA and Contribution to Infectivity in a Rat Model of Experimental Endocarditis. Infect. Immun. 2005, 73, 990–998. [Google Scholar] [CrossRef]
- Atwood, D.N.; Loughran, A.J.; Courtney, A.P.; Anthony, A.C.; Meeker, D.G.; Spencer, H.J.; Gupta, R.K.; Lee, C.Y.; Beenken, K.E.; Smeltzer, M.S. Comparative impact of diverse regulatory loci on Staphylococcus aureus biofilm formation. MicrobiologyOpen 2015, 4, 436–451. [Google Scholar] [CrossRef]
- Waters, N.R.; Samuels, D.J.; Behera, R.K.; Livny, J.; Rhee, K.Y.; Sadykov, M.R.; Brinsmade, S.R. A spectrum of CodY activities drives metabolic reorganization and virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 2016, 101, 495–514. [Google Scholar] [CrossRef] [PubMed]
- Stenz, L.; Francois, P.; Whiteson, K.; Wolz, C.; Linder, P.; Schrenzel, J. The CodY pleiotropic repressor controls virulence in gram-positive pathogens. FEMS Immunol. Med. Microbiol. 2011, 62, 123–139. [Google Scholar] [CrossRef] [PubMed]
- Majerczyk, C.D.; Sadykov, M.R.; Luong, T.T.; Lee, C.; Somerville, G.A.; Sonenshein, A.L. Staphylococcus aureus CodY Negatively Regulates Virulence Gene Expression. J. Bacteriol. 2008, 190, 2257–2265. [Google Scholar] [CrossRef] [PubMed]
- Majerczyk, C.D.; Dunman, P.M.; Luong, T.T.; Lee, C.Y.; Sadykov, M.R.; Somerville, G.A.; Bodi, K.; Sonenshein, A.L. Direct Targets of CodY in Staphylococcus aureus. J. Bacteriol. 2010, 192, 2861–2877. [Google Scholar] [CrossRef]
- Roux, A.; Todd, D.A.; Velázquez, J.V.; Cech, N.B.; Sonenshein, A.L. CodY-Mediated Regulation of the Staphylococcus aureus Agr System Integrates Nutritional and Population Density Signals. J. Bacteriol. 2014, 196, 1184–1196. [Google Scholar] [CrossRef]
- Mlynek, K.D.; Bulock, L.L.; Stone, C.J.; Curran, L.J.; Sadykov, M.R.; Bayles, K.W.; Brinsmade, S.R. Genetic and Biochemical Analysis of CodY-Mediated Cell Aggregation in Staphylococcus aureus Reveals an Interaction between Extracellular DNA and Polysaccharide in the Extracellular Matrix. J. Bacteriol. 2020, 202, e00593-19. [Google Scholar] [CrossRef]
- Rom, J.S.; Atwood, D.N.; Beenken, K.E.; Meeker, D.G.; Loughran, A.J.; Spencer, H.J.; Lantz, T.L.; Smeltzer, M.S. Impact of Staphylococcus aureus regulatory mutations that modulate biofilm formation in the USA300 strain LAC on virulence in a murine bacteremia model. Virulence 2017, 8, 1776–1790. [Google Scholar] [CrossRef] [PubMed]
- Mlynek, K.D.; Sause, W.E.; Moormeier, D.E.; Sadykov, M.R.; Hill, K.R.; Torres, V.J.; Bayles, K.W.; Brinsmade, S.R. Nutritional Regulation of the Sae Two-Component System by CodY in Staphylococcus aureus. J. Bacteriol. 2018, 200, e00012-18. [Google Scholar] [CrossRef] [PubMed]
- Conlon, K.M.; Humphreys, H.; O’Gara, J.P. icaR Encodes a Transcriptional Repressor Involved in Environmental Regulation of ica Operon Expression and Biofilm Formation in Staphylococcus epidermidis. J. Bacteriol. 2002, 184, 4400–4408. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.-M.; Zhou, C.; Lindgren, J.K.; Galac, M.R.; Corey, B.; Endres, J.E.; Olson, M.E.; Fey, P.D. Transcriptional Regulation of icaADBC by both IcaR and TcaR in Staphylococcus epidermidis. J. Bacteriol. 2019, 201, e00524-18. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, K.K.; Pier, D.B.; Goldmann, D.A.; Pier, G.B. The Teicoplanin-Associated Locus Regulator (TcaR) and the Intercellular Adhesin Locus Regulator (IcaR) Are Transcriptional Inhibitors of the ica Locus in Staphylococcus aureus. J. Bacteriol. 2004, 186, 2449–2456. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-M.; Jeng, W.-Y.; Ko, T.-P.; Yeh, Y.-J.; Chen, C.K.-M.; Wang, A.H.-J. Structural study of TcaR and its complexes with multiple antibiotics from Staphylococcus epidermidis. Proc. Natl. Acad. Sci. USA 2010, 107, 8617–8622. [Google Scholar] [CrossRef]
- Schwartbeck, B.; Birtel, J.; Treffon, J.; Langhanki, L.; Mellmann, A.; Kale, D.; Kahl, J.; Hirschhausen, N.; Neumann, C.; Lee, J.C.; et al. Dynamic in vivo mutations within the ica operon during persistence of Staphylococcus aureus in the airways of cystic fibrosis patients. PLoS Pathog. 2016, 12, e1006024. [Google Scholar] [CrossRef]
- Jefferson, K.K.; Cramton, S.E.; Götz, F.; Pier, G. Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of IcaR. Mol. Microbiol. 2003, 48, 889–899. [Google Scholar] [CrossRef]
- Yu, L.; Hisatsune, J.; Hayashi, I.; Tatsukawa, N.; Sato’O, Y.; Mizumachi, E.; Kato, F.; Hirakawa, H.; Pier, G.B.; Sugai, M. A Novel Repressor of the ica Locus Discovered in Clinically Isolated Super-Biofilm-Elaborating Staphylococcus aureus. mBio 2017, 8, e02282-16. [Google Scholar] [CrossRef]
- You, Y.; Xue, T.; Cao, L.; Zhao, L.; Sun, H.; Sun, B. Staphylococcus aureus glucose-induced biofilm accessory proteins, GbaAB, influence biofilm formation in a PIA-dependent manner. Int. J. Med. Microbiol. 2014, 304, 603–612. [Google Scholar] [CrossRef]
- Tiwari, N.; López-Redondo, M.; Miguel-Romero, L.; Kulhankova, K.; Cahill, M.P.; Tran, P.M.; Kinney, K.J.; Kilgore, S.H.; Al-Tameemi, H.; Herfst, C.A.; et al. The SrrAB two-component system regulates Staphylococcus aureus pathogenicity through redox sensitive cysteines. Proc. Natl. Acad. Sci. USA 2020, 117, 10989–10999. [Google Scholar] [CrossRef] [PubMed]
- Park, M.K.; Myers, R.A.M.; Marzella, L. Oxygen Tensions and Infections: Modulation of Microbial Growth, Activity of Antimicrobial Agents, and Immunologic Responses. Clin. Infect. Dis. 1992, 14, 720–740. [Google Scholar] [CrossRef] [PubMed]
- Pragman, A.A.; Yarwood, J.M.; Tripp, T.J.; Schlievert, P.M. Characterization of Virulence Factor Regulation by SrrAB, a Two-Component System in Staphylococcus aureus. J. Bacteriol. 2004, 186, 2430–2438. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, M.; Bastian, M.; Cramton, S.E.; Ziegler, K.; Pragman, A.A.; Bragonzi, A.; Memmi, G.; Wolz, C.; Schlievert, P.; Cheung, A.; et al. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol. Microbiol. 2007, 65, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Quoc, P.H.T.; Genevaux, P.; Pajunen, M.; Savilahti, H.; Georgopoulos, C.; Schrenzel, J.; Kelley, W.L. Isolation and Characterization of Biofilm Formation-Defective Mutants of Staphylococcus aureus. Infect. Immun. 2007, 75, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Mashruwala, A.A.; Van De Guchte, A.; Boyd, J.M. Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. eLife 2017, 6, e23845. [Google Scholar] [CrossRef] [PubMed]
- Kinkel, T.L.; Roux, C.M.; Dunman, P.M.; Fang, F.C. The Staphylococcus aureus SrrAB Two-Component System Promotes Resistance to Nitrosative Stress and Hypoxia. mBio 2013, 4, e00696-13. [Google Scholar] [CrossRef] [PubMed]
- Lamret, F.; Varin-Simon, J.; Velard, F.; Terryn, C.; Mongaret, C.; Colin, M.; Gangloff, S.C.; Reffuveille, F. Staphylococcus aureus Strain-Dependent Biofilm Formation in Bone-Like Environment. Front. Microbiol. 2021, 12, 714994. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Jeong, D.-W.; Liu, Q.; Yeo, W.-S.; Vogl, T.; Skaar, E.P.; Chazin, W.J.; Bae, T. Calprotectin Increases the Activity of the SaeRS Two Component System and Murine Mortality during Staphylococcus aureus Infections. PLoS Pathog. 2015, 11, e1005026. [Google Scholar] [CrossRef]
- Liu, Q.; Yeo, W.; Bae, T. The SaeRS Two-Component System of Staphylococcus aureus. Genes 2016, 7, 81. [Google Scholar] [CrossRef]
- Jeong, D.-W.; Cho, H.; Jones, M.B.; Shatzkes, K.; Sun, F.; Ji, Q.; Liu, Q.; Peterson, S.N.; He, C.; Bae, T. The auxiliary protein complex SaePQ activates the phosphatase activity of sensor kinase SaeS in the SaeRS two-component system of Staphylococcus aureus. Mol. Microbiol. 2012, 86, 331–348. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Yu, C.; Sun, J.; Liu, H.; Landwehr, C.; Holmes, D.; Ji, Y. Inactivation of a Two-Component Signal Transduction System, SaeRS, Eliminates Adherence and Attenuates Virulence of Staphylococcus aureus. Infect. Immun. 2006, 74, 4655–4665. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.E.; Nygaard, T.K.; Ackermann, L.; Watkins, R.L.; Zurek, O.W.; Pallister, K.B.; Griffith, S.; Kiedrowski, M.R.; Flack, C.E.; Kavanaugh, J.S.; et al. Staphylococcus aureus Nuclease Is an SaeRS-Dependent Virulence Factor. Infect. Immun. 2013, 81, 1316–1324. [Google Scholar] [CrossRef] [PubMed]
- Mrak, L.N.; Zielinska, A.K.; Beenken, K.E.; Mrak, I.N.; Atwood, D.N.; Griffin, L.M.; Lee, C.Y.; Smeltzer, M.S. saeRS and sarA Act Synergistically to Repress Protease Production and Promote Biofilm Formation in Staphylococcus aureus. PLoS ONE 2012, 7, e38453. [Google Scholar] [CrossRef] [PubMed]
- Cue, D.; Junecko, J.M.; Lei, M.G.; Blevins, J.S.; Smeltzer, M.; Lee, C.Y. SaeRS-Dependent Inhibition of Biofilm Formation in Staphylococcus aureus Newman. PLoS ONE 2015, 10, e0123027. [Google Scholar] [CrossRef] [PubMed]
- Mashruwala, A.A.; Gries, C.M.; Scherr, T.D.; Kielian, T.; Boyd, J.M. SaeRS Is Responsive to Cellular Respiratory Status and Regulates Fermentative Biofilm Formation in Staphylococcus aureus. Infect. Immun. 2017, 85, e00157-17. [Google Scholar] [CrossRef]
- Wittekind, M.A.; Frey, A.; Bonsall, A.E.; Briaud, P.; Keogh, R.A.; Wiemels, R.E.; Shaw, L.N.; Carroll, R.K. The novel protein ScrA acts through the SaeRS two-component system to regulate virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 2022, 117, 1196–1212. [Google Scholar] [CrossRef]
- Ranjit, D.K.; Endres, J.L.; Bayles, K.W. Staphylococcus aureus CidA and LrgA Proteins Exhibit Holin-Like Properties. J. Bacteriol. 2011, 193, 2468–2476. [Google Scholar] [CrossRef] [PubMed]
- Rice, K.C.; Bayles, K.W. Molecular Control of Bacterial Death and Lysis. Microbiol. Mol. Biol. Rev. 2008, 72, 85–109. [Google Scholar] [CrossRef]
- Rice, K.C.; Mann, E.E.; Endres, J.L.; Weiss, E.C.; Cassat, J.E.; Smeltzer, M.S.; Bayles, K.W. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2007, 104, 8113–8118. [Google Scholar] [CrossRef] [PubMed]
- Leroy, S.; Lebert, I.; Andant, C.; Micheau, P.; Talon, R. Investigating Extracellular DNA Release in Staphylococcus xylosus Biofilm In Vitro. Microorganisms 2021, 9, 2192. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-J.; Dunman, P.M.; Projan, S.J.; Bayles, K.W. Characterization of the Staphylococcus aureus CidR regulon: Elucidation of a novel role for acetoin metabolism in cell death and lysis. Mol. Microbiol. 2006, 60, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, D.F.; Brunskill, E.W.; Bayles, K.W. Analysis of Genetic Elements Controlling Staphylococcus aureus lrgAB Expression: Potential Role of DNA Topology in SarA Regulation. J. Bacteriol. 2000, 182, 4822–4828. [Google Scholar] [CrossRef] [PubMed]
- Sharma-Kuinkel, B.K.; Mann, E.E.; Ahn, J.-S.; Kuechenmeister, L.J.; Dunman, P.M.; Bayles, K.W. The Staphylococcus aureus LytSR Two-Component Regulatory System Affects Biofilm Formation. J. Bacteriol. 2009, 191, 4767–4775. [Google Scholar] [CrossRef] [PubMed]
- Moormeier, D.E.; Endres, J.L.; Mann, E.E.; Sadykov, M.R.; Horswill, A.R.; Rice, K.C.; Fey, P.D.; Bayles, K.W. Use of Microfluidic Technology To Analyze Gene Expression during Staphylococcus aureus Biofilm Formation Reveals Distinct Physiological Niches. Appl. Environ. Microbiol. 2013, 79, 3413–3424. [Google Scholar] [CrossRef]
- Lehman, M.K.; Bose, J.L.; Sharma-Kuinkel, B.K.; Moormeier, D.E.; Endres, J.L.; Sadykov, M.R.; Biswas, I.; Bayles, K.W. Identification of the amino acids essential for LytSR-mediated signal transduction in Staphylococcus aureus and their roles in biofilm-specific gene expression. Mol. Microbiol. 2015, 95, 723–737. [Google Scholar] [CrossRef] [PubMed]
- Windham, I.H.; Chaudhari, S.S.; Bose, J.L.; Thomas, V.C.; Bayles, K.W. SrrAB Modulates Staphylococcus aureus Cell Death through Regulation of cidABC Transcription. J. Bacteriol. 2016, 198, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Fournier, B.; Hooper, D.C. A New Two-Component Regulatory System Involved in Adhesion, Autolysis, and Extracellular Proteolytic Activity of Staphylococcus aureus. J. Bacteriol. 2000, 182, 3955–3964. [Google Scholar] [CrossRef] [PubMed]
- Crosby, H.A.; Schlievert, P.M.; Merriman, J.A.; King, J.M.; Salgado-Pabón, W.; Horswill, A.R. The Staphylococcus aureus Global Regulator MgrA Modulates Clumping and Virulence by Controlling Surface Protein Expression. PLoS Pathog. 2016, 12, e1005604. [Google Scholar] [CrossRef]
- Burgui, S.; Gil, C.; Solano, C.; Lasa, I.; Valle, J. A Systematic Evaluation of the Two-Component Systems Network Reveals That ArlRS Is a Key Regulator of Catheter Colonization by Staphylococcus aureus. Front. Microbiol. 2018, 9, 342. [Google Scholar] [CrossRef] [PubMed]
- Trotonda, M.P.; Tamber, S.; Memmi, G.; Cheung, A.L. MgrA Represses Biofilm Formation in Staphylococcus aureus. Infect. Immun. 2008, 76, 5645–5654. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Jin, Z.; Sun, B. MgrA Negatively Regulates Biofilm Formation and Detachment by Repressing the Expression of psm Operons in Staphylococcus aureus. Appl. Environ. Microbiol. 2018, 84, e01008-18. [Google Scholar] [CrossRef]
- Jenal, U.; Reinders, A.; Lori, C. Cyclic di-GMP: Second messenger extraordinaire. Nat. Rev. Genet. 2017, 15, 271–284. [Google Scholar] [CrossRef]
- Ha, D.-G.; O’Toole, G.A. c-di-GMP and its Effects on Biofilm Formation and Dispersion: A Pseudomonas Aeruginosa Review. Microbiol. Spectr. 2015, 3, MB-0003-2014. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J. Biofilm Dispersal: Mechanisms, Clinical Implications, and Potential Therapeutic Uses. J. Dent. Res. 2010, 89, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.-Q.; Fan, Y.-Z.; Song, X.; Liu, X.-X.; Xia, Y.-J.; Ai, L.-Z. The second messenger c-di-AMP mediates bacterial exopolysaccharide biosynthesis: A review. Mol. Biol. Rep. 2020, 47, 9149–9157. [Google Scholar] [CrossRef] [PubMed]
- Hengge, R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009, 7, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Corrigan, R.M.; Abbott, J.C.; Burhenne, H.; Kaever, V.; Gründling, A. c-di-AMP Is a New Second Messenger in Staphylococcus aureus with a Role in Controlling Cell Size and Envelope Stress. PLoS Pathog. 2011, 7, e1002217. [Google Scholar] [CrossRef]
- DeFrancesco, A.S.; Masloboeva, N.; Syed, A.K.; DeLoughery, A.; Bradshaw, N.; Li, G.-W.; Gilmore, M.S.; Walker, S.; Losick, R. Genome-wide screen for genes involved in eDNA release during biofilm formation by Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2017, 114, E5969–E5978. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, Y.; Zhu, F.; Cheung, A.L.; Wang, G.; Bai, G.; Proctor, R.A.; Yeaman, M.R.; Bayer, A.S.; Xiong, Y.Q. New Mechanistic Insights into Purine Biosynthesis with Second Messenger c-di-AMP in Relation to Biofilm-Related Persistent Methicillin-Resistant Staphylococcus aureus Infections. mBio 2021, 12, e0208121. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Kambara, K.; Meyer, H.; Stenz, L.; Bonetti, E.-J.; Girard, M.; Lalk, M.; Francois, P.; Schrenzel, J. GdpS contributes to Staphylococcus aureus biofilm formation by regulation of eDNA release. Int. J. Med. Microbiol. 2014, 304, 284–299. [Google Scholar] [CrossRef]
- Fechter, P.; Caldelari, I.; Lioliou, E.; Romby, P. Novel aspects of RNA regulation in Staphylococcus aureus. FEBS Lett. 2014, 588, 2523–2529. [Google Scholar] [CrossRef] [PubMed]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Desgranges, E.; Marzi, S.; Moreau, K.; Romby, P.; Caldelari, I. Noncoding RNA. Microbiol. Spectr. 2019, 7, 7-2. [Google Scholar] [CrossRef] [PubMed]
- Ghaz-Jahanian, M.A.; Khodaparastan, F.; Berenjian, A.; Jafarizadeh-Malmiri, H. Influence of Small RNAs on Biofilm Formation Process in Bacteria. Mol. Biotechnol. 2013, 55, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Romilly, C.; Lays, C.; Tomasini, A.; Caldelari, I.; Benito, Y.; Hammann, P.; Geissmann, T.; Boisset, S.; Romby, P.; Vandenesch, F. A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in Staphylococcus aureus. PLoS Pathog. 2014, 10, e1003979. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; Nair, M. The small RNA RsaF regulates the expression of secreted virulence factors in Staphylococcus aureus Newman. J. Microbiol. 2021, 59, 920–930. [Google Scholar] [CrossRef]
- Ibberson, C.B.; Parlet, C.P.; Kwiecinski, J.; Crosby, H.A.; Meyerholz, D.K.; Horswill, A.R. Hyaluronan Modulation Impacts Staphylococcus aureus Biofilm Infection. Infect. Immun. 2016, 84, 1917–1929. [Google Scholar] [CrossRef]
- Kim, S.; Reyes, D.; Beaume, M.; Francois, P.; Cheung, A. Contribution of teg49 Small RNA in the 5′ Upstream Transcriptional Region of sarA to Virulence in Staphylococcus aureus. Infect. Immun. 2014, 82, 4369–4379. [Google Scholar] [CrossRef]
- Manna, A.C.; Leo, S.; Girel, S.; González-Ruiz, V.; Rudaz, S.; Francois, P.; Cheung, A.L. Teg58, a small regulatory RNA, is involved in regulating arginine biosynthesis and biofilm formation in Staphylococcus aureus. Sci. Rep. 2022, 12, 14963. [Google Scholar] [CrossRef]
- Manna, A.C.; Kim, S.; Cengher, L.; Corvaglia, A.; Leo, S.; Francois, P.; Cheung, A.L. Small RNA teg49 Is Derived from a sarA Transcript and Regulates Virulence Genes Independent of SarA in Staphylococcus aureus. Infect. Immun. 2018, 86, e00635-17. [Google Scholar] [CrossRef] [PubMed]
- Kathirvel, M.; Buchad, H.; Nair, M. Enhancement of the pathogenicity of Staphylococcus aureus strain Newman by a small noncoding RNA SprX1. Med. Microbiol. Immunol. 2016, 205, 563–574. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Q.; Tang, X.; Dong, W.; Sun, N.; Yuan, W. A Review of Biofilm Formation of Staphylococcus aureus and Its Regulation Mechanism. Antibiotics 2023, 12, 12. https://doi.org/10.3390/antibiotics12010012
Peng Q, Tang X, Dong W, Sun N, Yuan W. A Review of Biofilm Formation of Staphylococcus aureus and Its Regulation Mechanism. Antibiotics. 2023; 12(1):12. https://doi.org/10.3390/antibiotics12010012
Chicago/Turabian StylePeng, Qi, Xiaohua Tang, Wanyang Dong, Ning Sun, and Wenchang Yuan. 2023. "A Review of Biofilm Formation of Staphylococcus aureus and Its Regulation Mechanism" Antibiotics 12, no. 1: 12. https://doi.org/10.3390/antibiotics12010012
APA StylePeng, Q., Tang, X., Dong, W., Sun, N., & Yuan, W. (2023). A Review of Biofilm Formation of Staphylococcus aureus and Its Regulation Mechanism. Antibiotics, 12(1), 12. https://doi.org/10.3390/antibiotics12010012






