Genomic Analysis of a mcr-9.1-Harbouring IncHI2-ST1 Plasmid from Enterobacter ludwigii Isolated in Fish Farming
Abstract
:1. Introduction
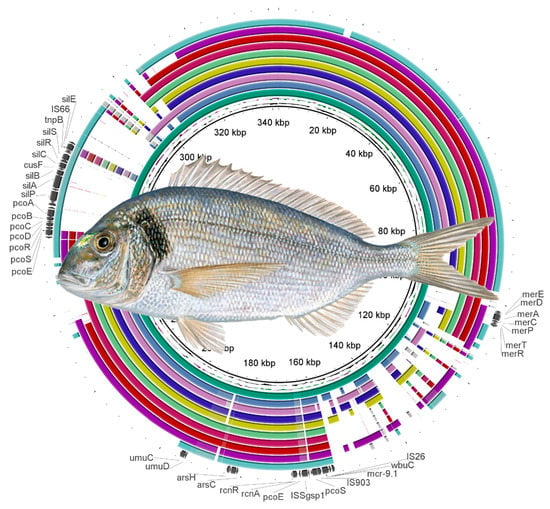
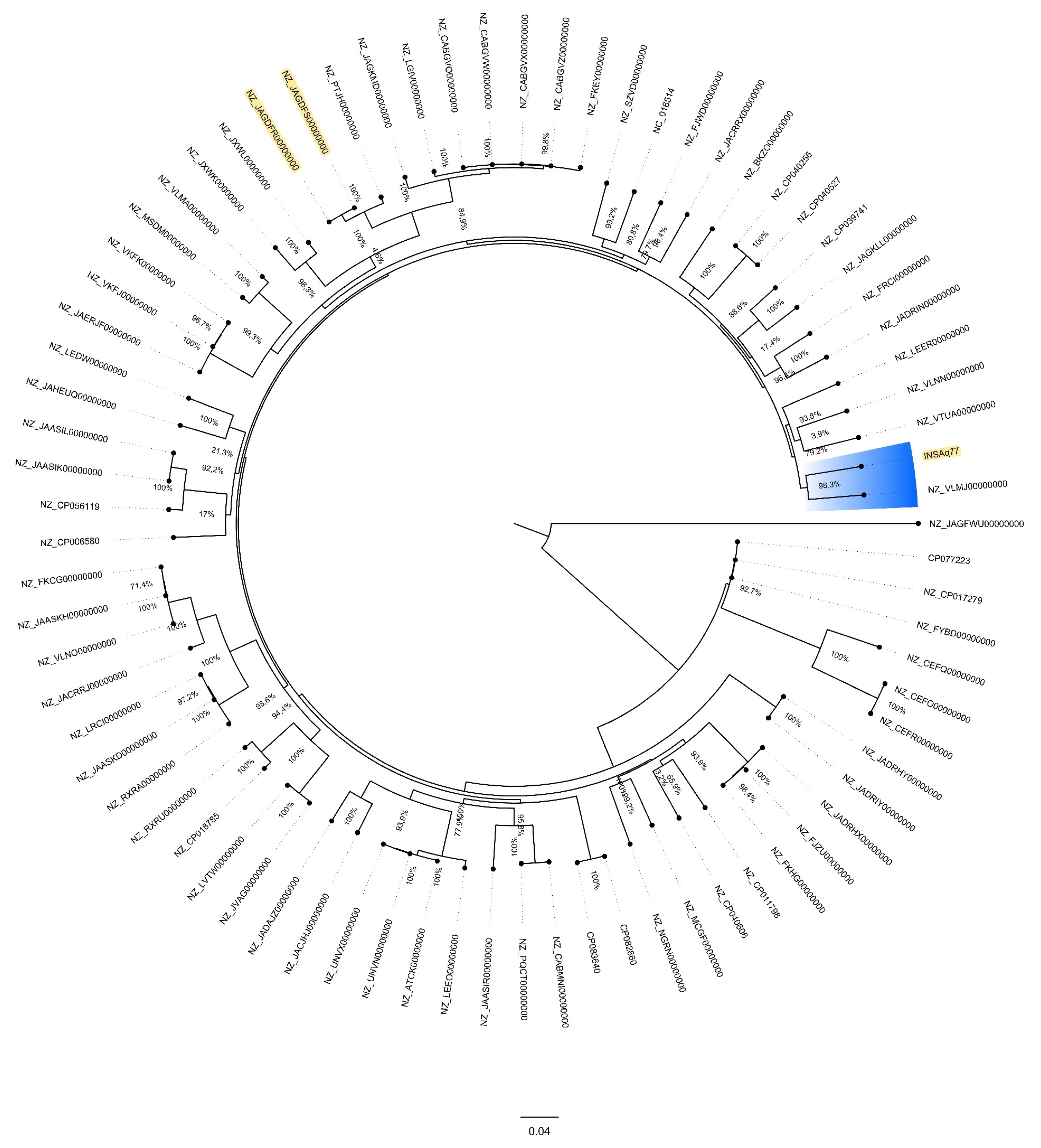
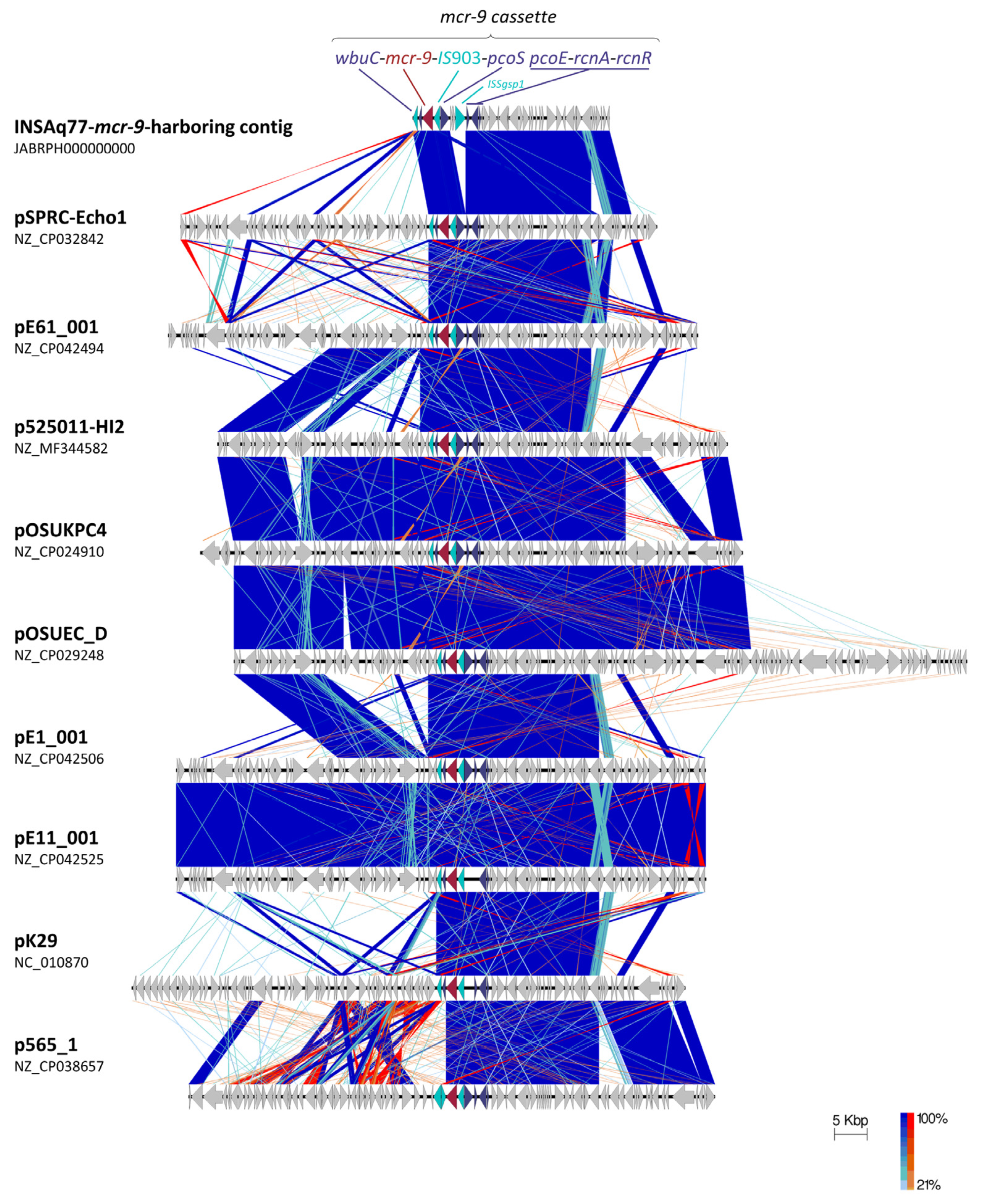
2. Results and Discussion
3. Materials and Methods
3.1. Study Design and Bacterial Identification
3.2. Antimicrobial Susceptibility Testing
3.3. Whole-Genome Sequencing
3.4. Genome Annotation and Analysis
3.5. Phylogenomic Analyses of E. ludwigii Genomes
3.6. Resistome, Virulome and Mobilome Analysis
3.7. Plasmid Characterization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kempf, I.; Jouy, E.; Chauvin, C. Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 2016, 48, 598–606. [Google Scholar] [CrossRef]
- El-Sayed Ahmed, M.A.E.G.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef] [PubMed]
- Andrade, F.F.; Silva, D.; Rodrigues, A.; Pina-Vaz, C. Colistin update on its mechanism of action and resistance, present and future challenges. Microorganisms 2020, 8, 1716. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, H.; Liu, Y.H.; Feng, Y. Towards Understanding MCR-like Colistin Resistance. Trends Microbiol. 2018, 26, 794–808. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Abbas, M.; Rehman, M.U.; Wang, M.; Jia, R.; Chen, S.; Liu, M.; Zhu, D.; Zhao, X.; Gao, Q.; et al. Updates on the global dissemination of colistin-resistant Escherichia coli: An emerging threat to public health. Sci. Total Environ. 2021, 799, 149280. [Google Scholar] [CrossRef]
- Ling, Z.; Yin, W.; Shen, Z.; Wang, Y.; Shen, J.; Walsh, T.R. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J. Antimicrob. Chemother. 2020, 75, 3087–3095. [Google Scholar] [CrossRef] [PubMed]
- Carroll, L.M.; Gaballa, A.; Guldimann, C.; Sullivan, G.; Henderson, L.O.; Wiedmanna, M. Identification of Novel Mobilized Colistin Resistance Gene mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium Isolate. MBio 2019, 10, e00853-19. [Google Scholar] [CrossRef]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes. Infect. 2020, 9, 508–516. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, R.; Schwarz, S.; Wu, C.; Shen, J.; Walsh, T.R.; Wang, Y. Farm animals and aquaculture: Significant reservoirs of mobile colistin resistance genes. Environ. Microbiol. 2020, 22, 2469–2484. [Google Scholar] [CrossRef] [Green Version]
- Cabello, F.C.; Godfrey, H.P. Aquaculture, exaptation, and the origin of mcr-positive colistin resistance. Antimicrob. Agents Chemother. 2018, 62, 13–14. [Google Scholar] [CrossRef] [PubMed]
- Cabello, F.C.; Tomova, A.; Ivanova, L.; Godfrey, H.P. Aquaculture and mcr colistin resistance determinants. MBio 2017, 8, 4–5. [Google Scholar] [CrossRef] [PubMed]
- Lei, T.; Zhang, J.; Jiang, F.; He, M.; Zeng, H.; Chen, M.; Wu, S.; Wang, J.; Ding, Y.; Wu, Q. First detection of the plasmid-mediated colistin resistance gene mcr-1 in virulent Vibrio parahaemolyticus. Int. J. Food Microbiol. 2019, 308, 108290. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Cao, Y.; Yu, P.; Huang, R.; Wang, J.; Wen, Q.; Zhi, C.; Zhang, Q.; Liu, J.-H. Detection of mcr-1 Gene among Escherichia coli Isolates from Farmed Fish and Characterization of mcr-1-Bearing IncP Plasmids. Antimicrob. Agents Chemother. 2018, 62, e02378-17. [Google Scholar] [CrossRef] [PubMed]
- Hoa, T.T.T.; Nakayama, T.; Huyen, H.M.; Harada, K.; Hinenoya, A.; Phuong, N.T.; Yamamoto, Y. Extended-spectrum beta-lactamase-producing Escherichia coli harbouring sul and mcr-1 genes isolates from fish gut contents in the Mekong Delta, Vietnam. Lett. Appl. Microbiol. 2020, 71, 78–85. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Kawahara, R.; Harada, K.; Teruya, S.; Nakayama, T.; Motooka, D.; Nakamura, S.; Do Nguyen, P.; Kumeda, Y.; Van Dang, C.; et al. The presence of colistin resistance gene mcr-1 and -3 in ESBL producing Escherichia coli isolated from food in Ho Chi Minh City, Vietnam. FEMS Microbiol. Lett. 2018, 365, fny100. [Google Scholar] [CrossRef]
- Lozano-Leon, A.; Garcia-Omil, C.; Dalama, J.; Rodriguez-Souto, R.; Martinez-Urtaza, J.; Gonzalez-Escalona, N. Detection of colistin resistance mcr-1 gene in Salmonella enterica serovar Rissen isolated from mussels, Spain, 2012- to 2016. Euro Surveill. 2019, 24, 1900200. [Google Scholar] [CrossRef]
- Hassan, J.; Eddine, R.Z.; Mann, D.; Li, S.; Deng, X.; Saoud, I.P.; Kassem, I.I. The mobile colistin resistance gene, mcr-1.1, is carried on Incx4 plasmids in multidrug resistant E. coli isolated from rainbow trout aquaculture. Microorganisms 2020, 8, 1636. [Google Scholar] [CrossRef]
- Kalová, A.; Gelbíčová, T.; Overballe-Petersen, S.; Litrup, E.; Karpíšková, R. Characterisation of colistin-resistant Enterobacterales and Acinetobacter strains carrying mcr genes from asian aquaculture products. Antibiotics 2021, 10, 838. [Google Scholar] [CrossRef]
- Xu, C.; Lv, Z.; Shen, Y.; Liu, D.; Fu, Y.; Zhou, L.; Liu, W.; Chen, K.; Ye, H.; Xia, X.; et al. Metagenomic insights into differences in environmental resistome profiles between integrated and monoculture aquaculture farms in China. Environ. Int. 2020, 144, 106005. [Google Scholar] [CrossRef]
- Li, Y.; Dai, X.; Zeng, J.; Gao, Y.; Zhang, Z.; Zhang, L. Characterization of the global distribution and diversified plasmid reservoirs of the colistin resistance gene mcr-9. Sci. Rep. 2020, 10, 8113. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Feng, Y.; Zong, Z. Precise Species Identification for Enterobacter: A Genome Sequence-Based Study with Reporting of Two Novel Species, Enterobacter quasiroggenkampii sp. nov. and Enterobacter quasimori sp. nov. mSystems 2020, 5, e00527-20. [Google Scholar] [CrossRef] [PubMed]
- Mateos, M.; Hernández-García, M.; Del Campo, R.; Martínez-García, L.; Gijón, D.; Morosini, M.I.; Ruiz-Garbajosa, P.; Cantón, R. Emergence and Persistence over Time of Carbapenemase-Producing Enterobacter Isolates in a Spanish University Hospital in Madrid, Spain (2005-2018). Microb. Drug Resist. 2021, 27, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Mezzatesta, M.L.; Gona, F.; Stefani, S. Enterobacter cloacae complex: Clinical impact and emerging antibiotic resistance. Future Microbiol. 2012, 7, 887–902. [Google Scholar] [CrossRef]
- Hoffmann, H.; Stindl, S.; Stumpf, A.; Mehlen, A.; Monget, D.; Heesemann, J.; Schleifer, K.H.; Roggenkamp, A. Description of Enterobacter ludwigii sp. nov., a novel Enterobacter species of clinical relevance. Syst. Appl. Microbiol. 2005, 28, 206–212. [Google Scholar] [CrossRef]
- Davin-Regli, A.; Lavigne, J.P.; Pagès, J.M. Enterobacter spp.: Update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin. Microbiol. Rev. 2019, 32, e00002-19. [Google Scholar] [CrossRef]
- Ali, A.; Sultan, I.; Mondal, A.H.; Siddiqui, M.T.; Gogry, F.A.; Haq, Q.M.R. Lentic and effluent water of Delhi-NCR: A reservoir of multidrug-resistant bacteria harbouring blaCTX-M, blaTEM and blaSHV type ESBL genes. J. Water Health 2021, 19, 592–603. [Google Scholar] [CrossRef]
- Preena, P.G.; Dharmaratnam, A.; Raj, N.S.; Raja, S.A.; Nair, R.R.; Swaminathan, T.R. Antibiotic-resistant Enterobacteriaceae from diseased freshwater goldfish. Arch. Microbiol. 2021, 203, 219–231. [Google Scholar] [CrossRef]
- Lee, K.E.; Adhikari, A.; Kang, S.M.; You, Y.H.; Joo, G.J.; Kim, J.H.; Kim, S.J.; Lee, I.J. Isolation and characterization of the high silicate and phosphate solubilizing novel strain Enterobacter ludwigii GAK2 that promotes growth in rice plants. Agronomy 2019, 9, 144. [Google Scholar] [CrossRef]
- Porto de Souza Vandenberghe, L.; Marcela Blandon Garcia, L.; Rodrigues, C.; Cândido Camara, M.; Vinícius de Melo Pereira, G.; de Oliveira, J.; Ricardo Soccol, C. Potential applications of plant probiotic microorganisms in agriculture and forestry. AIMS Microbiol. 2017, 3, 629–648. [Google Scholar] [CrossRef]
- Börjesson, S.; Greko, C.; Myrenås, M.; Landén, A.; Nilsson, O.; Pedersen, K. A link between the newly described colistin resistance gene mcr-9 and clinical Enterobacteriaceae isolates carrying blaSHV-12 from horses in Sweden. J. Glob. Antimicrob. Resist. 2020, 20, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.H.; Li, C.; Hsu, C.H.; Ayers, S.; Borenstein, S.; Mukherjee, S.; Tran, T.T.; McDermot, P.F.; Zhao, S. The mcr-9 gene of Salmonella and Escherichia coli is not associated with colistin resistance in the United States. Antimicrob. Agents Chemother. 2020, 64, e00573-20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, D.; Quan, J.; Hua, X.; Yu, Y. mcr-1 facilitated selection of high-level colistin-resistant mutants in Escherichia coli. Clin. Microbiol. Infect. 2019, 25, 517.e1–517.e4. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.Q.; Liu, Y.Y.; Wu, R.; Xun, H.; Sun, J.; Li, J.; Feng, Y.; Liu, J.H. Impact of mcr-1 on the Development of High Level Colistin Resistance in Klebsiella pneumoniae and Escherichia coli. Front. Microbiol. 2021, 12, 9666782. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.; Beisken, S.; Lueftinger, L.; Weinmaier, T.; Klein, M.; Bacher, J.; Patel, R.; von Haeseler, A.; Posch, A.E. Species identification and antibiotic resistance prediction by analysis of whole-genome sequence data by use of ARESdb: An analysis of isolates from the unyvero lower respiratory tract infection trial. J. Clin. Microbiol. 2020, 58, e00273-20. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Schwarz, S.; Kehrenberg, C.; Doublet, B.; Cloeckaert, A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 2004, 28, 519–542. [Google Scholar] [CrossRef]
- Li, J.; Zhang, H.; Ning, J.; Sajid, A.; Cheng, G.; Yuan, Z.; Hao, H. The nature and epidemiology of OqxAB, a multidrug efflux pump. Antimicrob. Resist. Infect. Control 2019, 8, 44. [Google Scholar] [CrossRef]
- Fernández, L.; Hancock, R.E.W. Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2012, 25, 661–681. [Google Scholar] [CrossRef]
- Kümmerle, N.; Feucht, H.H.; Kaulfers, P.M. Plasmid-mediated formaldehyde resistance in Escherichia coli: Characterization of resistance gene. Antimicrob. Agents Chemother. 1996, 40, 2276–2279. [Google Scholar] [CrossRef] [Green Version]
- Cloete, T.E. Resistance mechanisms of bacteria to antimicrobial compounds. Int. Biodeterior. Biodegrad. 2003, 51, 277–282. [Google Scholar] [CrossRef]
- Taylor, D.E. Bacterial tellurite resistance. Trends Microbiol. 1999, 7, 111–115. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Mikalová, L.; Bosák, J.; Hříbková, H.; Dědičová, D.; Benada, O.; Šmarda, J.; Šmajs, D. Novel temperate phages of Salmonella enterica subsp. salamae and subsp. diarizonae and their activity against pathogenic S. enterica subsp. enterica isolates. PLoS ONE 2017, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Jakhetia, R.; Talukder, K.A.; Verma, N.K. Isolation, characterization and comparative genomics of bacteriophage SfIV: A novel serotype converting phage from Shigella flexneri. BMC Genom. 2013, 14, 677. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Fang, Y.; Feng, Y.; Lyu, N.; Chen, L.; Li, J.; Xu, X.; Zhu, B.; Hu, Y. Discovery of mcr-3.1 gene carried by a prophage located in a conjugative IncA/C2 plasmid from a Salmonella Choleraesuis clinical isolate. J. Infect. 2021, 82, 414–451. [Google Scholar] [CrossRef]
- Zhang, C.; Feng, Y.; Liu, F.; Jiang, H.; Qu, Z.; Lei, M.; Wang, J.; Zhang, B.; Hu, Y.; Ding, J.; et al. A Phage-Like IncY Plasmid Carrying the mcr-1 Gene in Escherichia coli from a Pig Farm in China. Antimicrob. Agents Chemother. 2017, 61, e02035-16. [Google Scholar] [CrossRef]
- Wang, X.; Wood, T.K. Cryptic prophages as targets for drug development. Drug Resist. Updat. 2016, 27, 30–38. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Kizny Gordon, A.; Phan, H.T.T.; Lipworth, S.I.; Cheong, E.; Gottlieb, T.; George, S.; Peto, T.E.A.; Mathers, A.J.; Walker, A.S.; Crook, D.W.; et al. Genomic dynamics of species and mobile genetic elements in a prolonged blaIMP-4-associated carbapenemase outbreak in an Australian hospital. J. Antimicrob. Chemother. 2020, 75, 873–882. [Google Scholar] [CrossRef]
- Kieffer, N.; Royer, G.; Decousser, J.-W.; Bourrel, A.-S.; Palmieri, M.; Ortiz De La Rosa, J.-M.; Jacquier, H.; Denamur, E.; Nordmann, P.; Poirel, L. mcr-9, an Inducible Gene Encoding an Acquired Phosphoethanolamine Transferase in Escherichia coli, and Its Origin. Antimicrob. Agents Chemother. 2019, 63, e00965-19. [Google Scholar] [CrossRef]
- Bitar, I.; Papagiannitsis, C.C.; Kraftova, L.; Chudejova, K.; Mattioni Marchetti, V.; Hrabak, J. Detection of Five mcr-9 -Carrying Enterobacterales Isolates in Four Czech Hospitals. mSphere 2020, 5, e01008-20. [Google Scholar] [CrossRef] [PubMed]
- Khedher, M.B.; Baron, S.A.; Riziki, T.; Ruimy, R.; Raoult, D.; Diene, S.M.; Rolain, J.M. Massive analysis of 64,628 bacterial genomes to decipher water reservoir and origin of mobile colistin resistance genes: Is there another role for these enzymes? Sci. Rep. 2020, 10, 5970. [Google Scholar] [CrossRef] [PubMed]
- Xiaomin, S.; Yiming, L.; Yuying, Y.; Zhangqi, S.; Yongning, W.; Shaolin, W. Global impact of mcr-1-positive Enterobacteriaceae bacteria on “one health”. Crit. Rev. Microbiol. 2020, 46, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Salgueiro, V.; Manageiro, V.; Bandarra, N.M.; Reis, L.; Caniça, M. Bacterial Diversity and Antibiotic Susceptibility of Sparus aurata from Aquaculture. Microorganisms 2020, 8, 1343. [Google Scholar] [CrossRef] [PubMed]
- European Commission Directorate-General for Health and Food Safety; Braak, K.; Schrijver, R.; Bergevoet, R. Welfare of Farmed Fish: Common Practices during Transport and at Slaughter: Executive Summary; Publications Office of European Union: Luxembourg, 2017. [Google Scholar] [CrossRef]
- Ciufo, S.; Kannan, S.; Sharma, S.; Badretdin, A.; Clark, K.; Turner, S.; Brover, S.; Schoch, C.L.; Kimchi, A.; DiCuccio, M. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018, 68, 2386–2392. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Varani, A.M.; Siguier, P.; Gourbeyre, E.; Charneau, V.; Chandler, M. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 2011, 12, R30. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Cherak, Z.; Loucif, L.; Moussi, A.; Rolain, J.M. Epidemiology of mobile colistin resistance (mcr) genes in aquatic environments. J. Glob. Antimicrob. Resist. 2021, 27, 51–62. [Google Scholar] [CrossRef]

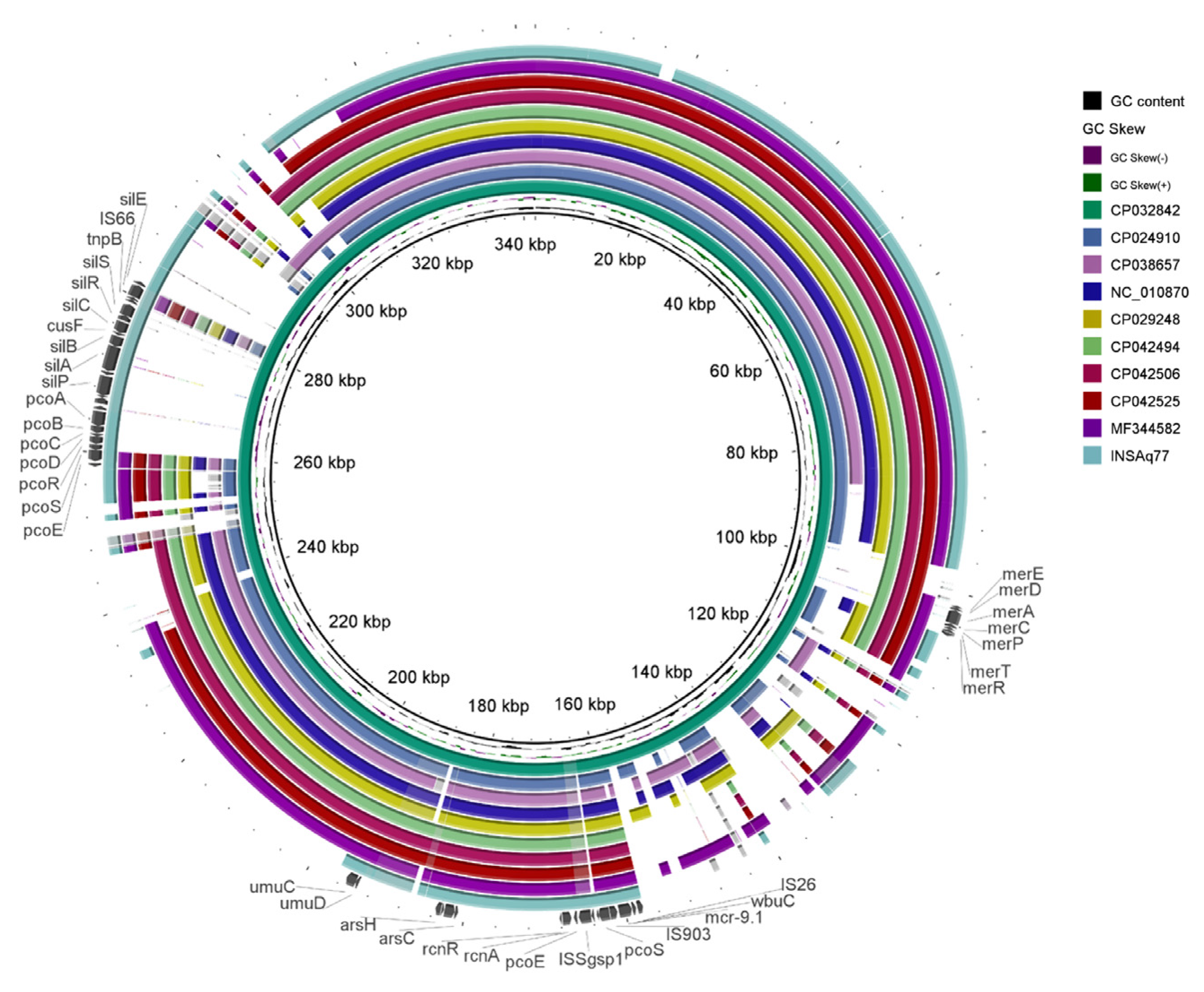
| Contig | RGI Criteria | ARO Term | Detection Criteria Model | AMR Gene Family | Drug Class | Resistance Mechanism | % Identity Matching Region | % Length Reference Sequence |
|---|---|---|---|---|---|---|---|---|
| INSAq77p_155 | Perfect | mcr-9.1 | protein homolog | MCR phosphoethanolamine transferase | peptide antibiotic | antibiotic target alteration | 100.0 | 100.0 |
| INSAq77p_4 | Strict | CRP | protein homolog | resistance-nodulation-cell division (RND) antibiotic efflux pump | macrolide antibiotic, fluoroquinolone antibiotic, penam | antibiotic efflux | 99.1 | 100.0 |
| INSAq77p_10 | Strict | ACT-12 | protein homolog | ACT beta-lactamase | carbapenem, cephalosporin, cephamycin, penam | antibiotic inactivation | 98.7 | 100.0 |
| INSAq77p_1 | Strict | FosA2 | protein homolog | fosfomycin thiol transferase | fosfomycin | antibiotic inactivation | 98.6 | 100.0 |
| INSAq77p_4 | Strict | Escherichia coli EF-Tu mutants (R234F) | protein variant | elfamycin-resistant EF-Tu | elfamycin antibiotic | antibiotic target alteration | 98.5 | 96.3 |
| INSAq77p_82 | Strict | baeR | protein homolog | resistance-nodulation-cell division (RND) antibiotic efflux pump | aminoglycoside antibiotic, aminocoumarin antibiotic | antibiotic efflux | 95.8 | 100.0 |
| INSAq77p_37 | Strict | H-NS | protein homolog | major facilitator superfamily (MFS) antibiotic efflux pump, resistance-nodulation-cell division (RND) antibiotic efflux pump | macrolide antibiotic, fluoroquinolone antibiotic, cephalosporin, cephamycin, penam, tetracycline antibiotic | antibiotic efflux | 95.6 | 100.0 |
| INSAq77p_47 | Strict | msbA | protein homolog | ATP-binding cassette (ABC) antibiotic efflux pump | nitroimidazole antibiotic | antibiotic efflux | 94.7 | 100.0 |
| INSAq77p_3 | Strict | emrR | protein homolog | major facilitator superfamily (MFS) antibiotic efflux pump | fluoroquinolone antibiotic | antibiotic efflux | 94.3 | 100.0 |
| INSAq77p_25 | Strict | Escherichia coli UhpT mutant (E350Q) | protein variant | antibiotic-resistant UhpT | fosfomycin | antibiotic target alteration | 93.7 | 100.0 |
| INSAq77p_21 | Strict | marA | protein homolog | resistance-nodulation-cell division (RND) antibiotic efflux pump, General Bacterial Porin with reduced permeability to beta-lactams | fluoroquinolone antibiotic, monobactam, carbapenem, cephalosporin, glycylcycline, cephamycin, penam, tetracycline antibiotic, rifamycin antibiotic, phenicol antibiotic, triclosan, penem | antibiotic efflux, reduced permeability to antibiotic | 93.6 | 99.2 |
| INSAq77p_3_7 | Strict | Klebsiella pneumoniae KpnH | protein homolog | major facilitator superfamily (MFS) antibiotic efflux pump | macrolide antibiotic, fluoroquinolone antibiotic, aminoglycoside antibiotic, carbapenem, cephalosporin, penam, peptide antibiotic, penem | antibiotic efflux | 92.2 | 100.6 |
| INSAq77p_11 | Strict | oqxA | protein homolog | resistance-nodulation-cell division (RND) antibiotic efflux pump | fluoroquinolone antibiotic, glycylcycline, tetracycline antibiotic, diaminopyrimidine antibiotic, nitrofuran antibiotic | antibiotic efflux | 91.1 | 100.0 |
| INSAq77p_21 | Strict | Escherichia coli marR mutant conferring antibiotic resistance | protein overexpression | resistance-nodulation-cell division (RND) antibiotic efflux pump | fluoroquinolone antibiotic, cephalosporin, glycylcycline, penam, tetracycline antibiotic, rifamycin antibiotic, phenicol antibiotic, triclosan | antibiotic target alteration, antibiotic efflux | 91.0 | 100.0 |
| INSAq77p_34 | Strict | Klebsiella pneumoniae KpnF | protein homolog | major facilitator superfamily (MFS) antibiotic efflux pump | macrolide antibiotic, aminoglycoside antibiotic, cephalosporin, tetracycline antibiotic, peptide antibiotic, rifamycin antibiotic | antibiotic efflux | 89.0 | 100.0 |
| INSAq77p_3 | Strict | rsmA | protein homolog | resistance-nodulation-cell division (RND) antibiotic efflux pump | fluoroquinolone antibiotic, diaminopyrimidine antibiotic, phenicol antibiotic | antibiotic efflux | 85.3 | 100.0 |
| INSAq77p_13 | Strict | Escherichia coli ampH beta-lactamase | protein homolog | ampC-type beta-lactamase | cephalosporin, penam | antibiotic inactivation | 85.2 | 100.8 |
| INSAq77p_34 | Strict | Klebsiella pneumoniae KpnE | protein homolog | major facilitator superfamily (MFS) antibiotic efflux pump | macrolide antibiotic, aminoglycoside antibiotic, cephalosporin, tetracycline antibiotic, peptide antibiotic, rifamycin antibiotic | antibiotic efflux | 82.0 | 83.3 |
| INSAq77p_11 | Strict | adeF | protein homolog | resistance-nodulation-cell division (RND) antibiotic efflux pump | fluoroquinolone antibiotic, tetracycline antibiotic | antibiotic efflux | 60.9 | 99.2 |
| INSAq77p_1 | Strict | Haemophilus influenzae PBP3 mutant (D350N, S357N) | protein variant | Penicillin-binding protein mutations conferring resistance to beta-lactam antibiotics | cephalosporin, cephamycin, penam | antibiotic target alteration | 53.1 | 96.4 |
| INSAq77p_9 | Strict | adeF | protein homolog | resistance-nodulation-cell division (RND) antibiotic efflux pump | fluoroquinolone antibiotic, tetracycline antibiotic | antibiotic efflux | 41.2 | 97.9 |
| Plasmid (bp) | Strain | Isolation Source/Country/Year | Identity (%) | Query Cover (%) | pMLST b | Acquired Antibiotic and Desinfectant Resistance Genes c | GenBank Acc. No. |
|---|---|---|---|---|---|---|---|
| INSAq77 IncHI2 (30,314) a | E. ludwigii INSAq77 | Seabream (Sparus aurata)/Portugal/2018 | - | - | DLST1 | mcr-9.1 | JABRPH000000000 |
| pSPRC-Echo1 (339,920) | E. hormaechei C15117 | Burns unit/Australia/2007 | 99.99% | 99.0% | DLST1 | aac(6’)-IIc, aph(3’’)-Ib-type, aph(6)-Id, blaSHV-12, blaTEM-1B, catA2-type, dfrA19, mcr-9, qacE, qnrA1-type, sul1, sul2, tet(D) | NZ_CP032842 |
| p525011-HI2 (354,045) | C. freundii 525011 | unknown/China/2017 | 100.00% | 97.0% | untyped, Nearest STs: 7,1,4,15 | aac(3)-IId-type, aac(6’)-aph(2’’), aadA5, armA, blaTEM-1B, catA2-type, dfrA1-type, mcr-9, mph(E), msr(E), qacE, qnrA1-type, sul1, sul2 | NZ_MF344582 |
| pOSUKPC4 (351,806) | E. hormaechei OSUKPC4_L | Animal/USA/2016 | 100.00% | 98.0% | DLST1 | aadA1, aph(3’’)-Ib-type, aph(6)-Id, blaKPC-4, blaOXA-129, dfrA21, mcr-9, qacE, sul1, tet(B) | NZ_CP024910 |
| pOSUEC_D (354,256) | E. hormaechei OSUVMCKPC4-2 | Animal/USA/2016 | 100.00% | 98.0% | DLST1 | aadA1, aph(3’’)-Ib-type, aph(6)-Id, blaKPC-4, blaOXA-129, dfrA21, mcr-9, qacE, sul1, tet(B) | NZ_CP029248 |
| pK29 (269,674) | K. pneumoniae NK29 | Human/Taiwan/2001 | 100.00% | 98.0% | DLST1 | aadA2, blaCMY-8, blaCTX-M-62-type, catB2, mcr-9, qacE, sul1 | NC_010870 |
| pE1_001 (357,530) | L. adecarboxylata E1 | Burns Unit Shower/Australia/2012 | 100.00% | 98.0% | DLST1 | formA-type, mcr-9 | NZ_CP042506 |
| pE11_001 (339,433) | C. freundii E11 | Burns Unit Shower/Australia/2012 | 100.00% | 98.0% | DLST1 | formA-type, mcr-9 | NZ_CP042525 |
| pE61_001 (357,530) | L. adecarboxylata E61 | Burns Unit Shower/Australia/2014 | 100.00% | 98.0% | DLST1 | formA-type, mcr-9 | NZ_CP042494 |
| p565_1 (263,189) | C. freundii 565 | Human Stool/Spain/2014 | 99,68% | 95,0% | DLST1 | aac(6’)-Ib-cr, aadA1, aadA2b-type, blaCTX-M-9, blaSHV-12, blaVIM-1, catA1-type, dfrA16, mcr-9-type, qacE, qnrA1-type, sul1 | NZ_CP038657 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manageiro, V.; Salgueiro, V.; Rosado, T.; Bandarra, N.M.; Ferreira, E.; Smith, T.; Dias, E.; Caniça, M. Genomic Analysis of a mcr-9.1-Harbouring IncHI2-ST1 Plasmid from Enterobacter ludwigii Isolated in Fish Farming. Antibiotics 2022, 11, 1232. https://doi.org/10.3390/antibiotics11091232
Manageiro V, Salgueiro V, Rosado T, Bandarra NM, Ferreira E, Smith T, Dias E, Caniça M. Genomic Analysis of a mcr-9.1-Harbouring IncHI2-ST1 Plasmid from Enterobacter ludwigii Isolated in Fish Farming. Antibiotics. 2022; 11(9):1232. https://doi.org/10.3390/antibiotics11091232
Chicago/Turabian StyleManageiro, Vera, Vanessa Salgueiro, Tânia Rosado, Narcisa M. Bandarra, Eugénia Ferreira, Terry Smith, Elsa Dias, and Manuela Caniça. 2022. "Genomic Analysis of a mcr-9.1-Harbouring IncHI2-ST1 Plasmid from Enterobacter ludwigii Isolated in Fish Farming" Antibiotics 11, no. 9: 1232. https://doi.org/10.3390/antibiotics11091232
APA StyleManageiro, V., Salgueiro, V., Rosado, T., Bandarra, N. M., Ferreira, E., Smith, T., Dias, E., & Caniça, M. (2022). Genomic Analysis of a mcr-9.1-Harbouring IncHI2-ST1 Plasmid from Enterobacter ludwigii Isolated in Fish Farming. Antibiotics, 11(9), 1232. https://doi.org/10.3390/antibiotics11091232