Impact of Skin Disinfection on Cutaneous Microbiota, before and after Peripheral Venous Catheter Insertion
Abstract
1. Introduction
2. Results
2.1. Study Population
2.2. No Alteration in the Diversity of the Skin Microbiota Was Associated with Antiseptic Procedure or Type of PVC, According to the Results of Blood or PVC Cultures
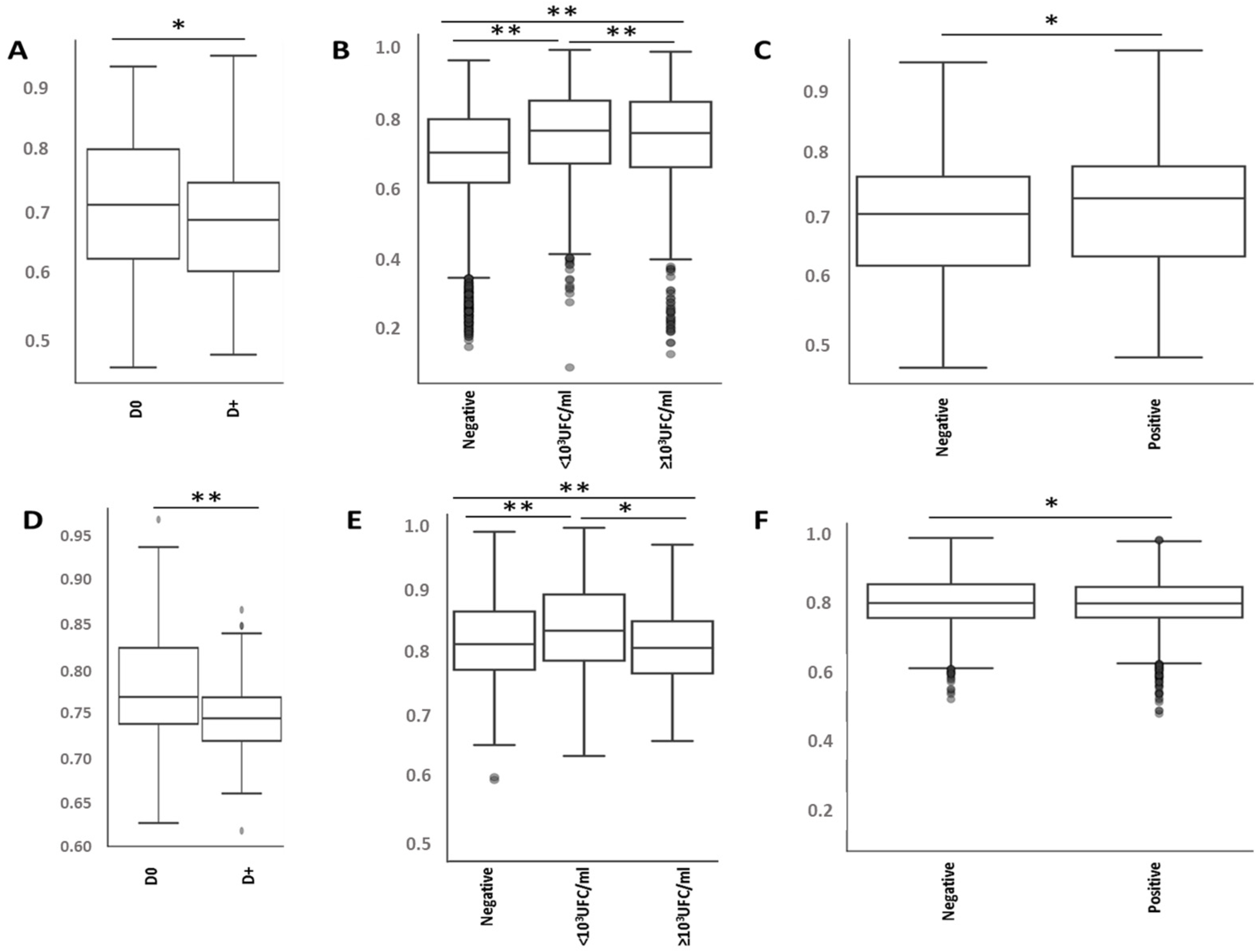
2.3. Antiseptic Procedures and PVC Types Are Associated with the Modification of Specific Bacteria Representations
2.4. Antiseptic Procedures and PVC Types Are Associated with Modification of the Predicted Metabolic Pathways
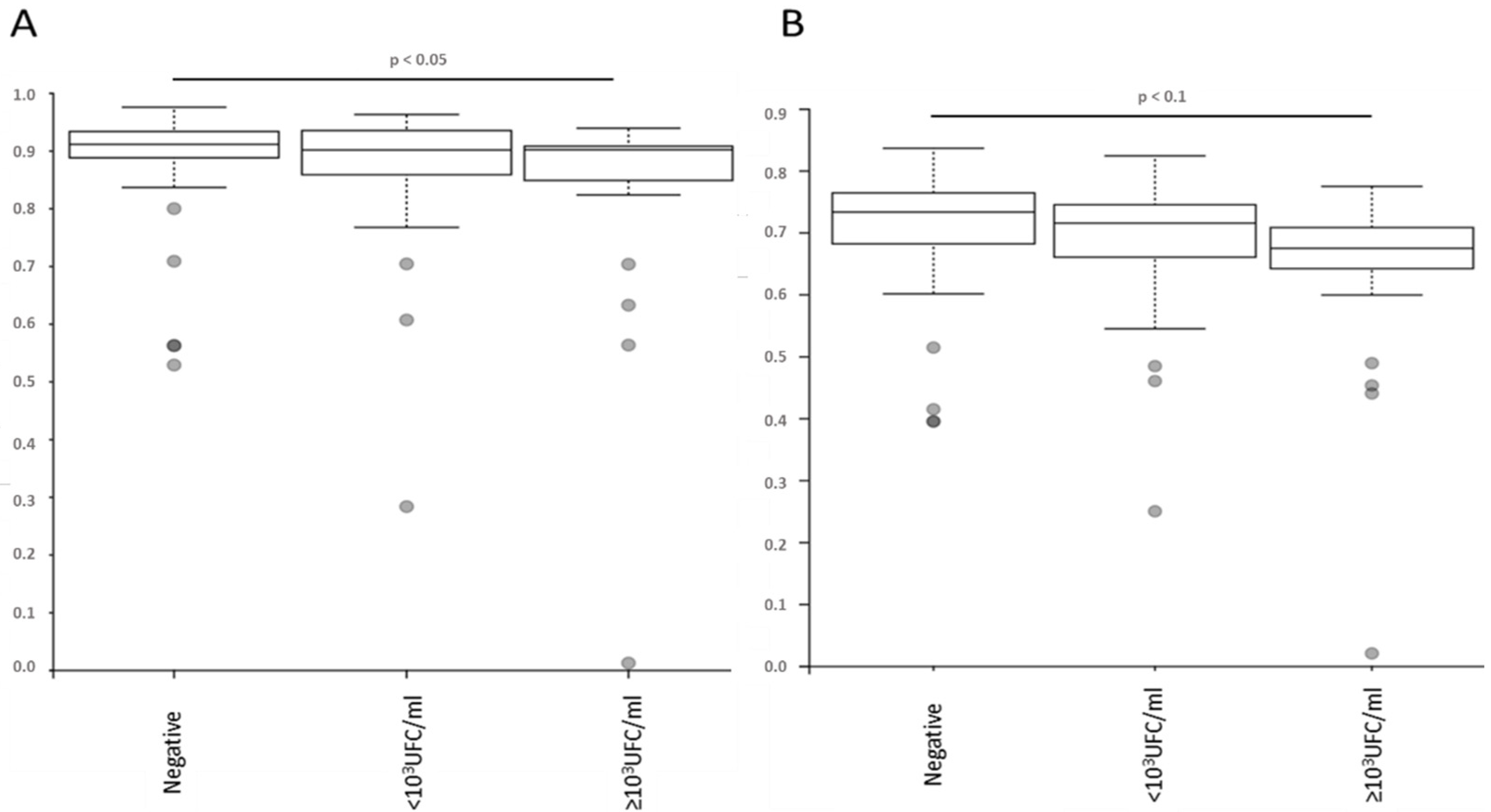
2.5. Bacterial Microbiota before PVC Is Not Predictive of Complications but Is Different in Diversity and Composition
3. Discussion
4. Materials and Methods
4.1. Selected Samples
4.2. Sample Preparation
4.3. Bioinformatic Analyses
4.4. Ethical Considerations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grice, E.A.; Segre, J.A. The Skin Microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from Human Skin Commensal Bacteria Protect against Staphylococcus Aureus and Are Deficient in Atopic Dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef] [PubMed]
- Scharschmidt, T.C.; Vasquez, K.S.; Pauli, M.L.; Leitner, E.G.; Chu, K.; Truong, H.-A.; Lowe, M.M.; Sanchez Rodriguez, R.; Ali, N.; Laszik, Z.G.; et al. Commensal Microbes and Hair Follicle Morphogenesis Coordinately Drive Treg Migration into Neonatal Skin. Cell Host Microbe 2017, 21, 467–477.e5. [Google Scholar] [CrossRef]
- Zipperer, A.; Konnerth, M.C.; Laux, C.; Berscheid, A.; Janek, D.; Weidenmaier, C.; Burian, M.; Schilling, N.A.; Slavetinsky, C.; Marschal, M.; et al. Human Commensals Producing a Novel Antibiotic Impair Pathogen Colonization. Nature 2016, 535, 511–516. [Google Scholar] [CrossRef]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial Community Variation in Human Body Habitats across Space and Time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; NISC Comparative Sequencing Program; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef]
- Beausoleil, C.M.; Paulson, D.S.; Bogert, A.; Lewis, G.S. In Vivo Evaluation of the Persistant and Residual Antimicrobial Properties of Three Hand-Scrub and Hand-Rub Regimes in a Simulated Surgical Environment. J. Hosp. Infect. 2012, 81, 283–287. [Google Scholar] [CrossRef]
- Carty, N.; Wibaux, A.; Ward, C.; Paulson, D.S.; Johnson, P. Antimicrobial Activity of a Novel Adhesive Containing Chlorhexidine Gluconate (CHG) against the Resident Microflora in Human Volunteers. J. Antimicrob. Chemother. 2014, 69, 2224–2229. [Google Scholar] [CrossRef][Green Version]
- Kampf, G.; Kramer, A. Epidemiologic Background of Hand Hygiene and Evaluation of the Most Important Agents for Scrubs and Rubs. Clin. Microbiol. Rev. 2004, 17, 863–893. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, G.; Russell, A.D. Antiseptics and Disinfectants: Activity, Action, and Resistance. Clin. Microbiol. Rev. 1999, 12, 147–179. [Google Scholar] [CrossRef]
- O’Grady, N.P.; Alexander, M.; Burns, L.A.; Dellinger, E.P.; Garland, J.; Heard, S.O.; Lipsett, P.A.; Masur, H.; Mermel, L.A.; Pearson, M.L.; et al. Guidelines for the Prevention of Intravascular Catheter-Related Infections. Clin. Infect. Dis. 2011, 52, e162–e193. [Google Scholar] [CrossRef]
- Huang, S.S.; Septimus, E.; Kleinman, K.; Moody, J.; Hickok, J.; Avery, T.R.; Lankiewicz, J.; Gombosev, A.; Terpstra, L.; Hartford, F.; et al. Targeted versus Universal Decolonization to Prevent ICU Infection. N. Engl. J. Med. 2013, 368, 2255–2265. [Google Scholar] [CrossRef] [PubMed]
- Huskins, W.C.; Huckabee, C.M.; O’Grady, N.P.; Murray, P.; Kopetskie, H.; Zimmer, L.; Walker, M.E.; Sinkowitz-Cochran, R.L.; Jernigan, J.A.; Samore, M.; et al. Intervention to Reduce Transmission of Resistant Bacteria in Intensive Care. N. Engl. J. Med. 2011, 364, 1407–1418. [Google Scholar] [CrossRef]
- Guenezan, J.; Marjanovic, N.; Drugeon, B.; Neill, R.O.; Liuu, E.; Roblot, F.; Palazzo, P.; Bironneau, V.; Prevost, F.; Paul, J.; et al. Chlorhexidine plus Alcohol versus Povidone Iodine plus Alcohol, Combined or Not with Innovative Devices, for Prevention of Short-Term Peripheral Venous Catheter Infection and Failure (CLEAN 3 Study): An Investigator-Initiated, Open-Label, Single Centre, Randomised-Controlled, Two-by-Two Factorial Trial. Lancet Infect. Dis. 2021, 21, 1038–1048. [Google Scholar] [CrossRef]
- Drugeon, B.; Pichon, M.; Marjanovic, N.; Mousse, S.; Seguin, S.; Raynaud, C.; Rahoui, A.; Frasca, D.; Mimoz, O.; Guenezan, J. Peripheral Venous Catheter Colonisation after Skin Disinfection with 0.5% Aqueous Sodium Hypochlorite, Preceded or Not by One Application of 70% Ethanol (DACLEAN): A Single Centre, Randomised, Open-Label, Pilot Study. J. Hosp. Infect. 2021, 120, 123–126. [Google Scholar] [CrossRef]
- Lin, Z.; Farooqui, A.; Li, G.; Wong, G.K.; Mason, A.L.; Banner, D. Next-Generation Sequencing and Bioinformatic Approaches to Detect and Analyze Influenza Virus in Ferrets. J. Infect. Dev. Ctries. 2014, 8, 498–509. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wiemken, T.L.; Ericsson, A.C. Chlorhexidine Gluconate Does Not Result in Epidermal Microbiota Dysbiosis in Healthy Adults. Am. J. Infect. Control 2021, 49, 769–774. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, I.J.; Wright, E.M.; Tagg, J.R.; Jain, R.; Hale, J.D.F. Skin Microbiome-The Next Frontier for Probiotic Intervention. Probiotics Antimicrob. Proteins 2021, 14, 630–647. [Google Scholar] [CrossRef]
- SanMiguel, A.J.; Meisel, J.S.; Horwinski, J.; Zheng, Q.; Bradley, C.W.; Grice, E.A. Antiseptic Agents Elicit Short-Term, Personalized, and Body Site-Specific Shifts in Resident Skin Bacterial Communities. J. Investig. Dermatol. 2018, 138, 2234–2243. [Google Scholar] [CrossRef] [PubMed]
- Naik, S.; Bouladoux, N.; Wilhelm, C.; Molloy, M.J.; Salcedo, R.; Kastenmuller, W.; Deming, C.; Quinones, M.; Koo, L.; Conlan, S.; et al. Compartmentalized Control of Skin Immunity by Resident Commensals. Science 2012, 337, 1115–1119. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Conlan, S.; Polley, E.C.; Segre, J.A.; Kong, H.H. Shifts in Human Skin and Nares Microbiota of Healthy Children and Adults. Genome Med. 2012, 4, 77. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.L.; Park, M.; NISC Comparative Sequencing Program; Kong, H.H.; Segre, J.A. Temporal Stability of the Human Skin Microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef] [PubMed]
- Langevin, S.; Pichon, M.; Smith, E.; Morrison, J.; Bent, Z.; Green, R.; Barker, K.; Solberg, O.; Gillet, Y.; Javouhey, E.; et al. Early Nasopharyngeal Microbial Signature Associated with Severe Influenza in Children: A Retrospective Pilot Study. J. Gen. Virol. 2017, 98, 2425–2437. [Google Scholar] [CrossRef]
- Crane, J.K.; Hohman, D.W.; Nodzo, S.R.; Duquin, T.R. Antimicrobial Susceptibility of Propionibacterium Acnes Isolates from Shoulder Surgery. Antimicrob. Agents Chemother. 2013, 57, 3424–3426. [Google Scholar] [CrossRef]
- Lee, M.J.; Pottinger, P.S.; Butler-Wu, S.; Bumgarner, R.E.; Russ, S.M.; Matsen, F.A. Propionibacterium Persists in the Skin despite Standard Surgical Preparation. J. Bone Jt. Surg. 2014, 96, 1447–1450. [Google Scholar] [CrossRef] [PubMed]
- Pichon, M.; Burucoa, C.; Evplanov, V.; Favalli, F. Efficacy of Three Povidone Iodine Formulations against Cutibacterium Acnes Assessed through In Vitro Studies: A Preliminary Study. Antibiotics 2022, 11, 665. [Google Scholar] [CrossRef]
- Christensen, G.J.M.; Scholz, C.F.P.; Enghild, J.; Rohde, H.; Kilian, M.; Thürmer, A.; Brzuszkiewicz, E.; Lomholt, H.B.; Brüggemann, H. Antagonism between Staphylococcus Epidermidis and Propionibacterium Acnes and Its Genomic Basis. BMC Genom. 2016, 17, 152. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Park, S.; Um, S.; Kim, S.; Lee, J.; Jang, J.; Jeong, H.-O.; Shin, J.; Kang, J.; Lee, S.; et al. Microbiome of Saliva and Plaque in Children According to Age and Dental Caries Experience. Diagnostics 2021, 11, 1324. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.I.; Niang, E.H.A.; Sarr, M.; Durand, G.; Tall, M.L.; Caputo, A.; Raoult, D.; Fournier, P.-E.; Fenollar, F. Fenollaria Timonensis Sp. Nov., A New Bacterium Isolated from Healthy Human Fresh Stool. Curr. Microbiol. 2020, 77, 3780–3786. [Google Scholar] [CrossRef]
- Machet, L.; Machet, M.C.; Estève, E.; Delarbre, J.M.; Pelucio-Lopes, C.; Pruvost, F.; Lorette, G. Actinomyces meyeri cutaneous actinomycosis with pulmonary localization. Ann. Derm. Venereol. 1993, 120, 896–899. [Google Scholar] [PubMed]
- Moretti, E.W.; Ofstead, C.L.; Kristy, R.M.; Wetzler, H.P. Impact of Central Venous Catheter Type and Methods on Catheter-Related Colonization and Bacteraemia. J. Hosp. Infect. 2005, 61, 139–145. [Google Scholar] [CrossRef][Green Version]
- Holland, K.T.; Greenman, J.; Cunliffe, W.J. Growth of Cutaneous Propionibacteria on Synthetic Medium; Growth Yields and Exoenzyme Production. J. Appl. Bacteriol. 1979, 47, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Messager, S.; Goddard, P.A.; Dettmar, P.W.; Maillard, J.-Y. Determination of the Antibacterial Efficacy of Several Antiseptics Tested on Skin by an “ex-Vivo” Test. J. Med. Microbiol. 2001, 50, 284–292. [Google Scholar] [CrossRef]
- Boisson, M.; Corbi, P.; Kerforne, T.; Camilleri, L.; Debauchez, M.; Demondion, P.; Eljezi, V.; Flecher, E.; Lepelletier, D.; Leprince, P.; et al. Multicentre, Open-Label, Randomised, Controlled Clinical Trial Comparing 2% Chlorhexidine-70% Isopropanol and 5% Povidone Iodine-69% Ethanol for Skin Antisepsis in Reducing Surgical-Site Infection after Cardiac Surgery: The CLEAN 2 Study Protocol. BMJ Open 2019, 9, e026929. [Google Scholar] [CrossRef]
- Integrative HMP (iHMP) Research Network Consortium The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [CrossRef] [PubMed]
- Rickard, C.M.; Webster, J.; Wallis, M.C.; Marsh, N.; McGrail, M.R.; French, V.; Foster, L.; Gallagher, P.; Gowardman, J.R.; Zhang, L.; et al. Routine versus Clinically Indicated Replacement of Peripheral Intravenous Catheters: A Randomised Controlled Equivalence Trial. Lancet 2012, 380, 1066–1074. [Google Scholar] [CrossRef]
- Tuffaha, H.W.; Rickard, C.M.; Webster, J.; Marsh, N.; Gordon, L.; Wallis, M.; Scuffham, P.A. Cost-Effectiveness Analysis of Clinically Indicated versus Routine Replacement of Peripheral Intravenous Catheters. Appl. Health Econ. Health Policy 2014, 12, 51–58. [Google Scholar] [CrossRef]
- Olivier, R.C.; Wickman, M.; Skinner, C.; Ablir, L. The Impact of Replacing Peripheral Intravenous Catheters When Clinically Indicated on Infection Rate, Nurse Satisfaction, and Costs in CCU, Step-Down, and Oncology Units. Am. J. Infect. Control 2021, 49, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Yung, D.B.Y.; Sircombe, K.J.; Pletzer, D. Friends or Enemies? The Complicated Relationship between Pseudomonas Aeruginosa and Staphylococcus Aureus. Mol. Microbiol. 2021, 116, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Bouslimani, A.; Porto, C.; Rath, C.M.; Wang, M.; Guo, Y.; Gonzalez, A.; Berg-Lyon, D.; Ackermann, G.; Moeller Christensen, G.J.; Nakatsuji, T.; et al. Molecular Cartography of the Human Skin Surface in 3D. Proc. Natl. Acad. Sci. USA 2015, 112, E2120–E2129. [Google Scholar] [CrossRef]
- Ursell, L.K.; Clemente, J.C.; Rideout, J.R.; Gevers, D.; Caporaso, J.G.; Knight, R. The Interpersonal and Intrapersonal Diversity of Human-Associated Microbiota in Key Body Sites. J. Allergy Clin. Immunol. 2012, 129, 1204–1208. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Abu-Ali, G.; Huttenhower, C. The Healthy Human Microbiome. Genome Med. 2016, 8, 51. [Google Scholar] [CrossRef]
- Castelino, M.; Eyre, S.; Moat, J.; Fox, G.; Martin, P.; Ho, P.; Upton, M.; Barton, A. Optimisation of Methods for Bacterial Skin Microbiome Investigation: Primer Selection and Comparison of the 454 versus MiSeq Platform. BMC Microbiol. 2017, 17, 23. [Google Scholar] [CrossRef]
- Gachet, C.; Prat, M.; Burucoa, C.; Grivard, P.; Pichon, M. Spermatic Microbiome Characteristics in Infertile Patients: Impact on Sperm Count, Mobility, and Morphology. J. Clin. Med. 2022, 11, 1505. [Google Scholar] [CrossRef]
- Chang, H.-W.; Yan, D.; Singh, R.; Liu, J.; Lu, X.; Ucmak, D.; Lee, K.; Afifi, L.; Fadrosh, D.; Leech, J.; et al. Alteration of the Cutaneous Microbiome in Psoriasis and Potential Role in Th17 Polarization. Microbiome 2018, 6, 154. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2--Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Faith, D.P. The Role of the Phylogenetic Diversity Measure, PD, in Bio-Informatics: Getting the Definition Right. Evol. Bioinform. 2007, 2, 277–283. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and Qualitative Beta Diversity Measures Lead to Different Insights into Factors That Structure Microbial Communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of Composition of Microbiomes: A Novel Method for Studying Microbial Composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef] [PubMed]

| Allocation Group | Chlorhexidine-Alcohol (n = 76; 45.8%) | Povidone-Iodine-Alcohol (n = 90; 54.2%) | |
|---|---|---|---|
| Sex (Male; n; x% of the respective category) | 35 (46.1) | 48 (53.3) | |
| Age category in years (n; x%) | |||
| 18–30 | 5 (6.6) | 3 (3.3) | |
| 31–45 | 4 (5.3) | 8 (8.9) | |
| 46–60 | 8 (10.5) | 10 (11.1) | |
| 61–75 | 23 (30.3) | 16 (17.8) | |
| 76–90 | 28 (36.8) | 38 (42.2) | |
| >90 | 8 (10.5) | 15 (16.7) | |
| Innovative PVC type (n; %) | 35 (46.1) | 43 (47.8) | |
| Local inflammation (n; x%) | 1 (1.3) | - | |
| Positive PVC culture (n; %) | 33 (43.4) | 43 (47.7) | |
| ≥103 UFC per mL (n; % of the positive culture) | 1 (3.0) | 23 (53.5) | |
| <103 UFC per mL (n; % of the positive culture) | 32 (97.0) | 20 (46.5) | |
| Positive blood culture (n; % of patients sampled for blood culture) * | 33 (43.4) | 43 (47.8) | |
| Other infection (n; %) | 3 (3.9) | 3 (3.3) | |
| Time elapsed between catheter insertion and removal (mean; SEM) | 2.30 (0.23) | 2.50 (0.16) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prat, M.; Guenezan, J.; Drugeon, B.; Burucoa, C.; Mimoz, O.; Pichon, M. Impact of Skin Disinfection on Cutaneous Microbiota, before and after Peripheral Venous Catheter Insertion. Antibiotics 2022, 11, 1209. https://doi.org/10.3390/antibiotics11091209
Prat M, Guenezan J, Drugeon B, Burucoa C, Mimoz O, Pichon M. Impact of Skin Disinfection on Cutaneous Microbiota, before and after Peripheral Venous Catheter Insertion. Antibiotics. 2022; 11(9):1209. https://doi.org/10.3390/antibiotics11091209
Chicago/Turabian StylePrat, Manon, Jeremy Guenezan, Bertrand Drugeon, Christophe Burucoa, Olivier Mimoz, and Maxime Pichon. 2022. "Impact of Skin Disinfection on Cutaneous Microbiota, before and after Peripheral Venous Catheter Insertion" Antibiotics 11, no. 9: 1209. https://doi.org/10.3390/antibiotics11091209
APA StylePrat, M., Guenezan, J., Drugeon, B., Burucoa, C., Mimoz, O., & Pichon, M. (2022). Impact of Skin Disinfection on Cutaneous Microbiota, before and after Peripheral Venous Catheter Insertion. Antibiotics, 11(9), 1209. https://doi.org/10.3390/antibiotics11091209








